Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most frequent malignancies worldwide. miR-193a-3p acts as an oncogene or tumor suppressor in different cancers. However, the functional role and regulatory mechanism of miR-193a-3p in ESCC remain to be elucidated. Our results demonstrated that miR-193a-3p expression was significantly upregulated and associated with advanced TNM stage, recurrence, and poor prognosis in ESCC patients. miR-193-3p targeted ALKBH5 and suppressed its expression. ALKBH5 inhibited miR-193a-3p expression in turn. ALKBH5 affected the primary miR-193a-3p processing by negatively regulating its m6A modification. These findings suggested a positive feedback regulation between miR-193a-3p and ALKBH5 in ESCC cells. Moreover, the functional assays indicated that the miR-193-3p-ALKBH5 feedback loop promoted the proliferation, migration and invasion ability of ESCC cells in vitro, and facilitated tumor growth and metastasis in vivo. Collectively, our current study identified a novel positive feedback regulation between miR-193a-3p and ALKBH5 in ESCC, which may be helpful to gain insight into ESCC pathogenesis and provide novel therapeutic target for ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer (EC) is the eighth most frequent malignancy and sixth leading cause of cancer-related death around the world [1]. The main histological forms of EC include esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. ESCC is observed to be the most common type of EC. Even though the great improvement of ESCC therapy has been achieved in the past decades, such as esophagectomy, radiotherapy and chemotherapy, the prognosis of patients with ESCC remains poor. The 5-year overall survival rate is still less than 15% [2]. Hence, it is of great importance to reveal the underlying mechanism of ESCC pathogenesis and explore an effective therapeutic target for ESCC.
MicroRNAs (miRNAs) are short RNA molecules with 19–25 nucleotides in length. miRNAs bind to the 3′-untranslated region (3′UTR) of target mRNAs, inducing mRNA degradation or suppressing mRNA translation [3]. Increasing studies demonstrated that each tumor type has a distinct miRNA signature that distinguishes it from normal tissues and other cancer types [4]. In ESCC, some miRNAs have been indicated to play important roles as oncogenes or tumor suppressors to regulate ESCC proliferation, apoptosis, invasion, chemotherapy, or radiotherapy sensitivity, such as miRNA-106b, miR-130a, miR-148a, microRNA-133b and miR-338 [5,6,7]. miR-193a-3p acts as an oncogene or tumor suppressor in different cancers. For instance, miR-193a-3p targets CCND1 to suppress the pathogenesis of hepatocellular carcinoma [8]. Nevertheless, miR-193a-3p promotes radio-resistance of nasopharyngeal cancer cells by targeting SRSF2 and hypoxia signaling pathway [9]. Recently, the upregulation of miR-193a-3p has been reported in ESCC [10, 11]. However, the functional role and target mRNAs of miR-193a-3p in ESCC cells remains to be elucidated.
N6-methyladenosine (m6A) is the most abundant internal chemical modification of RNA in eukaryotes [12]. Methyltransferase-like 3 (METTL3), METTL14 and Wilms’ tumor 1-associated protein (WTAP) form the core methyltransferase complex that catalyzes the m6A methylation, while fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) act as demethylases to induce the m6A demethylation. Emerging studies revealed that m6A modification may affect thousands of mRNAs or non-coding RNAs. Recently, m6A modification is found to be involved in primary miRNA processing in cancer cells. METTL14 increases the expression of miR-126 by facilitating the microprocessor protein DGCR8 binding to primary miR-126 (pri-miR-126) in an m6A-dependent manner in liver cancer cells [13]. In bladder cancer, METTL3 interacts with the DGCR8 and increases the m6A methylation of pri-miR-221/222, thus enhancing pri-miR-221/222 maturation [14]. However, whether m6A modification modulate the expression of miR-193a-3p is still unknown. In the present study, we investigated the expression level of miR-193a-3p in ESCC tissues and analyze its clinical significance. Then, the underlying regulatory mechanism of miR-193a-3p and its functional role was further explored in ESCC.
