Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that participate in transcriptional and post-transcriptional regulation of gene expression. miRNAs have numerous roles in cellular function including embryonic development. Human embryonic stem cells (hESCs) are capable of self-renewal and can differentiate into most of cell types including cardiomyocytes (CMs). These characteristics of hESCs make them considered as an important model for studying human embryonic development and tissue specific differentiation. In this study, we tried to demonstrate the profile of miRNA expression in cardiac differentiation from hESCs. To induce differentiation, we differentiated hESCs into CMs by direct differentiation method and characterized differentiated cells. To analyze the expression of miRNAs, we distinguished (days 4, 8, 12, 16, 20, 24, 28) and isolated RNAs from each differentiation stage. miRNA specific RT-qPCR was performed and the expression profile of miR-1, -30d, -133a, -143, -145, -378a, -499a was evaluated. The expression of all miRs was up-regulated at day 8. miR-143 and -145 expression was also up-regulated at the later stage of differentiation. Only miR-378a expression returned to undifferentiated hESC levels at the other stages of differentiation. In conclusion, we elucidated the expression profile of miRNAs during differentiation into cardiomyocytes from hESCs. Our findings demonstrate the expression of miRNAs was stage-dependent during differentiation and suggest that the differentiation into CMs can be regulated by miRNAs through direct or indirect pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Human embryonic stem cells (hESCs) have an ability to self-renew and can differentiate into many cell types including cardiomyocytes (CMs) [1]. These abilities of hESCs make them considered as important model for human development and tissue specific differentiation [2, 3]. Especially, intractable heart diseases require cell-based therapy utilizing stem cell-derived products.
To induce cardiac lineage differentiation, many growth factors such as bone morphogenetic proteins (BMPs) have been treated to hESCs in two-week differentiation protocols [4,5,6,7,8]. In spite of many previously reported protocols, the efficiency stills needs to be improved in terms of retrieving self-beating CM clusters. Recently, epigenetic control of cell differentiation has been considered to have a significant role in addition to genetic machinery [9].
Small non-coding RNA molecules such as microRNAs (miRNAs, miRs) play an important part in transcriptional and post-transcriptional regulation of gene expression [10,11,12]. miRNAs are conserved in vertebrate and known to have numerous roles in cellular function including embryonic development [13,14,15]. In pluripotent stem cells, microRNAs play numerous roles in cell lineage determination, maintenance of pluripotency, and cell cycle regulation [16]. The roles of miRNAs in cardiac differentiation include maintenance of normal heart structure, regulation of heart morphogenesis, electrophysiological contraction, and regulation of cell cycles [17,18,19,20]. They also contribute to regulation of apoptosis in cardiomyocytes [21, 22], which is related to myocardial infarction, cardiomyopathy and cardiac hypertrophy [23,24,25,26].
In this study, we tried to demonstrate the profile of miRNA expression in cardiac differentiation from hESCs at various stages. We selected 7 candidate miRNAs using database that are involved in early mesoderm induction, heart development and regulation of cardiac function. This attempt is critical, considering dynamic changes of differentiation-inducing factor expression along the time course of differentiation. If successfully searched, the identified candidate miRNAs can be co-treated with conventional cardiac lineage differentiation-inducing factors to enhance the differentiation efficiency.
2 Materials and methods
2.1 Ethics
The use of human embryonic stem (hES) cell line was approved by the Institutional Review Board of the Institute of Reproductive Medicine and Population (IRMP), Medical Research Center, Seoul National University (219932-201412-LR-12-01-01).
2.2 hESC culture
The hES cell line, SNUhES3 was provided from the Institute of IRMP [1] and maintained as previously described [27]. Briefly, undifferentiated hESC colonies were mechanically dissected and transferred onto freshly prepared, mitomycin C (MMC, Sigma-Aldrich, St. Louis, MO, USA)-treated STO (CRL-1503, ATCC, Manassas, VA, USA) feeder layer. The cells were passaged every 7 days by mechanical dissociation method. The medium was consisted of DMEM/F12 (Invitrogen, Waltham, MA, USA), 20% knockout serum replacement (KO-SR, Invitrogen), 1% nonessential amino acids (Invitrogen), 50 µg/mL streptomycin (Invitrogen), 50 U/mL penicillin (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 4 ng/mL basic fibroblast growth factor (bFGF, Invitrogen). The medium was changed daily.
