Abstract
Undifferentiated pleomorphic sarcoma (UPS), previously termed malignant fibrous histiocytoma, is one of the most aggressive sarcomas with no identifiable line of differentiation. Although the molecular mechanism of oncogenesis in UPS has not been clarified, radiation exposure is considered to be a risk factor in the development of UPS. In the treatment of UPS, surgical treatment remains the most important modality. While chemotherapy is considered in unresectable or metastatic cases, UPS is known to be refractory to conventional chemotherapy, leading to an unfavorable prognosis. To improve the clinical outcome of this condition, novel treatment methods are urgently needed. Patient-derived cell lines are essential tools in preclinical studies. However, owing to the rarity of UPS, only four UPS cell lines are publicly available. Thus, we established a novel UPS cell line, NCC-UPS3-C1, using a surgically resected tumor from a patient with radiation-associated UPS. NCC-UPS3-C1 cells had multiple genomic deletions including the tumor suppressor genes CDKN2A and CDKN2B. NCC-UPS3-C1 cells demonstrated constant growth, spheroid formation, and aggressive invasion ability. We also conducted a screening test using 214 drugs and identified that the histone deacetylase inhibitor, romidepsin, is highly effective on NCC-UPS3-C1 cells. Thus, we concluded that the NCC-UPS3-C1 cell line is a useful tool in preclinical studies for UPS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Undifferentiated pleomorphic sarcoma (UPS), previously termed malignant fibrous histiocytoma (MFH), is a mesenchymal neoplasm with no identifiable line of differentiation when analyzed by presently available technology [1, 2]. UPS accounts for approximately 10% of all soft tissue sarcomas and represents one of the most common sarcomas diagnosed in elderly patients [2, 3]. Notably, irradiation history is considered to be a risk factor in the development of UPS, and 25–30% of radiation-associated soft tissue sarcomas are UPS [4,5,6,7]. Although UPS most often arises from the extremities, radiation-associated UPS frequently develops in the trunk, reflecting the sites of radiation therapy [3, 5, 8]. Genetic analysis revealed complex karyotypes and genomic profiles of UPS. Although the specific genetic aberration of UPS has not been determined, frequent deletions and rearrangements involving CDKN2A and CDKN2B in 9p, PTEN in 10q, RB1 in 13q, and TP53 in 17p were reported [1, 9,10,11,12,13]. However, the molecular mechanism of the oncogenesis in UPS is still unknown [2]. In the treatment of UPS, surgical treatment remains the most important modality. However, complete resection is sometimes difficult to achieve due to the location of the tumor, which may be in the vicinity of a nerve or in the retroperitoneal area. When there is failure of complete tumor resection and the margin of the resected tumor is positive, the local recurrence rate may increase until approximately 70% [14]. Besides the high local recurrence rate, the metastatic rate is also high at 30–35%, resulting in an unfavorable 5-year overall survival of 60–70% [3, 8, 14, 15]. Although chemotherapy is considered in unresectable or metastatic cases, UPS is known to be refractory to conventional chemotherapy [16, 17]. Therefore, new therapeutic methods for UPS patients are necessary.
Patient-derived cell lines are essential tools in preclinical studies. Although long-term cultured cell lines have altered their genetic traits, short-term cultured cell lines maintained the characteristics of the original tumor [18, 19]. Taking advantage of this feature, genomic profiling and drug screening test using a large number of cell lines have been conducted recently [20,21,22,23]. In common cancers, hundreds of cell lines have been screened, and novel candidate drugs have been discovered. However, these screening studies had not been adequately conducted in sarcomas due to the rarity and paucity of sarcoma cell lines [24, 25]. Using Cellosaurus [26], we examined cell lines annotated as UPS or MFH and found only four cell lines publicly available (Supplementary Table 1). There are newly reported UPS cell lines that have not yet been included in Cellosaurus, but these cell lines are not deposited in public cell banks either [27, 28]. This makes it difficult to conduct screening studies in UPS and hinders the development of novel therapeutic drugs.
In this study, we established a novel UPS cell line, NCC-UPS3-C1, from surgically resected UPS tumor tissue. We characterized the NCC-UPS3-C1 cells and conducted a drug screening test to identify promising candidates for the treatment of UPS.