Materials and methods
Tissue samples collection
The ESCC and matched normal esophageal tissues were collected from 60 patients with ESCC in Luoyang Central Hospital Affiliated to Zhengzhou University. All patients had not received radiotherapy, chemotherapy, or biotherapy before curative resection. The tissue samples were stored at − 80 ℃ until used. The Ethics Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University approved the study protocols and the use of these tissue samples (approval number: 201908-06). Informed consent was obtained from these patients.
Cell culture
The human ESCC cell lines (KYSE-150 and ECA109) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 ℃ in 5% CO2. Cells were tested routinely for mycoplasma.
Transfection
For transfection, miR-193a-3p mimics or inhibitors (Genepharma), ALKBH5, FTO, METTL3, METTL14 and WTAP siRNAs (Genepharma), pCMV and pCMV-ALKBH5 were transfected into ESCC cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. After 48 h, the cells were used for further experiments. The siRNA sequences were already well-established and had been used in published study without off-target effects [15,16,17]. The target sequences of indicated siRNAs were shown as follows: siALKBH5: ACAAGTACTTCTTCGGCGA; siFTO: ATAGCCGCTGCTTGTGAGA; siMETTL3: CCTTATTCGAGAGGTGTCA; siMETTL14: GCTGGACTTGGGATGATATTA; siWTAP: GGAGGTAGTGGTTACGTAAAT.
RNA immunoprecipitation (RIP)
RIP assay was carried out using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the the manufacturer’s instrument. Normal rabbit IgG (Millipore) and an anti-ALKBH5 antibody (Millipore) or an anti- DGCR8 antibody (Abcam) was used for RIP assay.
Methylated RNA immunoprecipitation (MeRIP)
MeRIP assay was performed using Magna MeRIP™ m6A Kit (Millipore, USA) according to the manufacturer’s protocol. In brief, RNA is chemically fragmented into approximately 100 nucleotides fragments followed by magnetic immunoprecipitation with an anti-m6A antibody (Abcam). After immunoprecipitation, isolated and purified RNA fragments were subjected with qRT-PCR detection.
RNA isolation and quantitative Real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen). For mRNA detection, cDNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, China). For miRNA detection, Mir-X miRNA First-Strand Synthesis Kit was used according to the manufacturer’s instruction. qRT-PCR for mRNA and miRNA detection was performed on LightCycler 96 (Roche, USA). GAPDH and U6 were used as an internal reference for mRNA and miRNA detection, respectively. All primers used in this study are as follows: ALKBH5-F: GGAGATGGACAAGGAAGAGAAC; ALKBH5-R: CTCTGAGGACTCGTATGACTTG; GAPDH-F: GTCAACGGATTTGGTCGTATTG; GAPDH-R: TGTAGTTGAGGTCAATGAAGGG. For miR-193a-3p detection, the entire sequence of miR-193a-3p was used as miRNA specific 5′ primer, while 3′ primer for qRT-PCR is the mRQ 3′ Primer supplied with the kit. The U6 primer was also contained in the Mir-X miRNA First-Strand Synthesis Kit.
CCK-8 assay
2 × 103 cells per well were seeded in 96-well plates. Every 24 h until 96 h, 10 μl CCK-8 solution (Dojindo) was added into per well and then incubated for additional 1.5 h. Cell proliferation was measured with a microplate reader (Bio-rad, USA) at an absorbance of 450 nm.
Transwell assay
For migration detection, cells were suspended in serum-free culture medium and then seeded into the transwell upper chamber. For invasion detection, cells were suspended in serum-free culture medium and then seeded into the transwell upper chamber pre-coated with matrigel. The lower chamber was filled with medium containing 10% FBS. After incubation for 24 h, the cells on the upper membrane surface were removed. Cells which passed through the membrane were fixed with 4% paraformaldehyde and stained with 1% crystal violet and then counted under a microscope (Olympus, Japan).
