Abstract
Purpose
Cardiotocography (CTG), which measures uterine contraction (UC) and fetal heart rate (FHR), is a crucial tool for assessing fetal health during pregnancy. However, traditional computerized cardiotocography (cCTG) approaches have non-negligible calibration errors in feature extraction and heavily rely on the expertise and prior experience to define diagnostic features from CTG or FHR signals. Although previous works have studied deep learning methods for extracting CTG or FHR features, these methods still neglect the clinical information of pregnant women.
Methods
In this paper, we proposed a multimodal deep learning architecture (MMDLA) for intelligent antepartum fetal monitoring that is capable of performing automatic CTG feature extraction, fusion with clinical data and classification. The multimodal feature fusion was achieved by concatenating high-level CTG features, which were extracted from preprocessed CTG signals via a convolution neural network (CNN) with six convolution layers and five fully connected layers, and the clinical data of pregnant women. Eventually, light gradient boosting machine (LGBM) was implemented as fetal status assessment classifier. The effectiveness of MMDLA was evaluated using a dataset of 16,355 cases, each of which includes FHR signal, UC signal and pertinent clinical data like maternal age and gestational age.
Results
With an accuracy of 90.77% and an area under the curve (AUC) value of 0.9201, the multimodal features performed admirably. The data imbalance issue was also effectively resolved by the LGBM classifier, with a normal-F1 value of 0.9376 and an abnormal-F1 value of 0.8223.
Conclusion
In summary, the proposed MMDLA is conducive to the realization of intelligent antepartum fetal monitoring.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In antepartum care, cardiotocography (CTG) is the principal way to monitor fetal health, with continuous and simultaneous recording of fetal heart rate (FHR) and uterine contraction (UC) signals [1]. It enables obstetricians to further evaluate the fetal health and aids in the early diagnosis of abnormalities such as congenital heart malformations, fetal distress, or hypoxia [1, 2]. Additionally, it is non-invasive, clinically safe, inexpensive and has a high diagnostic value [3].
However, there is substantial subjectivity and limited reproducibility associated with obstetricians’ visual CTG interpretation in traditional clinical fetal monitoring. Hence, computerized cardiotocography (cCTG) was developed and exhibited certain advantages. The cCTG, in particular, can compute features that are problematic for obstetricians’ eyes to distinguish, such as short-term variation (STV) and long-term variability (LTV) [4]. Nonetheless, the effectiveness of these features has been clinically validated in assessing fetal status [5]. Therefore, cCTG interpretation has been crucial in non-stress testing (NST) for antepartum fetal monitoring [6].
Nowadays, the widely available of digital CTG data and the emergence of artificial intelligence (AI) technologies present great opportunities for substantial increasing the accuracy and scalability of cCTG analysis. Two approaches to automatic CTG evaluation based on machine learning (ML) have been developed: (1) feature extraction and classification based on traditional ML algorithms, and (2) CTG signal-based deep learning model for end-to-end CTG classification. Both have accomplished great outcomes [2].
Most of the current automated fetal monitoring methods involve complicated computer-based feature extraction and categorization using modern classifiers. Saleem et al. [7] constructed a robust automated classifier of fetal dynamics employing time-frequency (TF) features of fetal rate signals. Zeng et al. [8] addressed the CTG signal non-stationarity and the class imbalance by exploiting linear FHR features and TF features for training a cost-sensitive support vector machine classifier. Thippa et al. [9] trained classifiers based on principle component analysis (PCA) and linear discriminant analysis (LDA) utilizing nine CTG features and found the PCA-based classifier demonstrated superior performance than the LDA-based classifier. With a focus on the uncertainty and fuzziness of the CTG signal interpretation, Fei et al. [10] adopted nine CTG features after feature selection to establish an adaptive neural fuzzy inference system based on fuzzy C-means clustering (FCM-ANFIS). These methods all effectively enhanced the discrimination performance and assisted obstetrics in arriving at the discrimination results faster.