Materials and methods
Patient history
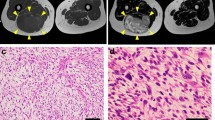
The patient donor was a 55-year-old female with undifferentiated pleomorphic sarcoma. The patient previously visited a hospital with the chief complaint of a mass on her left buttock. The patient has a history of being treated for cervical cancer and received radiation therapy. During that time, the buttock was included in the radiation field. Magnetic resonance imaging detected a soft tissue tumor in the left gluteus maximus with a maximum diameter of approximately 7 cm (Fig. 1a, b). As the result of needle biopsy indicated a malignancy, the patient was referred to the National Cancer Center Hospital (Tokyo, Japan). Open biopsy was performed, and the tumor was suspected to be a radiation-associated sarcoma. Since there was no distant metastasis, wide resection was performed. Pathologically, the tumor consisted of intersecting fascicles of pleomorphic spindle cells (Fig. 1c, d). The tumor showed very rare reactivity to cytokeratin AE1/AE3, whereas it was negative for MDM2, S100 protein, desmin, myogenin, CD34, and ERG. H3K27me3 expression was retained (Supplementary Fig. 1). No lines of differentiation were appreciated (Fig. 1c, d). A part of the resected tumor acquired from the operation was used to establish the cell line described in this study. The ethical committee of the National Cancer Center approved the use of clinical materials for this study, and a written informed consent was obtained from the patient donor.
Clinical and pathological data. Magnetic resonance imaging showing a soft tissue tumor in the left gluteus maximus, with A low intensity in T1-weighted image and B low intensity in the center and isointensity in the periphery in T2-weighted image. Yellow arrows indicate the tumor. C, D H&E staining showing the tumor consisting of intersecting fascicles of pleomorphic spindle cells with no lines of differentiation
Histological analysis
Histological examination was performed on 4-μm-thick sections from a paraffin-embedded tumor sample. The sections were deparaffinized and stained with hematoxylin and eosin (H&E).
Immunohistochemistry
Immunohistochemistry was performed using the deparaffinized tumor sample. The sections were exposed to 3% hydrogen peroxide for 15 min to block any endogenous peroxidase activity and were then washed in deionized water for 2–3 min. The preparations were subjected to heat-induced epitope retrieval. Primary antibodies against cytokeratin (AE1/AE3, prediluted; Dako, Glostrup, Denmark), MDM2 (IF2, 1:100; Thermo Fisher Scientific, Waltham, MA, USA), S100 protein (Polyclonal, prediluted; Dako), desmin (D33, prediluted; Dako), myogenin (F5D, prediluted; Dako), CD34 (QBEnd-10, 1:100; Dako), ERG (EPR3864, 1:2500; Abcam, Cambridge, UK), and H3K27me3 (C36B11, 1:200; Cell Signaling Technology, Danvers, MA, USA) were used. The slides were incubated for 1 h at room temperature, and subsequently labeled with peroxidase (EnVision system, Dako).
Cell culture
The surgically resected tumor tissue was used to establish the cell line as previously described [29]. Briefly, the cells were maintained in DMEM/F12 (Gibco, Grand Island, NY, USA) supplemented with 5% heat-inactivated fetal bovine serum (FBS; Gibco), 100 μg/mL penicillin and 100 μg/mL streptomycin (Nacalai Tesque, Kyoto, Japan), 0.4 µg/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO, USA), 5 ng/mL EGF (Sigma-Aldrich), 10 ng/mL bFGF (Sigma-Aldrich), 5 µg/mL insulin (Sigma-Aldrich), and 10 µM Y-27632 (Selleck Chemicals, Houston, TX, USA: Rock inhibitor) at 37 °C in a humidified atmosphere with 5% CO2. The cells were maintained for more than 6 months under tissue culture conditions and were passaged more than 20 times.
Authentication and quality control of the established cell line
The established cell line was authenticated by examining short tandem repeats (STRs) at 10 loci using the GenePrint 10 system (Promega, Madison, WI, USA) according to the manufacturer’s instructions and the procedure we previously described [29].
Mycoplasma contamination was examined using the e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Gyeonggi-do, Korea) according to the manufacturer’s instructions. Briefly, the extracted DNA of the cell line was heated at 95 °C for 10 min, and then the DNA sequence unique to mycoplasma was amplified. The amplified DNA was separated by gel electrophoresis. Finally, the gel was stained with Midori Green Advance stain (Nippon Genetics, Tokyo, Japan) and analyzed using an Amersham Imager 600 system (GE Healthcare Biosciences, Little Chalfont, UK).