Luciferase reporter assay
The wild-type (WT) or mutant (MUT) 3′UTR of ALKBH5 was inserted into luciferase reporter pmirGLO (pmirGLO-ALKBH5-WT or pmirGLO-ALKBH5-MUT) plasmid. Control or miR-193a-3p mimics were co-transfected with ALKBH5-WT or ALKBH5-MUT into cells by Lipofectamine 3000 reagent as the manufacturer’s protocol. 48 h later, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (Promega, USA) and calculated by normalizing the firefly luminescence to the renilla luminescence.
Animal studies
KYSE-150 cells were infected with lentiviral particles expressing control or ALKBH5 shRNA and/or miR-193a-3p inhibitor (FulenGen, Guangzhou, China). Stable cells were selected with 2 μg/ml puromycin for two weeks. Male athymic BALB/c nude mice (4–5 weeks old) were used. Subcutaneous tumor growth assays were used for growth assay. A total of 5 × 106 cells was subcutaneously injected into the nude mice (n = 3). The tumor volume was measured. The mice were sacrificed 4 weeks after injection. The tumor tissues were isolated and weighed. A tail vein injection model was used for metastasis assays. A total of 5 × 105 cells were injected into tail vein of nude mice (n = 3). After 8 weeks, mice were sacrificed, and the lung tissues were isolated and then subjected to H&E staining. The pulmonary metastasis nodules were counted under a microscope. The animal studies were approved by the Institutional Animal Care and Use Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University.
Statistical analysis
Data were expressed as mean ± SD and compared using the Student t test. The correlation between miR-193a-3p and clinico-pathological features of patients with ESCC was analyzed by the χ2 test. The correlation between miR-193a-3p and ALKBH5 expression was analyzed by Pearson statistic. Kaplan–Meier method and log-rank test were used to evaluate the relationship between miR-193a-3p expression and overall survival time of patients with ESCC. All of the experiments were repeated three times.
Results
Elevation of miR-193a-3p expression correlates with poor prognosis of patients with ESCC
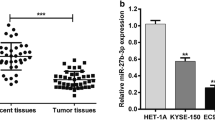
We first detected the miR-193a-3p expression in 60 pairs of ESCC and matched normal esophageal tissues. The results of qRT-PCR assays indicated that miR-193a-3p was significantly increased in ESCC tissue samples compared with that in the normal esophageal tissues (Fig. 1a). The ESCC patients were divided into two groups according to the median expression of miR-193a-3p in ESCC tissues. Statistical associations between miR-193a-3p and clinico-pathological characteristics were observed, including TNM stage and recurrence (Table 1). We further determined whether miR-193a-3p expression correlated with prognosis of ESCC patients. Kaplan–Meier survival analysis and log-rank test demonstrated that high miR-193a-3p expression in ESCC tissues was markedly associated with a lower rate of overall survival (Fig. 1b). These data demonstrated that the overexpression of miR-193a-3p may be involved in the progression of ESCC and serve as a prognostic indicator of ESCC patients.
Overexpression of miR-193a-3p in ESCC tissues predicts poor prognosis of patients with ESCC. a qRT-PCR analysis of the miR-193a-3p expression in 60 pairs of ESCC and matched normal esophageal tissues. The expression level was calculated by the formula 2−△CT. b Kaplan–Meier survival analysis and log-rank test was used to evaluate the correlation between miR-193a-3p expression and overall survival of patients with ESCC. The median of miR-193a-3p expression in ESCC tissues was used as cutoff
miR-193a-3p targets ALKBH5
Using Targetscan and StarBase online analysis tool, RNA demethylase ALKBH5 is a potential target of miR-193a-3p (Fig. 2a). To validate this, a dual-luciferase reporter assay was carried out. We found that transfection with miR-193a-3p mimics markedly decreased the luciferase activity of pmirGLO-ALKBH5-WT in KYSE-150 and ECA109 cells, while mutation of the ALKBH5-binding site reversed this decrease (Fig. 2b). The effect of miR-193a-3p on the expression of ALKBH5 in ESCC cells was further evaluated. The results of qRT-PCR and western blot indicated that overexpression of miR-193a-3p downregulated ALKBH5 expression (Fig. 2c–e), while transfection of miR-193a-3p inhibitors elevated ALKBH5 expression in KYSE-150 and ECA109 cells (Fig. 2fh). Furthermore, ALKBH5 was found to be highly expressed and negatively correlated with miR-193a-3p in ESCC tissues (Fig. 2i, j). Taken together, miR-193a-3p directly targets ALKBH5 and represses its expression in ESCC cells.
