Abstract
Microbial indicators are routinely applied to test the efficacy of the drinking water treatment process. Clostridium perfringens spores and faecal coliforms are being suggested as indicators in order to evaluate the efficacy of the drinking water treatment process. Serial dilutions of Escherichia coli and extracted C. perfringens spores were prepared and filtered using 0.2-µm cellulose nitrate filter membranes. After filtration, elution and conventional cultivation methods were performed for C. perfringens and E. coli. Dark and pinkish colonies in the attributed media confirmed the presence of C. perfringens and E. coli, respectively. Cultivation method showed the limit of detection 102 and 104 CFU/mL for C. perfringens and E. coli, respectively. In addition, PCR assays showed the limit of detection 102 CFU/mL for both C. perfringens and E. coli. Elution of the spores/bacteria followed by the molecular method is a rapid and easy method that has similar results with traditional cultivation method, and decreases both cost and time for the assessment of the quality of water treatment process.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Waterborne microbial infections are considered as one of the most important public health concerns in both developing and developed countries. However, it seems that approximately 4.0% of all deaths and 5.7% of the total disease burden all over the world can be caused by waterborne pathogens (Prüss et al. 2002). According the previously published studies, faecal coliforms like E. coli and Enterococci are the indicators that have been commonly used to evaluate the efficacy of water treatment process. Almost all faecal coliforms are more susceptible to the presence of chlorine in comparison with cysts/oocysts of parasites and enteric human viruses, particularly in the aquatic conditions (Lipp et al. 2001; Tree et al. 2003; Wéry et al. 2008; Ryzinska-Paier et al. 2011). Nevertheless, despite the nonspecific faecal origin (Scott et al. 2002; Simpson et al. 2002), multiplying in environments (Solo-Gabriele et al. 2000; John et al. 2009), unknown contamination sources (Field et al. 2003) and low specificity of faecal coliforms as indicators for the assessment of the drinking water treatment process (Hörman et al. 2004; Byamukama et al. 2005; Savichtcheva and Okabe 2006; Girones et al. 2010; Parasidis et al. 2013), faecal coliforms, such as E. coli, together with C. perfringens spores could be good indicators to measure the efficacy of the water treatment process (Payment and Franco 1993). C. perfringens forms spores that increase the chance of prolonged detection in the environment in case of faecal contamination. Therefore, evaluation of the presence of spores of C. perfringens can help to assess the quality of the water treatment process (Araujo et al. 2004). Payment and colleague reported that C. perfringens spores are resistant facing routine chlorination and could be useful for the assessment of rough wall enteric microbes such as cysts/oocysts and ova of parasites and also viruses with faecal origin, in water resources (Payment and Franco 1993). Several studies have suggested different techniques to evaluate the efficacy of the drinking water treatment process (Araujo et al. 2004; Wohlsen et al. 2006; Girones et al. 2010; De Keuckelaere et al. 2013). Approximately, all studies have described the filtration of treated water followed by cultivation of microbes collected on top of the membrane on specific culture media. The necessity for specific culture media for distinct microorganisms and the time-consuming process of cultivation are known as the main limitations of conventional methods. Recently, an elution method has been described as sensitive method to recover enteric viruses from water samples (De Keuckelaere et al. 2013). However, this method has not been applied for evaluation of other microbial indicators from water samples. Therefore, it is hypothesized that an easy-to-use protocol can improve the pitfalls of the conventional methods, help simultaneous recovery of the faecal coliform and spore indicators, and save both time and cost of the water quality evaluation, as well. This study was performed in Foodborne and Waterborne Diseases Research Center, Shahid Behesshti University of Medical Sciences during June to December 2016. The current study aimed to evaluate the elution technique followed by the molecular method that was previously introduced to recover virus particles and compare it with the conventional culture-dependent method to recover microbial indicators from treated water.
Materials and methods
Samples preparation and serial dilution
Clostridium perfringens: briefly, a previously proven C. perfringens strains (RIGLD-2) (Ganji et al. 2017) was cultivated on egg yolk agar medium (Merck, Germany) supplemented with 7% sheep blood and 0.04% neomycin. The cultured plate was incubated at 37 °C for at least 48–96 h under anaerobic conditions (Anoxomat, MART Microbiology, The Netherlands). The plate of egg yolk agar medium with pure colonies was placed at room temperature for 10 days to induce sporulation process. In order to confirm sporulation of C. perfringens, gram staining was performed on pure colonies before and after incubation of the colonies at room temperature. C. perfringens colonies were washed three times by phosphate-buffered saline (PBS) with pH 8, and then the plate was treated with buffer consisted of lysozyme (Sigma, USA) 50 μg/mL and 1 mM Tris-HCl, pH = 7.5 at 37 °C for 30 min. Afterwards, extracted spores were washed five times using sterile PBS and centrifuging at 12,000 rpm for 10 min. Finally, the extracted spores were diluted in defined volumes of sterile PBS.