Genetic analysis
Single nucleotide polymorphism (SNP) array genotyping was performed with the Infinium OmniExpressExome-8 version 1.4 BeadChip (Illumina, San Diego, CA, USA) as previously described [29]. The SNP array data were analyzed using the R version 4.0.3 (R Foundation for Statistical Computing, http://www.R-project.org) and DNAcopy package version 1.64.0 (Bioconductor, https://bioconductor.org/). Chromosome regions with copy numbers > 3 and < 1 were defined as amplifications and deletions, respectively. Genes that showed copy number alterations (CNAs) were annotated using the biomaRt package version 2.46.0 (Bioconductor) and “Cancer Gene Census” in the Catalogue Of Somatic Mutations In Cancer database (GRCh 37 version 91).
Cell proliferation assay
Cell proliferation assays were performed as previously described [29]. Briefly, the cells were seeded at a density of 2.5 × 104 cells/well in 24-well culture plates at day 0. The number of cells was counted every 24 h for 4 days. The doubling time was calculated based on the growth curve. All the experiments were conducted in triplicates.
Spheroid formation assay
Spheroid formation assay was performed as previously described [29]. Briefly, the obtained spheroids were prepared for paraffin sections using iPGell (Genostaff, Tokyo, Japan) according to the manufacturer’s instructions. Cell blocks were fixed with 10% formalin neutral buffer solution and embedded in paraffin. Four-micrometer-thick paraffin sections were prepared and stained with H&E.
Invasion assay using real-time cell analyzer
The invasion potential was assessed using a real-time cell analyzer (xCELLigence, Agilent, Santa Clara, CA, USA) as previously described [30]. MG63 osteosarcoma cells [31] (JCRB; Ibaraki Osaka, Japan) were used as controls. This assay was monitored every 15 min for 72 h and plotted as a function of time after seeding.
Assessment of tumorigenicity in nude mice
Tumorigenicity of the cells were assessed as previously described [29]. Briefly, we used female Balb/c nude mice purchased from CLEA Japan, Inc. (Tokyo, Japan). A 100 μL volume of cells in a 1:1 mixture of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was subcutaneously injected into the mice (1 × 106 cells). Subsequently, the tumor size was measured weekly. The tumor volumes were calculated according to the following formula: volume = (length × width2)/2. After 2 months, the tumors were surgically resected, and the specimens were stained with H&E. The animal experiment in this study was performed according to the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute.
Drug screening for the antiproliferative effects
Screening for the antiproliferative effects of 214 drugs including FDA-approved anticancer agents was conducted as previously described [29]. The drugs used in this screening test are listed in Supplementary Table 2.
The IC50 value, the concentration required to inhibit cell growth by 50% in comparison with the growth of the control cells, was evaluated in 24 anticancer agents which exhibited prominent antiproliferative effects in the screening test.
Results
Authentication of the established cell line
We authenticated the NCC-UPS3-C1 cell line by analyzing the STR pattern of 10 microsatellites (Table 1, Supplementary Fig. 2). Calculated based on the Tanabe algorithm [32], the STR match ratio between the NCC-UPS3-C1 cells and the corresponding original tumor was 83.3%. The score met the standard match threshold of 80% [33]. In addition, the STR patterns did not match those of any other cell line in the public cell banks examined using the cell line database, Cellosaurus [26]. These results confirm that the NCC-UPS3-C1 cells were not contaminated with other cells and were established from the original tumor.
We did not detect the DNA sequence unique to mycoplasma in the DNA of NCC-UPS3-C1 cells (data not shown).
Characterization of the cell line
SNP array analysis revealed multiple CNAs in NCC-UPS3-C1 cells. Partial allelic deletions were identified in chromosomes 1pq, 2p, 3q, 4p, 5q, 6p, 7q, 8p, 9pq, 10p, 11p, 12q, 15q, and 18pq. The deletion in chromosome 9p included the tumor suppressor genes CDKN2A and CDKN2B (Fig. 2, Supplementary Table 3).
NCC-UPS3-C1 cells are comprised of pleomorphic cells under 2D culture conditions (Fig. 3a, b). These cells exhibited constant growth, and the population doubling time was 61.6 h based on the growth curve (Fig. 3c). They also had the capability of spheroid formation. The H&E-stained spheroid section showed dense proliferation of pleomorphic cells with nuclear atypia (Fig. 3d). Real-time cell analyzer assay revealed a more aggressive invasion ability of NCC-UPS3-C1 cells compared to MG63 cells (Fig. 3e).