Overall, the above mentioned models depend mainly on the features, such as time-domain or frequency-domain features retrieved from CTG or FHR signals. They are severely confined by domain expertise to specify, and are collected by specialist equipment separately. Measurement errors that cannot be disregarded will arise during the secondary extraction of features [11, 12]. Consequently, deep learning models for CTG signal classification have emerged. Deep learning feature extraction no longer relies on artificial definition, but comes from automatic machine learning extraction. For example, Fergus et al. [13] utilized a one-dimensional convolutional neural network (1DCNN) and multi-layer perceptron (MLP) to potentially integrate and automatically extract features from time series Windows without manually making the feature extraction algorithms. A general framework for FHR feature extraction, selection, and classification based on a deep convolutional neural network (CNN) was proposed by Zhao et al. [14]. The short-term Fourier transform (STFT) of FHR signals was used by Cmert et al. [15] to construct a deep CNN architecture with a transfer learning approach for fetal hypoxia diagnosis. Ma’Sum et al. [16] introduced a deep learning method of hypoxia detection focused on FHR signals. Tang et al. [17] designed a neural network-based FHR signal classification method. They make good use of the deep learning model to extract the signal features directly and avoid the measurement errors of the secondary feature extraction. While the aforementioned CTG-based deep learning methods for fetal health monitoring disregarded the potential impact of maternal physiological factors on fetal growth and development, such as the gestational age and maternal age [18, 19].
Given the above gap in literature, we proposed a multimodal deep learning architecture (MMDLA) that can facilitate end-to-end learning of CTG signals by fusing pregnant women’s clinical features, such as maternal age and gestational age. A CNN with six convolutional layers and five fully connected layers was designed to automatically obtain the high-level features from the preprocessed CTG signals, and concatenated them with the preprocessed clinical data to accomplish multimodal feature fusion. Terminally, light gradient boosting machine (LGBM) acted as the fetal status assessment classifier for its receptiveness to unbalanced data [20].
The rest of the paper is organized as follows: in section II, an overview on CTG classification based on multimodal data is presented. Data description and exploration, data preprocessing and MMDLA for intelligent antepartum fetal monitoring are introduced conscientiously in section III. Section IV shows the experiment and results to verify the effectiveness of our proposed approach. Finally, we summarize the contribution of this paper and the plan for future work in section V.
Contributions In this paper, we proposed a framework to support multimodal medical data fusion for assisting doctors in preliminary screening. A summary of the contributions is as follows:
-
1.
We extracted high-level features from CTG signals by CNN, including both FHR signals and UC signals, rather than employing only FHR signals as in previous automatic CTG classification studies.
-
2.
We fused CTG signals with clinical data such as maternal age and gestational age to form multimodal features for classification.
-
3.
We utilized the classifier LGBM to alleviate the data-imbalanced problem, instead of the combination of fully connected layer and softmax function regularly employed in the multimodal networks.
Overview on CTG classification based on multimodal data
With the rapid development of computer technologies, healthcare facilities have entered the era of electronic medical records. The digitization of massive volumes of medical data provides an unprecedented opportunity for AI to improve medical care. These medical records reflect the intricacy of medical information in the real world study. It contains multiple modalities, such as demographic characteristics, clinical diagnosis text, blood test results, medical images, electrocardiograms and cardiotocographic signals. The goal of multimodal learning is to construct models that can efficiently process and link information utilizing various modalities [21], allowing for a better understanding and utilization of information from diverse modalities. Multimodal learning strategies have been widely applied in the medical and health domains, such as hybrid data exploration, brain tumor detection, and diagnosis of hepatocellular carcinoma [22,23,24]. Moreover, in antepartum fetal monitoring, some scholars have also employed multimodal data to conduct research on the automatic interpretation of CTG.
For instance, Qureshi et al. [25] employed four features of FHR, UC, fetal movements profile (FMP) and fetal heart sound (FHS) for segmentation of acceleration, deceleration and baseline reference parts. Petrizziello et al. [26] constructed a multimodal network to detect fetal compromise using the feature of FHR, uterine contractions and quality. Similarly, Improta et al. [27] fused the gestational age with other diagnostic CTG features for delivery type prediction. Chen et al. [28] proposed a deep-forest-based fetal monitoring model that categorizes CTG based on gestational age and other CTG diagnostic features. To monitor late intrauterine growth restriction, Pini et al. [29] combined time-domain, frequency-domain, time-frequency, and morphological aspects of FHR signals with gestational age, fetus gender and maternal age.
These studies employed time-domain and frequency-domain analysis of CTG or FHR recordings along with linear and nonlinear methodologies, and the clinical features of pregnant women were spliced directly with these CTG diagnostic features to generate multimodal data for classification, improving CTG interpretation for assessing fetal health. However, these multimodal approaches relied heavily on CTG for feature selection and definition, whose design, effectiveness, and interpretability were confined to the domain expertise. Alternatively, maternal physiological factors were ignored in some studies.