miR-193a-3p targets ALKBH5. a Binding site between miR-193a-3p and ALKBH5 3′ UTR predicted using the TargetScan. b Luciferase activity in KYSE-150 and ECA109 cells co-transfected with non-target negative control miRNAs (miR-NC) or miR-193a-3p and luciferase reporter vectors containing nothing, ALKBH5 3′UTR or mutant ALKBH5 3′UTR (ALKBH5-MUT). c KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p, and the miR-193a-3p expression was detected by qRT-PCR. d KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p, and the ALKBH5 mRNA expression was detected by qRT-PCR. e KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p, and the ALKBH5 protein expression was detected by western blot. f KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p inhibitors, and the miR-193a-3p expression was detected by qRT-PCR. g KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p inhibitors, and the ALKBH5 mRNA expression was detected by qRT-PCR. h KYSE-150 and ECA109 cells were transfected with miR-NC or miR-193a-3p inhibitors, and the ALKBH5 protein expression was detected by western blot. i qRT-PCR analysis of the ALKBH5 mRNA expression in 60 pairs of ESCC and matched normal esophageal tissues. j The correlation between miR-193a-3p and ALKBH5 mRNA expression in 60 ESCC tissues was analyzed. *p < 0.05, **p < 0.01
ALKBH5 downregulates miR-193a-3p expression
Then, we explored the regulatory mechanism of miR-193a-3p upregulation in ESCC. The negative correlation between miR-193a-3p and ALKBH5 prompted us to suspect whether ALKBH5 influenced the miR-193a-3p expression in turn. To validate this, KYSE-150 and ECA109 cells were transfected with siRNAs targeting ALKBH5. The results of qRT-PCR demonstrated that depletion of ALKBH5 induced the upregulation of miR-193a-3p expression (Fig. 3a, b). Conversely, miR-193a-3p expression was decreased after overexpression of ALKBH5, but not affected by catalytic-inactive mutant ALKBH5 (H204A) (Fig. 3c, d). However, neither overexpression nor knockdown of ALKBH5 affected the luciferase activity of the promoter of miR-193a-3p (Fig. 3e, f), indicating that ALKBH5 post-transcriptionally regulate miR-193a-3p expression. We also observed that knockdown of the other demethylase FTO did not show dramatic changes on miR-193a-3p expression (Supplemental Fig. 1a, b).