Characterization of NCC-UPS3-C1 cells. A, B NCC-UPS3-C1 cells showing pleomorphic cell morphology under 2D culture conditions. C Growth curve of NCC-UPS3-C1 cells. The Y-axis indicates the relative cell proliferation of NCC-UPS3-C1 cells, and the X-axis represents the day after seeding. D The H&E-stained spheroid section of NCC-UPS3-C1 cells showing dense proliferation of pleomorphic cells. E Real-time cell analyzer invasion assay showing the invasion ability of NCC-UPS3-C1 cells compared to that of MG63 osteosarcoma cells
Tumorigenesis in nude mice
NCC-UPS3-C1 cells transplanted into BALB/c nude mice formed small tumor masses under the described conditions (Supplementary Fig. 3a). The H&E-stained tumor section exhibited dense proliferation of pleomorphic cells (Supplementary Fig. 3b). Although viable tumor cells were observed in H&E sections, the tumor growth was not sufficient (Supplementary Fig. 3c).
Sensitivity to anticancer agents
The antiproliferative effect of 214 drugs against NCC-UPS3-C1 cells was examined (Supplementary Table 2). The cell viability after treatment with each agent at a fixed concentration of 10 µM was described in Supplementary Table 4.
The IC50 values of 24 drugs that showed prominent antiproliferative effects on NCC-UPS3-C1 in the screening test were determined. The IC50 values of 24 agents are presented in Supplementary Table 5, and the six agents with the lowest IC50 values are shown in Table 2. The growth curves that served as the basis for the calculation of the IC50 values are shown in Fig. 4 and Supplementary Fig. 4.
Discussion
UPS is a rare malignant tumor with a poor prognosis. Although systemic chemotherapy is often adapted in the advanced cases of UPS, the efficacy of conventional chemotherapy has not been clarified adequately [16, 17]. Considering that UPS accounts for a large number of patients with soft tissue sarcomas, the development of novel treatment method is urgently required. Patient-derived cell lines are essential tools in preclinical studies. Screening studies using a large number of cell lines were conducted in common cancers to reveal genomic profiling and sensitivity to the anticancer agents [20,21,22,23]. However, due to the paucity and rarity of UPS cell lines, the research on UPS is still behind in these fields [24]. Hence, we established a novel UPS cell line, NCC-UPS3-C1.
The NCC-UPS3-C1 cell line was established from the radiation-associated UPS that developed in the buttock. While sporadic UPS usually occurs in the extremities, radiation-associated UPS predominantly occurs in the trunk, including the buttock, reflecting the irradiation area of pre-radiation therapy [3, 5, 8]. Therefore, NCC-UPS3-C1 cell line was established from a typical radiation-associated UPS case. In addition, UPS is the most common histological subtype of radiation-associated sarcoma, accounting for 25–30% of the cases [4,5,6,7]. Thus, NCC-UPS3-C1 cell line will be useful in further studies of UPS and in understanding the characteristics of radiation-associated sarcoma.
NCC-UPS3-C1 cells showed multiple chromosomal deletions reflecting the complex genotype of UPS. Among the observed CNAs, the deletion of CDKN2A is frequently confirmed in UPS; however, it is not specific to UPS [9,10,11]. Notably, the deletion of CDKN2A is associated with poor prognosis of UPS, indicating a malignant potential of NCC-UPS3-C1 cell line [34]. In a recent study, it was reported that AMPD2 was associated with poor prognosis [35]. However, NCC-UPS3-C1 cells did not exhibit the amplification of AMPD2, reflecting the diversity of UPS.
NCC-UPS3-C1 cells displayed the typical morphology of pleomorphic sarcoma and demonstrated constant growth and capability of spheroid formation. Furthermore, NCC-UPS3-C1 cells exhibited more invasiveness than MG63 osteosarcoma cells. These in vitro characteristics reflected the malignant potential of the original tumor. Meanwhile, NCC-UPS3-C1 cells did not sufficiently grow in nude mice, indicating that these may not be suitable for xenograft experiment.