To handle the issues, we presented a MMDLA for intelligent antepartum fetal monitoring, which can fulfill end-to-end FHR and UC signal features extraction and effectively fuse them with pregnant women’s clinical data to achieve multimodal data.
Methods
Data description and exploration
From 2016 to 2018, collaborating hospitals collected antepartum CTG traces containing FHR and UC signals and recorded them via an SRF618A pro fetal monitor at 1.25 Hz (Fig. 1). Three obstetricians interpreted the CTG records for three rounds according to the antepartum fetal monitoring guideline of the 9th edition of Chinese Obstetrics and Gynecology [30]. There were 16355 cases in this dataset with consistent classification labels, including 11998, 4326 and 31 cases of normal, suspicious and pathological categories, respectively. Considering the scarcity of pathological cases, we classified pathological and suspicious case as the abnormal, as depicted in Fig. 2. This can realize the preliminary screening of abnormal cases.
Along with CTG signals, the described model includes clinical data from pregnant women. To explore the potential impact of maternal age and gestational age on fetal health, the classification results of the two characteristics were visualized in percent stacked bar chart (Figs. 3, 4). There were differences in the distribution of the maternal age and gestational age between normal and abnormal fetuses, indicating the possibility of introducing them as model inputs. Table 1 displays information about pregnant women.
Data preprocessing
Preprocessing of cardiotocographic (CTG) Signals
First, outliers at the beginning and end of the signal were directly deleted. When there was no sudden increase and decrease between the two signals, interpolation was not considered, and replaced with Nan. Isolated FHR values were also assigned to Nan and then analyzed. Let the first value of the outliers or missing segment be \(d_{st}, 1< st < N,\) and chose a segment \(d_{1:st}.\) Calculate the median number \(d_{med}\) of the segment, and replaced the \(d_{st}\) with \(d_{med}\).
The UC signal synchronizes with the FHR signal, therefore, it was interpolated or deleted simultaneously with the FHR signal. Let the start signal point of the outliers or missing segments be \(u_{st}, 1< st < N,\) then chose a segment \(u_{(st-60:st)},\) and updated \(u_{st}\) with median \(u_{med}.\) The remaining segments were processed in the same manner as described previously.
Acceleration and deceleration are significant parameters for fetal status evaluation in the clinical prenatal fetal stress test. Sencond, this work subtracted the baseline [31] to make them more noticeable. Finally, the FHR were decentralized, with the following decentralized formula:
where \({S_0}(t)\) is the pretreatment result, and B(t) is the fetal heart baseline.
After the above process, the CTG signals length of each case would be different. Here, we implemented signal segmentation and specified the last 15 min of the signal (1125 points) for further research. The pretreatment flow chart is displayed in Fig. 5.
Preprocessing of clinical data of pregnant women
The missing values of the pregnant women’s age characteristics were supplemented by the arithmetical average value. Thereafter, the zero-mean method was used to normalize the maternal and gestational age.
Dataset divisions
The signals were first randomly divided into the training group and the test group by a factor of 7:3, and then the training group was divided into a training set and a verification set by a factor of 8:2. The testing group functioned as a test set.
Multimodal deep learning architecture for intelligent antepartum fetal monitoring
We designed a MMDLA. It automatically retrieved high-level features from CTG signals and combined them with the clinical data of pregnant women, enabling intelligent monitoring of fetal health.
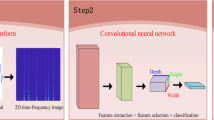
The CTG signals were first in the method preprocessed as shown in Fig. 5, and then fed into the model presented in Fig. 6. The model contained two stages: (1) acquisition of CTG high-level features, (2) formation of multimodal features, and output of classification results. The output of the final model is normal or abnormal, which can assist the doctor in performing an auxiliary preliminary screening. These two phases are described further below.
CNN-based high-level CTG feature extraction
Here, a convolutional neural network (CNN) was introduced to extract high-level features from preprocessed CTG signals, for it has strong feature learning ability. It also shares the convolution kernel, which allows signals to retain their original location relationship after convolution and can pay attention to subtle details. The convolutional layer was used to extract features from signal data, and the pooling layer was utilized for feature reduction. The fully connected layer can in charge of features selection and the batch normalization (BN) layer ensured that the input data distribution of each layer in the network was generally steady. Meanwhile, overfitting is a major issue in deep learning, hence, the dropout layer was employed to avoid it.