ALKBH5 downregulates miR-193a-3p expression. a KYSE-150 and ECA109 cells were transfected with negative control or ALKBH5 siRNAs, and the ALKBH5 mRNA expression was detected by qRT-PCR. b KYSE-150 and ECA109 cells were transfected with negative control or ALKBH5 siRNAs, and the miR-193a-3p expression was detected by qRT-PCR. c KYSE-150 and ECA109 cells were transfected with empty vector, ALKBH5 or catalytic-inactive mutant ALKBH5 (H204A), and the ALKBH5 mRNA expression was detected by qRT-PCR. d KYSE-150 and ECA109 cells were transfected with empty vector, ALKBH5 or catalytic-inactive mutant ALKBH5 (H204A), and the miR-193a-3p expression was detected by qRT-PCR. e Luciferase activity in KYSE-150 and ECA109 cells co-transfected with negative control or ALKBH5 siRNAs and luciferase reporter vector containing miR-193a-3p promoter. f Luciferase activity in KYSE-150 and ECA109 cells co-transfected with empty vector or ALKBH5 and luciferase reporter vector containing miR-193a-3p promoter. *p < 0.05, **p < 0.01
ALKBH5 suppresses the maturation of primary miR-193a-3p
m6A modification is important for primary miRNA processing [18]. To confirm whether ALKBH5-mediated m6A demethylation was involved in the maturation of primary miR-193a-3p (pri-miR-193a-3p), we detected the pri-miR-193a-3p levels in KYSE-150 and ECA109 cells transfected with ALKBH5 siRNAs. As shown in Fig. 4a, decrease of pri-miR-193a-3p occurred in ALKBH5-knockdown cells. In contrast, ALKBH5 overexpression resulted in the accumulation of pri-miR-193a-3p in KYSE-150 and ECA109 cells (Fig. 4b). Additionally, ALKBH5 could significantly associate with pri-miR-193a-3p as evidenced by RIP assay (Fig. 4c). The results of MeRIP assay indicated that the m6A level of pri-miR-193a-3p was increased by ALKBH5 knockdown (Fig. 4d), while ectopic expression of ALKBH5 reduced the m6A modification of pri-miR-193a-3p (Fig. 4e). Moreover, we found an increased level of pri-miR-193a-3p binding by DGCR8 immuno-precipitated in ALKBH5-knockdown cells (Fig. 4f), whereas ALKBH5 overexpression attenuated the interaction between pri-miR-193a-3p and DGCR8 (Fig. 4g). Our data indicated that ALKBH5 suppressed the recognition of pri-miR-193a-3p by DGCR8 and the subsequent processing to mature miR-193a-3p in an m6A-dependent manner.
ALKBH5 suppresses the maturation of primary miR-193a-3p. a The pri-miR-193a-3p expression in KYSE-150 and ECA109 cells transfected with negative control or ALKBH5 siRNAs was analyzed by qRT-PCR. b The pri-miR-193a-3p expression in KYSE-150 and ECA109 cells transfected with empty vector or ALKBH5 was analyzed by qRT-PCR. c The RIP assay using ALKBH5 antibody was performed in KYSE-150 and ECA109 cells to detect the interaction between pri-miR-193a-3p and ALKBH5. IgG was used as negative control. d The MeRIP assay was used to detect the m6A level of pri-miR-193a-3p in KYSE-150 and ECA109 cells transfected with negative control or ALKBH5 siRNAs. e The MeRIP assay was used to detect the m6A level of pri-miR-193a-3p in KYSE-150 and ECA109 cells transfected with empty vector or ALKBH5. f The RIP assay using DGCR8 antibody was performed in KYSE-150 and ECA109 cells with or without ALKBH5 knockdown to detect the interaction between pri-miR-193a-3p and DGCR8. g The RIP assay using DGCR8 antibody was performed in KYSE-150 and ECA109 cells with or without ALKBH5 overexpression to detect the interaction between pri-miR-193a-3p and DGCR8. h The KYSE-150 and ECA109 cells were transfected with siRNA targeting METTL3, METTL14 or WTAP, and then the miR-193a-3p was detected using qRT-PCR. i The KYSE-150 and ECA109 cells were transfected with siRNA targeting METTL3, METTL14 or WTAP, and then the pri-miR-193a-3p was detected using qRT-PCR. j The RIP assay using METTL3 antibody was performed in KYSE-150 and ECA109 cells to detect the interaction between pri-miR-193a-3p and METTL3. IgG was used as negative control. k The MeRIP assay was used to detect the m6A level of pri-miR-193a-3p in KYSE-150 and ECA109 cells transfected with control or METTL3 siRNA. l The RIP assay using DGCR8 antibody was performed in KYSE-150 and ECA109 cells with or without METTL3 knockdown to detect the interaction between pri-miR-193a-3p and DGCR8. *p < 0.05, **p < 0.01, ***p < 0.001
We then determined which RNA methyltransferase was responsible for the m6A modification of pri-miR-193a-3p. siRNAs against METTL3, METTL14 or WTAP were transfected into ESCC cells, and the results showed that only knockdown of METTL3 could increase pri-miR-193a-3p expression and decrease miR-193a-3p expression (Fig. 4h, i and Supplemental Fig. 2). RIP assay demonstrated that pri-miR-193a-3p was significantly enriched by METTL3 (Fig. 4j). Moreover, the m6A modification of pri-miR-193a-3p and the interaction between pri-miR-193a-3p and DGCR8 was suppressed METTL3 knockdown (Fig. 4k, l). Taken together, METTL3 and ALKBH5 oppositely regulates m6A modification of pri-miR-193a-3p.