NCC-UPS3-C1 cells exhibited high sensitivity to the actinomycin D, romidepsin, bortezomib, idarubicin, mitoxantrone, and homoharringtonine in a drug screening test using 214 drugs. These drugs demonstrated more antiproliferative effect than trabectedin and eribulin, which have been reported to be effective in UPS cell lines [28, 36]. It is worthy to note that NCC-UPS1-C1 cells also showed remarkable sensitivity to romidepsin [37]. Romidepsin is a histone deacetylase (HDAC) inhibitor, which is recommended to be used as a second-line therapy in patients with peripheral T-cell lymphoma [38]. Only a few studies have reported on the efficacy of HDAC inhibitor on UPS [39,40,41]. Rivera-Reyes et al. reported that HDAC inhibitor, vorinostat, affected the epigenetic modulators that demonstrated an antiproliferative effect on UPS [41]. Romidepsin may have shown anti-proliferative effects by a similar mechanism, warranting further study.
To summarize, we successfully established a novel UPS cell line, NCC-UPS3-C1, which had highly malignant characteristics in vitro reflecting radiation-associated UPS. Genetically, NCC-UPS3-C1 cells have multiple CNAs including the deletion of CDKN2A, indicating the complexity of UPS. Drug screening test revealed novel candidate drugs for the treatment of UPS. However, this study has limitations. Firstly, NCC-UPS3-C1 cell line cannot represent all UPSs due to the high heterogeneity of UPS. Secondly, although we identified candidate drugs in this study, dozens of UPS cell lines are needed to identify which drugs are effective at a statistically significant level. Thus, we conclude that continuous efforts to establish novel UPS cell lines are required and that these should be shared widely in the research community.
References
WHO classification of tumours of soft tissue and bone. 5th edn. Lyon: IARC; 2020.
Widemann BC, Italiano A. Biology and management of undifferentiated pleomorphic sarcoma, myxofibrosarcoma, and malignant peripheral nerve sheath tumors: state of the art and perspectives. J Clin Oncol. 2018;36:160–7.
Chen S, Huang W, Luo P, et al. Undifferentiated pleomorphic sarcoma: long-term follow-up from a large institution. Cancer Manag Res. 2019;11:10001–9.
Gladdy RA, Qin LX, Moraco N, et al. Do radiation-associated soft tissue sarcomas have the same prognosis as sporadic soft tissue sarcomas? J Clin Oncol. 2010;28:2064–9.
Dineen SP, Roland CL, Feig R, et al. Radiation-associated undifferentiated pleomorphic sarcoma is associated with worse clinical outcomes than sporadic lesions. Ann Surg Oncol. 2015;22:3913–20.
Joo MW, Kang YK, Ogura K, et al. Post-radiation sarcoma: a study by the Eastern Asian Musculoskeletal Oncology Group. PLoS ONE. 2018;13:e0204927.
Callesen LB, Safwat A, Rose HK, Sorensen FB, Baad-Hansen T, Aggerholm-Pedersen N. Radiation-induced sarcoma: a retrospective population-based study over 34 years in a single institution. Clin Oncol (R Coll Radiol). 2021;33:e232–8.
Vodanovich DA, Spelman T, May D, Slavin J, Choong PFM. Predicting the prognosis of undifferentiated pleomorphic soft tissue sarcoma: a 20-year experience of 266 cases. ANZ J Surg. 2019;89:1045–50.
Kresse SH, Ohnstad HO, Bjerkehagen B, Myklebost O, Meza-Zepeda LA. DNA copy number changes in human malignant fibrous histiocytomas by array comparative genomic hybridisation. PLoS ONE. 2010;5:e15378.
Carneiro A, Francis P, Bendahl PO, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest. 2009;89:668–75.
Simons A, Schepens M, Jeuken J, et al. Frequent loss of 9p21 (p16(INK4A)) and other genomic imbalances in human malignant fibrous histiocytoma. Cancer Genet Cytogenet. 2000;118:89–98.
Perot G, Chibon F, Montero A, et al. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177:2080–90.
Gibault L, Perot G, Chibon F, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223:64–71.
Kamat NV, Million L, Yao DH, et al. The outcome of patients with localized undifferentiated pleomorphic sarcoma of the lower extremity treated at Stanford University. Am J Clin Oncol. 2019;42:166–71.
Goldblum JR. An approach to pleomorphic sarcomas: can we subclassify, and does it matter? Mod Pathol. 2014;27(Suppl 1):S39-46.
Grimer R, Judson I, Peake D, Seddon B. Guidelines for the management of soft tissue sarcomas. Sarcoma. 2010;2010:506182.
Reichardt P. Soft tissue sarcomas, a look into the future: different treatments for different subtypes. Future Oncol. 2014;10:s19-27.