We constructed a CNN with six convolutional layers and five fully connected layers to extract high-level features from CTG signals in this work. The retrieved high-level signal features were then combined with two one-dimensional structured features, including maternal age and gestational age, to produce multimodal features. The parameter values displayed in Table 2 were found after multiple parameter modifications to optimize the CNN’s extraction of deep features from the signal. The CTG signal was fed into the model as input, and its processing in CNN is as follows:
The CTG signals \({t_i}\) were first input into the convolutional layer, and then output \({h_i}\). In this layer, the CTG features were retrieved using lots of convolution kernels.
where b is the bias.
Next, \({h_i}\) were fed into the average-pooling layer. Here, the layer averaged the vicinity of signals to achieve feature reduction and reduce the number of parameters.
Finally, the BN layer was utilized to do batch normalization before entering the convolutional layer next time.
The signals entered the global-average-pooling layer after passing through the sixth layer of BN, and the output is \({y_i}\). Eventually, it went through the five-layer connected layer to extract high-level features.
where L is the output of the fully connected layer, \(\partial\) is the weights, b is the bias.
Neurons that do not have extracted features or whose extracted features are negligible were discarded in the fully connected layer. Hence, after being selected, the CTG signals will generate important high-level features.
Multimodal feature fusion and LGBM-based fetal status assessment
High-level signal features are integrated with maternal age and gestational age after CNN-based deep learning to form multimodal features. Given the data-imbalanced challenge, the multimodal features were terminally input to the classifier named LGBM, rather than the combination of fully connected layer and softmax function that is commonly employed in the networks for classification. The parameters of the optimized LGBM are listed below in Table 3.
LGBM can handle data imbalance better than other classifiers by adjusting the class weight. And it adjustd the positive sample weight a to:
where \({N_n}\) is the number of negative samples, \({N_p}\) is the number of positive samples.
Intelligent antepartum fetal monitoring model. Stage 1: CTG signals were passed through CNN to obtain high-level features; Stage 2: Multi-modal features were formed by combining all CTG signals output by CNN, maternal age, and gestational age, and the features were then fed into the terminal classifier LGBM for classification. The model was made up of these two stages
Evaluation index
In order to verify the effectiveness of the model proposed in this paper, the confusion matrix was employed as the criterion for judging the classification model (Table 4). The confusion matrix of binary classification is defined as:
In this experiment, the following metrics were measured: accuracy (Acc), precision (Pre), recall, specificity (Spe), F1-score, kappa coefficient and Matthews correlation coefficient (MCC) and the area under curve (AUC). And the F1 of normal and abnormal category are respectively expressed as normal-F1 and abnormal-F1. The calculation methods of these indicators are as follows:
The horizontal and vertical coordinates of receiver operating characteristic (ROC) are respectively the false positive rate (FPR) and the true positive (TPR), and the formula is:
The AUC value is the area below the curve used to measure ROC. This value ranges from 0 to 1, and the larger the number, the better the classification effect.
Experiment and results
The experimental environment is originally introduced in this section. Then, three groups of comparative experiments are presented, together with related results analysis.
Experimental environment
In this paper, the experiments were implemented using Python deep learning framework Keras. Additionally, platform tools are: Python3.6.5, TensorFlow-GPU 1.9.0 and Keras 2.2.4. The machine’s CPU configuration is Intel (R) Xeon (R) E5-26700@2.60GHz, and the memory is 16 G. Furthermore, the graphics card is GeForce GTX 750Ti from NVIDIA and the operating system is Windows10 professional version 1709.
The performance of CNN-based high-level CTG feature extraction
The CNN structure design
Aiming to determine the optimum CNN structure to extract high-level features from CTG signals, nine alternative CNN architectures were examined. They were 6C2D (6 layers of convolutional layers, two layers of fully connected layers), 6C3D, 6C4D, 6C5D, 6C6D, 3C5D, 4C5D, 5C5D and 7C5D.
The results in Table 5 indicated that compared with other eight CNN structures, the adopted six-layer convolutional and five-layer fully connected structure (6C5D) exhibited the best discrimination performance. Its accuracy, specificity, F1-score, kappa coefficient and MCC were highest. Notably, it surpassed the worst-performing CNN architecture by 2% in accuracy, 3% in abnormal F1-score, and 4% in kappa coefficient and MCC. The high abnormal F1-score revealed a remarkable capacity to identify abnormal classes, and the high kappa coefficient proved that the discriminant result is highly consistent with the real result. In addition, the high MCC demonstrated that prediction performed effectively in all of four confusion matrix categories.