2.3 Differentiation into cardiac precursor cells from hESCs
Undifferentiated hESCs were treated with 2 mg/mL of collagenase type IV (Invitrogen) and incubated for 30 min at 37 °C and isolated colonies were mechanically disrupted and transferred to vitronectin (VTN-N, Invitrogen)-coated culture dish and media was switched to Essential 8 medium (Invitrogen). When colonies were fully grown, the colonies were washed with PBS (Invitrogen) and then medium was switched to differentiation medium consisted of RPMI 1640 (Invitrogen) and B27 supplement (Invitrogen). The cells were treated with 100 ng/mL of Activin A (R&D Systems, Minneapolis, MN, USA) for 24 h and 10 ng/mL of BMP2 (R&D Systems) for following 4 days. The growth factors were omitted from the differentiation day 5.
2.4 Immunofluorescence staining
In order to evaluate the localization of specific proteins, differentiated cells were washed with PBS and fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich) for 15 min at room temperature and blocked with 3% bovine serum albumin (BSA, Sigma-Aldrich) solution for 12 h at 4 °C. Then, the cells were incubated with each primary antibody (1:100), rabbit anti-human Nkx 2.5 (Abcam, Cambridge, UK), mouse anti-human ANP (SantaCruz Biotechnology, Dallas, TX, USA)and mouse anti-smooth muscle actin (SMA, SantaCruz Biotechnology), for overnight at 4 °C, and washed three times with TBS containing triton X100 (TBST, Sigma-Aldrich). Secondary antibodies, goat anti-rabbit 488 IgG (Molecular Probes, Grand Island, NY, USA) and goat anti-mouse 594 IgG (Molecular Probes) were added for 50 min at 37 °C and washed three times with TBST. Finally, 10 µg/mL of DAPI (Sigma-Aldrich) was treated and the images were assessed under fluorescence microscope (EVOS-FL, Life Technologies, Grand Island, NY, USA).
2.5 MicroRNA quantitative reverse transcription and polymerase chain reaction (qRT-PCR)
To evaluate the expression level of miRNAs, total RNAs were extracted from the samples using Trizol reagent (Invitrogen). cDNAs were synthesized from 0.05 µg total RNAs using NCode™ VILO™ miRNA cDNA Synthesis Kit (Invitrogen) and templates were used for the qRT-PCR reactions. Each specific primer (Table 1) and NCode™ EXPRESS SYBR® GreenER™ miRNA qRT-PCR premix (Invitrogen) were added to the cDNAs and amplified under the following conditions: incubation for 2 min at 50 °C and following 2 min at 95 °C, and followed by 40 cycles of 15 s at 95 °C, and 60 s at 60 °C. All the reactions were performed in triplicate and the Ct value was calculated based on the U6 expression.
2.6 Statistical analysis
The entire experiments were repeated at least three times. All data were expressed as means and standard deviations and compared using the Student’s t test. The differences were considered statistically significant when they were p < 0.05. Data were analyzed using the Statistical Package for the Social Sciences for Windows (version 12.0, SPSS Inc., Chicago, IL).
3 Results
3.1 Differentiation of hESCs into cardiac lineage cells
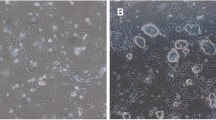
To induce cardiac lineage differentiation, a sequential culture of hESCs with on-feeder and feeder-free culture system was employed (Fig. 1). During feeder-free culture period, the differentiation into cardiac precursors was enhanced by eliminating the residual feeder cells and pre-secreted undifferentiated state-maintaining factors. Cardiac precursor cells showed beating as early as at days 13 and 14 (Suppl. movie). These beating cell clusters showed typical cardiomyocyte-like morphology (Fig. 2).