E. coli: 100 μL of the stock of E. coli (ATCC25923) was cultured on MacConkey Agar (Merck, Germany) and incubated for 24 h. The bacterial colonies were suspended in sterile PBS, and then the turbidity of bacterial suspension was adjusted equivalent to a 0.5 McFarland standard.
Serial dilutions for C. perfringens spores, E. coli colonies and a mixture of bacteria and spores were freshly prepared at defined amounts of sterile PBS to give final concentrations, 101, 102, 104, 106 and 108 colony-forming units (CFU)/mL.
Analysis of samples
All serially diluted mixtures, which were prepared as mentioned in the sample preparation section, were filtered using sterile 47-mm cellulose nitrate membrane (Sartorious, Goettingen, Germany) with pore size 0.2 µm and polysulfone in-line filter holder for 50-mm filters (Whatman). After that, one of the following procedures was performed for each of the filtered membranes.
Cultivation
Filtration of the diluted samples (separate suspensions of spores and bacteria) was done using sterile 0.2-µm cellulose nitrate filter membranes. Then, the filtered membrane was placed on the specific media (egg yolk agar and MacConkey agar for C. perfringens and E. coli, respectively). Cultivated egg yolk agar for each dilution was incubated in anaerobic condition, and all the cultivated media were examined for growth of bacteria every 48 h for a week. Furthermore, cultivated MacConkey agar media were incubated in aerobic condition at 37 °C for 24 h.
Elution
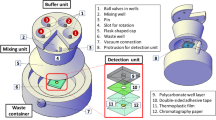
After filtration of each of the serially diluted mixtures (suspensions of spores and bacteria), filtered spores/bacteria were eluted as mentioned elsewhere (De Keuckelaere et al. 2013). Briefly, the membrane was rinsed in 300 mL 0.5 mM H2SO4 (pH 3.4). Then, 50 mL elution buffer (0.05 M KH2PO4, 1 M NaCl, 0.1% (v/v) Triton X-100, pH 9.2) was added to the membrane and remained on the filter membrane without pressure to soak the membranes for 10 min. Afterwards, the elution was filtered through the membranes to gather spores and bacteria. Then, the filtered elution was immediately counteracted using 1 M HCl. The eluted samples were centrifuged for 15 min at 3000 rpm, supernatant was discarded, and the pellet was collected in sterile tubes. Acquired pellets were introduced to DNA extraction kit. Figure 1 shows the schematic overview of the applied methods.
Molecular identification
DNA extraction
DNA of the pellets was extracted using Instagen kit (InstaGene TM Matrix, Bio-Rad, USA) according to manufacturer’s procedure. Extracted DNA was stored at − 20 °C for further investigations.
Polymerase chain reaction
For identification of C. perfringens, amplification of about 279-bp fragment of 16S ribosomal RNA (rRNA) gene was carried out in a final volume of 25 μL consisted of 2.5 µL of 10X PCR buffer, 0.5 µL of each primer (20 picomol), 1 µL of MgCl2 (50 mM), 0.5 µL of each deoxy ribonucleotide triphosphate (dNTP) (10 mM), 0.3 U of Taq DNA polymerase (5 U/mL) and 3 µL of DNA. PCRs were accomplished using primers forward, 5′-AAAGATGGCATCATCATTCAAC-3′, and reverse, 5′-TACCGTCATTATCTTCCCCAAA-3′, which were mentioned elsewhere (Wu et al. 2009) in a thermocycler (AG 22331; Eppendorf, Hamburg, Germany) as the following: initial denaturation for 10 min at 95 °C and then 30 cycles of 95 °C for 10 s, 52 °C for 1 min and 72 °C for 1 min, and final extension at 72 °C for 10 min.