Saito S, Morita K, Kohara A, et al. Use of BAC array CGH for evaluation of chromosomal stability of clinically used human mesenchymal stem cells and of cancer cell lines. Hum Cell. 2011;24:2–8.
Ben-David U, Beroukhim R, Golub TR. Genomic evolution of cancer models: perils and opportunities. Nat Rev Cancer. 2019;19:97–109.
Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23.
Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Hattori E, Oyama R, Kondo T. Systematic review of the current status of human sarcoma cell lines. Cells. 2019;8:157.
Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38.
Amm HM, DeVilliers P, Srivastava AR, Diniz MG, Siegal GP, MacDougall M. Mandibular undifferentiated pleomorphic sarcoma: molecular analysis of a primary cell population. Clin Exp Dent Res. 2020;6:495–505.
De Vita A, Recine F, Mercatali L, et al. Primary culture of undifferentiated pleomorphic sarcoma: molecular characterization and response to anticancer agents. Int J Mol Sci. 2017;18:2662.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-SS4-C1: a novel patient-derived cell line of synovial sarcoma. Hum Cell. 2021;34:998–1007.
Sin Y, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of a novel alveolar rhabdomyosarcoma cell line, NCC-aRMS1-C1. Hum Cell. 2020;33:1311–20.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–38.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
Bui NQ, Przybyl J, Trabucco SE, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019;9:12.
Orth MF, Gerke JS, Knosel T, et al. Functional genomics identifies AMPD2 as a new prognostic marker for undifferentiated pleomorphic sarcoma. Int J Cancer. 2019;144:859–67.
De Vita A, Recine F, Miserocchi G, et al. The potential role of the extracellular matrix in the activity of trabectedin in UPS and L-sarcoma: evidences from a patient-derived primary culture case series in tridimensional and zebrafish models. J Exp Clin Cancer Res. 2021;40:165.
Takai Y, Oyama R, Kito F, et al. Establishment and characterizationof cell line of undifferentiated pleomorphic sarcoma. Tiss Cult Res Commun. 2017;36:41–8.
Iyer SP, Foss FF. Romidepsin for the treatment of peripheral T-cell lymphoma. Oncologist. 2015;20:1084–91.
Becker M, Graf C, Tonak M, et al. Xenograft models for undifferentiated pleomorphic sarcoma not otherwise specified are essential for preclinical testing of therapeutic agents. Oncol Lett. 2016;12:1257–64.
Saitoh Y, Bureta C, Sasaki H, et al. The histone deacetylase inhibitor LBH589 inhibits undifferentiated pleomorphic sarcoma growth via downregulation of FOS-like antigen 1. Mol Carcinog. 2019;58:234–46.
Rivera-Reyes A, Ye S, Gloria EM, et al. YAP1 enhances NF-kappaB-dependent and independent effects on clock-mediated unfolded protein responses and autophagy in sarcoma. Cell Death Dis. 2018;9:1108.
Acknowledgements
We thank Drs. E. Kobayashi, S. Iwata, S. Fukushima, M. Nakagawa, S. Ozaki, C. Sato (Department of Musculoskeletal Oncology), Dr. T. Ushikusa (Department of Diagnostic Pathology), and the National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Ms. Y. Kuwata (Division of Rare Cancer Research). We appreciate the technical support provided by Ms. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for their help with English language editing and constructive comments on the manuscript. This research was received technical assistance from the Fundamental Innovative Oncology Core at the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (Grant Number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004-050).
Informed consent
Written informed consent for publication was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13577_2021_633_MOESM8_ESM.tif
Supplementary file8 Supplementary Fig. 3 Tumorigenesis in nude mice. (A) NCC-UPS3-C1 cells transplanted into BALB/c nude mice formed a small tumor mass under the described condition. The yellow circles indicate the tumor. (B) The H&E-stained tumor represents the dense proliferation of pleomorphic cells. (C) The graph showing estimated tumor volume. Bars represent the mean standard error (TIF 1337 KB)
13577_2021_633_MOESM9_ESM.tif
Supplementary file9 Supplementary Fig. 4 Growth curves for the IC50 value calculations of 24 anticancer agents (TIF 618 KB)
Rights and permissions
About this article
Cite this article
Tsuchiya, R., Yoshimatsu, Y., Noguchi, R. et al. Establishment and characterization of NCC-UPS3-C1: a novel patient-derived cell line of undifferentiated pleomorphic sarcoma. Human Cell 35, 384–391 (2022). https://doi.org/10.1007/s13577-021-00633-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00633-w