3.2 Expression of cardiac lineage proteins
To confirm the complete differentiation into cardiac cells, the expression of cardiac linage-specific and non-specific markers was evaluated. The expression of cardiac specific transcription factor Nkx 2.5 was confirmed at day 18 and the expression of cardiomyocyte-secreting hormone, ANP was also confirmed. The expression of SMA, non-cardiac, smooth muscle-specific protein, was not expressed in hESC-derived cardiac precursor cells (Fig. 3). Taken together, these results indicated that undifferentiated hESCs differentiated into cardiac lineage and the differentiated cells showed the characteristics of cardiac precursor cells.
3.3 MicroRNA expression profile of cardiac lineage differentiation
To analyze the expression profile of selected miRNAs during differentiation from hESCs into cardiac lineage cells, we evaluated the differentiated cells collected at the stages of every 4 days using miRNAs -specific qRT-PCR. Characteristically, the stages from day 4 to day 8 showed dynamic up-regulation of miRNAs, which may be a pre-requisite for differentiation into cardiac lineage (Figs. 4, 5, 6, 7). This expression pattern was in contrast to those of undifferentiated hESCs and of differentiated beating cardiomyocytes. Taken together, the expression of miRNA is correlated to the differentiation into cardiac precursor and those effects were stage-specific.
3.4 Expression profile of miR-1
miR-1 is known to be expressed during the entire stage of differentiation. This important miRNA was down-regulated at the induction stage (day 0 to day 4), and drastically up-regulated during the transition stage from early mesoderm to cardiac mesoderm (day 5 to day 12). Its expression was down-regulated till maturation (day 12 to day 23) and up-regulated again (day 24 to day 28) (Fig. 4).
3.5 Expression profile of miRs related to cardiac development
Cardiac development-relatedmiRs-30d, and -143 were selected based on the database search (www.miRbase.org). The expression of miR-143 was extremely up-regulated at differentiation day 8 and down-regulated till maturation stage. After the appearance of beating cardiomyocyte precursor cells, the expression level was up-regulated again and the expression level was highest at differentiation day 28. The expression of miR-30d was also up-regulated during differentiation, however, their highest level was observed at day 8, its expression level was maintained as low as that of undifferentiated hESCs, and then increased at the later stage of differentiation (Fig. 5).
3.6 Expression profile of miRNAs related to cardiomyocyte precursor differentiation
The expression patterns of cardiomyocyte precursor differentiation-related miR-133a and miR-499a was evaluated during differentiation. They were significantly up-regulated on day 8 and their expression showed an increase as high as 20-fold those of other evaluated miRNAs. The expression of miR-499a was increased again after maturation.
3.7 Expression profile of miRNAs related to regulation of functionality
The expression of functionality-related miR-145 was slightly down-regulated at day 4, was up-regulated at day 8 and was maintained the expression level until maturation at least twofold in comparison with undifferentiated hESCs. The expression was significantly up-regulated at day 28 and this phenomenon was not observed in the expression pattern of other miRNAs. The expression of miR-378a was also up-regulated at day 8 and then down-regulated as low as undifferentiated hESCs levels until maturation stage.
4 Discussion
To the best of our knowledge, this is the first report that elucidated the serial expression pattern of selected cardiomyocyte-associated miRNAs during the entire differentiation duration of hESCs into cardiac lineage, although there have been numerous reports on the selected miRNAs that regulate the differentiation and proliferation in human-derived cardiomyocyte progenitor cells [28], smooth muscle cell fate and plasticity [29].
The authors demonstrated the profile of miRNA expression at various stages during cardiac lineage differentiation of hESCs. This attempt was very important since differentiation-inducing factor expression is highly dynamic over the time course of differentiation. Through this study, many miRNAs were successfully searched (Table 2), and these miRNAs can be good candidates that can be co-treated with conventional cardiac lineage differentiation-inducing factors to enhance the differentiation efficiency along with mechanical stimuli [30].
Our data showed that the expression pattern of microRNAs was dynamic during cardiac lineage differentiation of hESCs. Analyzed miRNAs were significantly up-regulated at differentiation day 8, the transition stage to early mesoderm. Their expression was down-regulated when their cell fate was committed to cardiac lineage (Figs. 4, 5, 6, 7).
The expression of miR-1 was highly up-regulated at the transition stage into cardiac mesoderm in our study (Fig. 4). This is supported by the previous report thatmiR-1 is known to enhance the differentiation into cardiac cells by inhibiting differentiation into neuronal linages [18]. These results showed that miR-1 importantly contributed to the cardiac lineage differentiation in the employed cell line, SNUhES3.