Moreover, 95-bp fragment of 16S rRNA gene was amplified using primers forward, 5′-CATGCCGCGTGTATGAAGAA-3′, and reverse, 5′-CGGGTAACGTCAATGAGCAAA-3′, which were introduced previously (Huijsdens et al. 2002), to detect E. coli. The 25 µL PCR mixture consisted of 10X buffer, 1 U Taq polymerase, and 1.5 mM of MgCl2, 0.5 mM dNTPs and 0.5 µL 20 ρmol of each primer and 5 µL of DNA sample. PCR conditions were as follows: initial denaturation at 5 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 58 °C and 1.5 min at 72 °C, and final extension at 72 °C for 10 min.
Ten microlitres of amplified products was electrophoresed on 1.2% agarose gel, and then the PCR products were stained with ethidium bromide and visualized under UV light (Cleaver scientific Ltd, Warwickshire, UK).
Results and discussion
In order to motivate the sporulation of C. perfringens, cultured media were placed at room temperature for 10 days. The presence of C. perfringens spores was confirmed using gram staining before and after the sporulation period. Accordingly, microscopic examination showed gram-positive spore-forming bacilli of C. perfringens at the beginning of the sporulation process that after 72 h, spore production was started and after 10 days almost all vegetative forms were changed to the spore formation.
Cultivation method
Clostridium perfringens: All the cultured egg yolk media supplemented by Neomycin and 7% sheep blood were kept out at 37 °C and checked every 48 h for a week. Specific dark colonies of C. prefringens were considered as positive. The results showed that the limit of detection for C. perfringens spores filtered by 0.2-µm cellulose nitrate membrane was 102 spores/mL.
E. coli: Cultivation of the filtered membrane of E. coli was carried out on MacConkey agar media. The results showed that only dilution ≥ 104 CFU/mL successfully grew in MacConkey agar after 24 h. For further confirmation, expected fragments of 16S rRNA gene were amplified for both C. prefringens- and E. coli-positive colonies.
Elution method
C. prefringens: Amplification of 16S rRNA gene for the spores resulted from the elution process showed that the expected fragment of C. prefringens was amplified in dilutions with 102–108 spores/mL. The sample with 10 spores/mL was positive for neither cultivation method nor concentration method (Figs. 2a, b, 3a, b).
E. coli: The results showed that the 95-bp fragment of 16S rRNA gene of E. coli was only amplified for the eluted bacteria with bacterial concentration > 102 CFU/mL (Fig. 2a, b).
Numerous approaches are available to assess the microbial quality of water resources to reduce the risk of waterborne outbreaks (Tyagi et al. 2007). Evaluation of microbial indicators such as coliforms, E. coli, as well as C. perfringens spores have been introduced as a reliable measurement tool to monitor the efficacy of water treatment process, so far. It seems that these indicators with their morphology, ability to persist in the environment and resistance during treatment processes, are similar to harmful pathogens (Nappier et al. 2006). Moreover, because of this fact that coliform bacteria are easily detectable in water samples, they are useful indicators to assess the treatment process. The presence of faecal coliform bacteria as well as C. perfringens spores in treated water samples in more numbers than the allowed range (Lee et al. 2002) indicates both/either the possibility of pollution of the water supply system with the faeces and/or an insufficient disinfection process (Rompré et al. 2002).
In the current study, the conventional cultivation was evaluated and compared with elution followed by the molecular method for the detection of C. perfringens spores and E. coli in water samples in order to introduce a rapid and reliable technique to evaluate the efficacy of the water treatment process.
Recently, Keuckelaere and colleagues introduced and evaluated four different methods to concentrate the enteric viruses from water resources using filtration. This study showed that the elution method using electronegative cellulose nitrate filter membrane and Tr alk elution buffer was more effective to recover the enteric viruses from water sample (De Keuckelaere et al. 2013). In the current study, the aforementioned method with some modifications was used to recover the microbial indicators (E. coli and C. perfringens) from water samples. The findings were in agreement with the mentioned study and showed a consistency between C. perfringens spore counts in the filter membrane method followed by cultivation with those determined by the elution method followed by the molecular test. The analysis of culture and molecular method for detection of C. perfringens showed a sensitivity limit of 102 spores/mL. Additionally, the findings represented discordance between the results of the elution method followed by molecular methods and the conventional cultivation of filter membrane for E. coli. Interestingly, the limit of detection of elution followed by molecular methods was 102 CFU/mL, whereas the limit of detection of conventional cultivation of filter membrane was 104 CFU/mL.