As can be observed from Fig. 7, the ROC curve of the 6C5D structure was above those of most of the other eight structures. It can be concluded that the prediction performance of the 6C5D structure was better than other structures, and Fig. 8 further presented that the 6C5D structure had the highest accuracy.
In summary, the CNN structure with six convolutional layers and five fully connected layers was employed in the proposed model.
Comparison with the signal-based deep learning model
To verify the superiority of features extracted from CTG signals for CNN discrimination, four different deep learning models were selected for comparison. They were deep neural networks (DNN), recurrent neuron network (RNN), gated recurrent unit (GRU) and long short-term memory (LSTM).
Table 6 shows that, when compared to the previous four deep learning models, other deep learning methods were associated with the problem of high specificity and low recall, however the CNN utilized in this article can effectively alleviate this problem. Moreover, the accuracy, precision, recall, F1-score, kappa coefficient, and MCC had all been considerably improved and were all among the top. Overall, these results illustrated CNN’s superiority in extracting high-level CTG features, and the predicted results were highly consistent with the real results.
Figure 9 shows that the ROC curve of CNN was extremely higher than that of other deep learning models and it also demonstrated that CNN outperformed other models in terms of prediction performance. Furthermore, as shown in Fig. 10, CNN had the highest F1-score and showed good convergence during training. Hence, we chose a convolutional neural network to extract the CTG signal’s high-level features.
The performance of LGBM-based fetal status assessment
Misclassifying abnormal cases belonging to minority class as normal ones can result in missing optimal treatment time and even jeopardize fetal health. To evaluating the performance of the proposed LGBM classifier on imbalance-data, k-nearest neighbor (KNN), support vector machine (SVM), decision tree (DT) [32], random forest (RF), back-propagation (BP), Bayes, gradient boosting decision tree (GBDT) were compared.
It can be observed in Table 7 that when dealing with unbalanced data, other machine learning models might be affected by the data set and are more inclined to judge the case as a normal class with a large amount. As a result, specificity, precision and normal class F1-score were falsely high, while recall, abnormal F1-score, kappa coefficient and MCC were low. However, when dealing with unbalanced data, the LGBM used in the proposed model and the GBDT performed admirably. They had a high recall, specificity, normal F1-score, aberrant F1-score, kappa coefficient, and MCC. The high recall and abnormal F1-score revealed excellent abilities to distinguish abnormal classes and effectively solved the imbalance problem. Furthermore, the high kappa coefficient and MCC indicated that the predicted results and the real results were highly consistent and all four confusion matrix categories had outstanding results.
In short, the LGBM employed in this study demonstrated the best discrimination performance, improved accuracy by about 7%, and successfully solved the imbalance problem. In addition, it compensated for the lack of utilizing CNN, which requires data balance, to learn signals directly. The ROC curve of LGBM was higher than that of the terminal classifiers, as shown in Fig. 11. It also demonstrated LGBM’s excellent performance.
The effect of multimodal representations on antepartum fetal monitoring
The discriminatory capability of five different feature combinations was assessed in this experiment to validate the effect of multimodal representations on antepartum fetal monitoring. They were FHR, FHR and uterine constriction (FHR + UC), FHR, UC and maternal age (FHR + UC + Age), FHR, UC and gestational age (FHR + UC + GA) and the multimodal representation in the proposed model (FHR + UC + GA + Age).
The results, as shown in Table 8, indicated that the models trained on multimodal data performed significantly better than single-modal models. After taking into account clinical characteristics of pregnant women such as gestational age and maternal age, the discrimination performance improved markedly, and the accuracy rate increased by approximately 8%. Obviously, the accuracy, precision, recall, specificity, F1-score, kappa coefficient and MCC of multimodal data comprised of CTG signals and the clinical characteristics of pregnant women were all at their peak. According to the findings, the multimodal representation produced the best classification effect in this study, and the discriminative results were highly consistent with the real results.
Figure 12 illustrated that the ROC curve of the multimodal representation we adopted was above that of most other combinations. It was obvious that the prediction performance of FHR + UC + GA + Age was better than other combinations.