Interestingly, the expression pattern of miR-143 was different from those of other miRNAs (Fig. 5). It was highly expressed at day 8, down-regulated till day 16 and up-regulated again after maturation (day 20) and reached the highest at late stage (day 28). Targets of miR-143 include mesoderm induction gene, MIER3, myosin light chain interacting protein and potassium voltage-gated channel related genes [29]. These results imply that miR-143 may play an important role in cardiac differentiation andin maintaining the functionality of hESC-derived cardiac cells. Intriguingly, cardiac development-related miR-30d showed a high expression pattern at day 8, then down-regulated, and again up-regulated at day 24. It is known that miR-30d interacts with a major cardiac lineage transcription factor, MEF2C/D, which is amajor determinant into cardiac lineage, and with K + channel-related KCN family [31].
miR-133a and miR-499a also showed the highest expression at day 8 (Fig. 6). miR-133a is known to interact with myosin heavy chain (MHC), major structural protein of cardiomyocytes [32]. miR-499a is known to interact with Tbx3, a major determinant into cardiac lineage, that is expressed at patterning stage of cardiac development and restrictively expressed atrium and left ventricle at the later stage of development [33]. Our results may imply that they contribute to structural formation and ultimately the maintenance of hESC-derived cardiomyocytes.
The expression level of miR-145 was sustained after up-regulated at day 8 (Fig. 7). It has been reported to contribute to differentiation and functionality of hESC-derived CMs [34]. The expression of miR-378a was up-regulated at day 8 and down-regulated as low as undifferentiated cells. It is known to interact with BMP2 receptor that is involved in the signaling pathway that regulates the cardiac development [35]. Therefore, these miRNAs seem to play at the cardiac-specific differentiation.
This study has some limitations. Firstly, the observed data were retrieved from an XY cell line, therefore those from XX cell lines should be confirmed along with their differentiation efficiency. Secondly, the tested miRNAs were selected based on the database (www.miRbase.org), however, their overlapping interaction is currently unavailable. The number and combination of various miRNAs’ expression should be further investigated. Finally, the authors are planning to search and evaluate the target gene expression [36, 37] of analyzed miRNAs in the future investigations.
In conclusion, we elucidated the expression profile of miRNAs during hESC differentiation into cardiomyocytes. Our findings demonstrate the expression of miRNAs was stage-dependent during cardiac lineage differentiation and thus suggest that the differentiation into CMs can be regulated by miRNAs through direct or indirect pathway.
References
Oh SK, Kim HS, Ahn HJ, Seol HW, Kim YY, et al. Derivation and characterization of new human embryonic stem cell lines: SNUhES1, SNUhES2, and SNUhES3. Stem Cells. 2005;23:211–9.
Kim YY, Ku SY, Huh Y, Liu HC, Kim SH, et al. Anti-aging effects of vitamin C on human pluripotent stem cell-derived cardiomyocytes. Age. 2013;35:1545–57.
Kim YY, Ku JB, Liu HC, Ku SY, Kim SH, et al. Ginsenosides may enhance the functionality of human embryonic stem cell-derived cardiomyocytes in vitro. Reprod Sci. 2014;21:1312–8.
Kim YY, Ku SY, Jang J, Oh SK, Kim HS, et al. Use of long-term cultured embryoid bodies may enhance cardiomyocyte differentiation by BMP2. Yonsei Med J. 2008;49:819–27.
Kim YY, Ku SY, Liu HC, Cho HJ, Oh SK, et al. Cryopreservation of human embryonic stem cells derived-cardiomyocytes induced by BMP2 in serum-free condition. Reprod Sci. 2011;18:252–60.
Kim YY, Ku SY, Rosenwaks Z, Liu HC, Oh SK, et al. Red ginseng extract facilitates the early differentiation of human embryonic stem cells into mesendoderm lineage. Evid Based Complement Alternat Med 2011. doi:10.1155/2011/167376.
Adam AA, Takahashi Y, Katagiri S, Nagano M. In vitro culture of mouse preantral follicles using membrane inserts and developmental competence of in vitro ovulated oocytes. J Reprod Dev. 2004;50:579–86.