There are several methods like pour plates and membrane filter methods for a quantitative detection of C. perfringens spores in water resources (Mead 1985; Araujo et al. 2004; Maheux et al. 2013). Sartory and colleagues proved the selective role of elevated temperatures for the detection of C. perfringens in water samples (Sartory 1986). All of the applied techniques have been based on the sulphite reduction and stormy fermentation after anaerobic incubation. Although sulphite reduction has been introduced as the best test for the detection of C. perfringens, this method is not specific enough for the identification of C. perfringens in water samples.
Furthermore, several techniques for enumeration of coliform bacteria and E. coli like multiple-tube fermentation (MTF), the membrane filter (MF) technique, enzymatic procedures and direct determination of enzymatic activity by fluorimetry have been approved as common procedures for testing of water quality in many countries (Rompré et al. 2002; Mesquita and Noble 2013; Hojris et al. 2016).The main disadvantages of the conventional cultivation method are: (1) subculturing of C. perfringens colonies is time-consuming, (2) no specificity of morphological criteria of C. perfringens colony and (3) complex cultivation and further confirmation steps that are needed for faecal coliforms (Rompré et al. 2002). Therefore, the molecular methods have been described as appropriate tests for identification of the faecal coliform, E. coli, as well as spores of C. perfringens during water treatment process (Wohlsen et al. 2006; Deshmukh et al. 2016).
Signoretto showed that the PCR method due to its advantages is a valuable implement for identification of microbial indicators/pathogens in water resources that can generate abundant as well as important information about the common and emerging/re-emerging microbes (Signoretto and Canepari 2008). However, the filtration method is an acceptable method for detection of low counts of microbial indicators such as C. perfringens spores and E. coli in high volume of water samples. According the findings, elution followed by molecular method showed sufficient sensitivity, and specificity compared with the conventional cultivation. Therefore, concerning the reliable results, rapidity and the cost-effectiveness of the elution followed by molecular method, this approach could be introduced for evaluation of efficacy of water treatment process.
Conclusion
Elution of spore/bacteria followed by molecular method is a rapid and easy method that was able to detect 102 CFU/mL of both C. perfringens spores and E. coli. In addition, traditional cultivation method was only able to recover 102 and 104 CFU/mL from water samples for C. perfringens spores and E. coli, respectively. The current study suggested that elution method followed by molecular method is not only a sensitive technique for detection of virus particles in water samples as mentioned previously, but also has higher sensitivity for detection of microbial indicators during water treatment process in comparison with conventional methods. Furthermore, this method can decrease both cost and time (E. coli and spores of C. perfringens need at least 24 h and 3–4 days, respectively, to grow in specific media while elution followed by molecular technique needs only 3–4 h) in order to evaluate the efficacy of water treatment process.
References
Araujo M et al (2004) Enumeration of Clostridium perfringens spores in groundwater samples: comparison of six culture media. J Microbiol Methods 57:175–180
Byamukama D et al (2005) Discrimination efficacy of fecal pollution detection in different aquatic habitats of a high-altitude tropical country, using presumptive coliforms, Escherichia coli, and Clostridium perfringens spores. Appl Environ Microbiol 71:65–71
De Keuckelaere A et al (2013) Evaluation of viral concentration methods from irrigation and processing water. J Virol Methods 187:294–303
Deshmukh RA et al (2016) Recent developments in detection and enumeration of waterborne bacteria: a retrospective minireview. Microbiol Open 5:901–922. https://doi.org/10.1002/mbo3.383
Field KG, Bernhard AE, Brodeur TJ (2003) Molecular approaches to microbiological monitoring: fecal source detection. Environ Monit Assess 81:313–326
Ganji L et al (2017) Comparison of the detection limits of the culture and PCR methods for the detection of Clostridium difficile, Clostridium perfringens, Campylobacter jejuni, and Yersinia enterocolitica in human stool. Arch Pediatr Infect Dis 5:e38888. https://doi.org/10.5812/pedinfect.38888
Girones R et al (2010) Molecular detection of pathogens in water—the pros and cons of molecular techniques. Water Res 44:4325–4339