Discussion and conclusions
In this work, we have developed a novel MMDLA for intelligent antepartum fetal monitoring. Four groups of comparative tests had been conducted to evaluate the performance of the proposed MMDLA. The experimental results demonstrated that the novel model has achieved outstanding performance in fetal status assessment and all of the evaluation metrics have clearly improved significantly. Overall, the MMDLA proposed in this paper is advantageous for the realization of intelligent antepartum fetal monitoring.
There are three major contributions to this paper. First, CNN acquired the CTG high-level features end-to-end, avoiding measurement errors in secondary extraction the diagnostic of features. Furthermore, the physiological differences of pregnant women and the previously disregarded uterine contraction signal were taken into account to achieve multimodal features, allowing diverse modalities’inputs to be adequately incorporated into the model. Finally, to overcome the imbalance problem, the LGBM was employed as the fetal status assessment classifier.
However, there are some limitations. The clinical features selected in this study were maternal age and gestational age. Other clinical data, such as body mass index (BMI), weight change, and blood pressure, can be considered in the future study. In addition, the data in this study has a certain imbalance problem. We will consider generative adversarial networks (GANs) for data enhancement to alleviate this problem better.
References
Al-Yousif S, Jaenul A, Al-Dayyeni W, Alamoodi A, Saleh AH. Distributed under creative commons cc-by 4.0 open access a systematic review of automated pre-processing, feature extraction and classification of cardiotocography. J Comput Sci Technol. 2021. https://doi.org/10.7717/peerj-cs.452
Georgieva A, Abry P, Chudáček V, Djurić PM, Frasch MG, Kok R, Lear CA, Lemmens SN, Nunes I, Papageorghiou AT, et al. Computer-based intrapartum fetal monitoring and beyond: a review of the 2nd workshop on signal processing and monitoring in labor (October 2017, Oxford, UK). Acta Obstet Gynecol Scand. 2019;98(9):1207–17.
Kahankova R, Martinek R, Jaros R, Behbehani K, Behar JA. A review of signal processing techniques for non-invasive fetal electrocardiography. IEEE Rev Biomed Eng. 2019;13:51–73.
Behar J.A, Weiner Z, Warrick P. Special session on computational fetal monitoring. In: 2019 Computing in cardiology (CinC), 2019;1–4 . https://doi.org/10.23919/CinC49843.2019.9005927.
Pereira S, Ingram C, Gupta N, Singh M, Chandraharan E. Recognising fetal compromise in the cardiograph during the antenatal period: pearls and pitfalls. Asian J Med Health. 2020. https://doi.org/10.9734/AJMAH/2020/v18i930238.
Ibrahim HA, Elgzar WT, Saied E. The effect of different positions during non-stress test on maternal hemodynamic parameters, satisfaction, and fetal cardiotocographic patterns. Afr J Reprod Health. 2021;25(1):81–9.
Saleem S, Naqvi S.S, Manzoor T, Saeed A, Rehman N.U, Mirza J A strategy for classification of ‘vaginal vs. cesarean section’ delivery: bivariate empirical mode decomposition of cardiotocographic recordings. Front Physiol. 2019;10:246.
Zeng R, Lu Y.S, Long S, Wang C, Murphy L. Cardiotocography signal abnormality classification using time-frequency features and ensemble cost-sensitive svm classifier. Comput Biol Med. 2021;130:104218.
Thippa RG, Praveen K, Lakshmanna K, Kaluri R, Baker T. Analysis of dimensionality reduction techniques on big data. IEEE Access 2020;8:54776–54788.
Fei Y, Huang X, Chen Q, Chen J, Wei H. Automatic classification of antepartum cardiotocography using fuzzy clustering and adaptive neuro-fuzzy inference system. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM); 2020.
Hruban L, Spilka J, Chudacek V, Janku P. Agreement on intrapartum recordings between expert-obstetricians. J Eval Clin Pract. 2015;21(4):694–702.
Romano M, Bifulco P, Ruffo M, Improta G, Cesarelli M. Software for computerised analysis of cardiotocographic traces. Comput Methods Programs Biomed. 2016;124(C):121–37.
Fergus P, Chalmers C, Montanez C.C, Reilly D, Pineles B. Modelling segmented cardiotocography time-series signals using one-dimensional convolutional neural networks for the early detection of abnormal birth outcomes. IEEE Trans Emerg Top Comput Intell. 2020;5(6):882–92.