Kim YY, Ku SY, Kim YJ, Cho MS, Oh SK, et al. Effects of mycoplasma elimination agent on maintenance and cardiogenic potentials of human embryonic stem cells. Tissue Eng Regen Med. 2010;7:419–24.
Li Y, Du W, Zhao R, Hu J, Li H, et al. New insights into epigenetic modifications in heart failure. Front Biosci (Landmark Ed). 2017;22:230–47.
Kim YJ, Ku SY, Rosenwaks Z, Liu HC, Chi SW, et al. MicroRNA expression profiles are altered by gonadotropins and vitamin C status during in vitro follicular growth. Reprod Sci. 2010;17:1081–9.
Kim YJ, Ku SY, Kim YY, Liu HC, Chi SW, et al. MicroRNAs transfected into granulosa cells may regulate oocyte meiotic competence during in vitro maturation of mouse follicles. Hum Reprod. 2013;28:3050–61.
Kim YJ, Ku SY, Kim YY, Suh CS, Kim SH, et al. MicroRNA profile of granulosa cells after ovarian stimulation differs according to maturity of retrieved oocytes. Geburtshilfe Frauenheilkd. 2016;76:704–8.
Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–58.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25.
Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–97.
Ivey KN, Muth A, Arnold J, King FW, Yeh RF, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–29.
Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet. 2010;3:426–35.
Takaya T, Ono K, Kawamura T, Takanabe R, Kaichi S, et al. MicroRNA-1 and MicroRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J. 2009;73:1492–7.
Li Q, Song XW, Zou J, Wang GK, Kremneva E, et al. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci. 2010;123:2444–52.
Wang H, Cai J. The role of microRNAs in heart failure. Biochim Biophys Acta. 2016;S0925–4439(16):30328–3.
Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. Sci World J. 2007;7:155–66.
Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–74.
Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22.
He L, He X, Lim LP, de Stanchina E, Xuan Z, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4.
Oh SK, Kim HS, Park YB, Seol HW, Kim YY, et al. Methods for expansion of human embryonic stem cells. Stem Cells. 2005;23:605–9.
Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–68.
Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–32.
Shradhanjali A, Riehl BD, Kwon IK, Lim JY. Cardiomyocyte stretching for regenerative medicine and hypertrophy study. Tissue Eng Regen Med. 2015;12:398–409.
Hotchkiss A, Feridooni T, Zhang F, Pasumarthi KB. The effects of calcium channel blockade on proliferation and differentiation of cardiac progenitor cells. Cell Calcium. 2014;55:238–51.
Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J Mol Cell Cardiol. 2016;94:107–21.
Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–22.
Wang YS, Li SH, Guo J, Mihic A, Wu J, et al. Role of miR-145 in cardiac myofibroblast differentiation. J Mol Cell Cardiol. 2014;66:94–105.
de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–54.
Kim YY, Kim H, Ku S, Suk CS, Kim SH, et al. Effects of estrogen on intracellular calcium-related T-lymphocyte function. Tissue Eng Regen Med. 2016;13:270–3.
Kim YY, Kim YJ, Cho KM, Kim SH, Park KE, et al. The expression profile of angiotensin system on thawed murine ovaries. Tissue Eng Regen Med. 2016;13:724–32.
Acknowledgements
This study was supported by grants of Ministry of Science, ICT and Future Planning (2014R1A1A305264 and 2016R1E1A1A01943455). The authors would like to express sincere thanks to the technical assistance of Kyung Mee Cho and Mi Ae Lee.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest.
Ethical statement
The study was approved by the IRB of the IRMP, MRC, Seoul National University (219932-201412-LR-12-01-01).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary movie. Representative beating cluster of cardiomyocytes differentiated from human embryonic stem cells (MPG 5067 kb)
Rights and permissions
About this article
Cite this article
Kim, Y.Y., Min, H., Kim, H. et al. Differential MicroRNA Expression Profile of Human Embryonic Stem Cell-Derived Cardiac Lineage Cells. Tissue Eng Regen Med 14, 163–169 (2017). https://doi.org/10.1007/s13770-017-0051-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-017-0051-4