Hojris B et al (2016) A novel, optical, on-line bacteria sensor for monitoring drinking water quality. Sci Rep 6:23935. https://doi.org/10.1038/srep23935
Hörman A et al (2004) Campylobacter spp., Giardia spp., Cryptosporidium spp., noroviruses, and indicator organisms in surface water in southwestern Finland, 2000–2001. Appl Environ Microbiol 70:87–95
Huijsdens XW et al (2002) Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J Clin Microbiol 40:4423–4427
John P et al (2009) Persistence and growth of faecal culturable bacterial indicators in water column and sediments of Vidy Bay, Lake Geneva, Switzerland. J Environ Sci 21:62–69
Lee SH et al (2002) Surveillance for waterborne-disease outbreaks–United States, 1999–2000. MMWR Surveill Summ 51(8):1–47
Lipp EK, Farrah SA, Rose JB (2001) Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar Pollut Bull 42:286–293
Maheux AF et al (2013) Abilities of the mCP Agar method and CRENAME alpha toxin-specific real-time PCR assay to detect Clostridium perfringens spores in drinking water. Appl Environ Microbiol 79:7654–7661. https://doi.org/10.1128/AEM.02791-13
Mead G (1985) Selective and differential media for Clostridium perfringens. Int J Food Microbiol 2:89–98
Mesquita S, Noble RT (2013) Recent developments in monitoring of microbiological indicators of water quality across a range of water types. In: Wurbs R (ed) Water resources planning, development and management. Chap 2, Texas A&M University, College Station, TX, USA, pp 29–53
Nappier SP, Aitken MD, Sobsey MD (2006) Male-specific coliphages as indicators of thermal inactivation of pathogens in biosolids. Appl Environ Microbiol 72:2471–2475
Parasidis TA, Konstantinidis TG, Alexandropoulou IG (2013) Environmental monitoring of enteric viruses in wastewater. Virol Mycol 2:e106. https://doi.org/10.4172/2161-0517.1000e106
Payment P, Franco E (1993) Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl Environ Microbiol 59:2418–2424
Prüss A et al (2002) Estimating the burden of disease from water, sanitation, and hygiene at a global level. Environ Health Perspect 110:537–542
Rompré A et al (2002) Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J Microbiol Methods 49:31–54
Ryzinska-Paier G et al (2011) Acid phosphatase test proves superior to standard phenotypic identification procedure for Clostridium perfringens strains isolated from water. J Microbiol Methods 87:189–194
Sartory D (1986) Membrane filtration enumeration of faecal clostridia and Clostridium perfringens in water. Water Res 20:1255–1260
Savichtcheva O, Okabe S (2006) Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res 40:2463–2476
Scott TM et al (2002) Microbial source tracking: current methodology and future directions. Appl Environ Microbiol 68:5796–5803
Signoretto C, Canepari P (2008) Towards more accurate detection of pathogenic Gram-positive bacteria in waters. Curr Opin Biotechnol 19:248–253
Simpson JM, Santo Domingo JW, Reasoner DJ (2002) Microbial source tracking: state of the science. Environ Sci Technol 36:5279–5288
Solo-Gabriele HM et al (2000) Sources of Escherichia coli in a coastal subtropical environment. Appl Environ Microbiol 66:230–237
Tree JA, Adams MR, Lees DN (2003) Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl Environ Microbiol 69:2038–2043
Tyagi V et al (2007) Alternative microbial indicators of faecal pollution: current perspective. J Environ Health Sci Eng 3:205–216
Wéry N et al (2008) Behaviour of pathogenic and indicator bacteria during urban wastewater treatment and sludge composting, as revealed by quantitative PCR. Water Res 42:53–62
Wohlsen T et al (2006) Evaluation of an alternative method for the enumeration and confirmation of Clostridium perfringens from treated and untreated sewages. Lett Appl Microbiol 42:438–444
Wu J et al (2009) Detection and toxin typing of Clostridium perfringens in formalin-fixed, paraffin embedded tissue samples by PCR. J Clin Microbiol 47:807–810
Acknowledgements
This study was part of the proposal which was done in the Foodborne and Waterborne Diseases Research Center. This Project No. RIGLD 813 was financially supported by Research Institute for Gastroenterology and Liver Diseases, Shahid Behehsti University of Medical Sciences, Tehran, Iran. The authors would like to appreciate all colleagues of Foodborne and Waterborne Diseases Research Center for their laboratory cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Editorial responsibility: Binbin Huang
Rights and permissions
About this article
Cite this article
Azimirad, M., Tajeddin, E., Hasani, Z. et al. Development of a culture-independent method for rapid monitoring of microbial indicators in water samples. Int. J. Environ. Sci. Technol. 16, 3165–3170 (2019). https://doi.org/10.1007/s13762-018-1680-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-018-1680-4