Zhao Z, Deng Y, Zhang Y, Zhang Y, Zhang X, Shao L. DeepFHR: intelligent prediction of fetal Acidemia using fetal heart rate signals based on convolutional neural network. BMC Med Inform Decis Mak. 2019;19:286.
Cmert Z, Kocamaz AF. Fetal hypoxia detection based on deep convolutional neural network with transfer learning approach. Cham: Springer; 2018.
Ma’Sum MA, Intan P, Jatmiko W, Krisnadhi AA, Suarjaya I. Improving deep learning classifier for fetus hypoxia detection in cardiotocography signal. In: 2019 International workshop on big data and information security (IWBIS); 2019.
Haijing, Tg, Taoyi, Wang, Mengke, Li, Xu, Yang: The design and implementation of cardiotocography signals classification algorithm based on neural network. Comput Math Methods Med. 2018. https://doi.org/10.1155/2018/8568617.
Yavuz P, Taze M, Salihoglu O. The effect of adolescent and advanced-age pregnancies on maternal and early neonatal clinical data. J Maternal Fetal Neonatal Med. 2022;35(25):7399–405.
Ferguson KK, Sammallahti S, Rosen E, Dries MVD, Jaddoe VWV. Fetal growth trajectories among small for gestational age babies and child neurodevelopment. Epidemiology. 2021;32(5):664–671.
Ni X, Wang H, Shaolan LV, Xiong M. An ensemble classification model based on imbalanced data for aviation safety; 2021.
Xu Z, So DR, Dai AM. MUFASA: multimodal fusion architecture search for electronic health records; 2021. https://arxiv.org/abs/2102.02340.
Zhang Y, Sheng M, Liu X, Wang R, Lin W, Ren P, Wang X, Zhao E, Song W. A heterogeneous multi-modal medical data fusion framework supporting hybrid data exploration. Health Inf Sci Syst. 2022;10(1):1–11.
Jha M, Gupta R, Saxena R. A framework for in-vivo human brain tumor detection using image augmentation and hybrid features. Health Inf Sci Syst. 2022;10(1):1–12.
Menegotto AB, Becker CDL, Cazella SC. Computer-aided diagnosis of hepatocellular carcinoma fusing imaging and structured health data. Health Inf Sci Syst. 2021;9(1):1–11.
Zhong M, Yi H, Lai F, Liu M, Zeng R, Kang X, Xiao Y, Rong J, Wang H, Bai J. CTGNet: automatic analysis of fetal heart rate from cardiotocograph using artificial intelligence. Maternal Fetal Med. 2022. https://doi.org/10.1097/FM9.0000000000000147.
Petrozziello A, Redman CW, Papageorghiou AT, Jordanov I, Georgieva A. Multimodal convolutional neural networks to detect fetal compromise during labor and delivery. IEEE Access. 2019;7:112026–36.
Improta G, Ricciardi C, Amato F, D’Addio G, Romano M. Efficacy of machine learning in predicting the kind of delivery by cardiotocography. In: XV Mediterranean conference on medical and biological engineering and computing—MEDICON 2020;2019. p. 793–800.
Chen Y, Guo A, Chen Q, Quan B, Hao Z. Intelligent classification of antepartum cardiotocography model based on deep forest. Biomed Signal Process Control. 2021;67(2): 102555.
Pini N, Lucchini M, Esposito G, Tagliaferri S, Signorini MG. A machine learning approach to monitor the emergence of late intrauterine growth restriction. Front Artif Intell. 2021;4: 622616.
Xie B. X. Kong, Duan T. Gynecology and obstetrics, 9th edn. Beijing: People’s Health Publishing House; 2018. p. 54–56.
Gao W, Lu Y. Fetal heart baseline extraction and classification based on deep learning. In: 2019 International conference on information technology and computer application (ITCA); 2019.
Cömert Z, Şengür A, Budak Ü, Kocamaz AF. Prediction of intrapartum fetal hypoxia considering feature selection algorithms and machine learning models. Health Inf Sci Syst. 2019;7(1):17.
Acknowledgements
This work is supported by Natural Science Foundation of China Nos. 61976052 and 71804031, and Medical Scientific Research Foundation of Guangdong Province No. A2019428.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, Z., Wang, G., Xu, L. et al. Intelligent antepartum fetal monitoring via deep learning and fusion of cardiotocographic signals and clinical data. Health Inf Sci Syst 11, 16 (2023). https://doi.org/10.1007/s13755-023-00219-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-023-00219-w