The miR-193a-3p-ALKBH5 positive feedback loop facilitates ESCC proliferation and metastasis in vitro and in vivo
Here, we found that miR-193a-3p targeted ALKBH5, and ALKBH5 suppressed pri-miR-193a-3p processing to downregulate miR-193a-3p expression, suggesting a positive feedback loop between ALKBH5 and miR-193a-3p. Then, the functional role of miR-193a-3p-ALKBH5 positive feedback loop in the malignant phenotypes of ESCC cells was determined. The cell proliferation, migration and invasion was examined by CCK-8 and transwell assay, respectively. First, we observed that knockdown of miR-193a-3p significantly inhibited the proliferative, migratory and invasive ability of KYSE-150 and ECA109 cells, while depletion of ALKBH5 exerted opposite functions (Fig. 5a, b). Second, transfection of ALKBH5 siRNAs restored the reduced proliferation, migration and invasion induced by miR-193a-3p knockdown (Fig. 5a, b).
The miR-193a-3p-ALKBH5 positive feedback loop promotes the malignant phenotypes of ESCC cells in vitro. a The CCK-8 assay was performed to detect the effect of miR-193a-3p-ALKBH5 positive feedback loop on ESCC cells proliferation. b The function of miR-193a-3p-ALKBH5 positive feedback loop in the migration and invasion of ESCC cells was analyzed by transwell assay. *p < 0.05, **p < 0.01
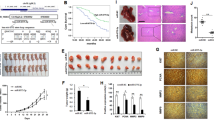
Finally, we confirmed the functional role of miR-193a-3p-ALKBH5 feedback regulation in ESCC growth and metastasis in vivo. Compared with those produced from control xenografts, tumor volumes and weight were decreased by miR-193a-3p knockdown but enhanced by depletion of ALKBH5 in KYSE-150 cells. Downregulation of ALKBH5 restored the growth inhibition by miR-193a-3p knockdown (Fig. 6a, b). Moreover, we found a significant decrease in the number of metastatic nodules in the lungs of mice in the miR-193a-3p knockdown group compared with the vector control group, while depletion of ALKBH5 expression showed opposite effect. ALKBH5 downregulation rescued the suppressive effect of miR-193a-3p knockdown (Fig. 6c). Taken together, our data indicated that the positive feedback loop between miR-193a-3p and ALKBH5 promotes ESCC progression.
The miR-193a-3p-ALKBH5 positive feedback loop facilitates ESCC growth and metastasis in vivo. a, b Subcutaneous tumor model of indicated stable KYSE-150 cells was constructed. The represent images were shown, and the tumor growth curve was measured (a). The weight of tumor tissues was detected (b). c Tail vein injection model of indicated stable KYSE-150 cells was constructed. Representative histological images of H&E staining for lung tissues were shown. The numbers pulmonary metastatic nodules were counted. *p < 0.05, **p < 0.01
Discussion
In the present study, we found that miR-193a-3p expression was significantly upregulated and associated with advanced TNM stage, recurrence, and poor prognosis in ESCC patients. Consistent with our results, previous studies also confirmed the overexpression of miR-193a-3p in ESCC tissues [10, 11], suggesting that miR-193a-3p functioned as an oncogene in ESCC cells. The function and targets of miR-193a-3p are different. In colorectal cancer, miR-193a-3p targets PLAU to suppress cell proliferation [19]. miR-193a-3p promotes radio-resistance of nasopharyngeal cancer cells by targeting SRSF2 [9]. miR-193a-3p positively modulates chemo-radiation resistance in EC cells via the PSEN1 [11]. Here, our data showed that miR-193a-3p promoted ESCC cell proliferation, migration and invasion. Furthermore, we identified ALKBH5 as a target of miR-193a-3p. A negative correlation between miR-193a-3p and ALKBH5 expression in ESCC tissues was also observed, supporting that miR-193a-3p could target ALKBH5. Results of rescue experiments indicated that ALKBH5 was responsible for ESCC cell phenotypes regulated by miR-193a-3p.
ALKBH5 is a member of the AlkB family and has an efficient demethylation activity toward m6A in mRNA. ALKBH5 affects mRNA export and RNA metabolism, and modulates spermatogenesis and apoptosis through p53 signaling pathway [20]. ALKBH5 was demonstrated to act as an oncogene or tumor suppressor in cancer cells. For example, overexpression of ALKBH5 enhances self-renewal and proliferation of glioblastoma stem cell by reducing m6A abundance on FOXM1 mRNA [21]. In non-small cell lung cancer cell, ALKBH5 suppressed cell proliferation and invasion via reducing m6A modification of YAP [22]. However, whether ALKBH5 regulates miRNA processing remains unclear in ESCC. Here, we found that ALKBH5 expression was decreased in ESCC tissues. Functional assays indicated that ALKBH5 acted as a tumor suppressor to attenuate proliferative, migratory and invasive ability of ESCC cells. Interestingly, ALKBH5 could significantly associate with pri-miR-193a-3p, thus downregulating the m6A modification of pri-miR-193a-3p. Additionally, ALKBH5 suppressed the binding of microprocessor protein DGCR8 to pri-miR-193a-3p to negatively modulate its maturation, resulting in a decrease of miR-193a-3p expression. However, the mechanism by which ALKBH5 affects the binding of DGCR8 to pri-miR-193a-3p remains unknown. METTL3 and METTL14 have been proved to associate with DGCR8 to regulate the maturation of primary miRNAs [13, 14]. Here, we validated that only METTL3 was involved in the m6A methylation of pri-miR-193a-3p, indicating that METTL3 and ALKBH5 oppositely regulates m6A modification of pri-miR-193a-3p. A recent study reported that ALKBH5 promotes ESCC proliferation and migration [23]. These results were contradictory with our findings. This research did not compare the differential expression pattern between ESCC and normal esophageal tissues and just detected the ALKBH5 expression in ESCC tissues. They also found that ALKBH5 binds to CDKN1A mRNA and decreases its m6A level in only one cell line, TE5. Here, we found that ALKBH5 targeted pri-miR-193-3p to suppress KYSE-150 and ECA109 proliferation, migration and invasion. We suspected that ALKBH5 targets different RNA to exert disparate function in different types of ESCC cell lines which has different genetic background. In addition, the results of TCGA database using GEPIA online tool demonstrated that the patients with high ALKBH5 expression showed longer overall survival time than those with low ALKBH5 expression (Supplemental Fig. 3), supporting our findings that ALKBH5 exerted as a tumor suppressor in ESCC.
Conclusion
In conclusion, our findings indicated that overexpression of miR-193a-3p in ESCC tissues was associated with advanced TNM stage, recurrence, and poor prognosis in ESCC patients. To our knowledge, this is the first report that miR-193-3p targeted ALKBH5, and ALKBH5 affected the pri-miR-193-3p processing by regulating its m6A modification, which suggested a positive feedback loop between miR-193-3p and ALKBH5 in ESCC cells. The functional assays indicated oncogenic role of miR-193-3p-ALKBH5 feedback loop. Our findings may be helpful to gain insight into ESCC pathogenesis and provide novel therapeutic target for ESCC.
Availability of data and materials
The data and materials used in this study are available.
Abbreviations
- EC:
-
Esophageal cancer
- ESCC:
-
Esophageal squamous cell carcinoma
- miRNAs:
-
MicroRNAs
- 3′UTR:
-
3′-Untranslated region
- m6A:
-
N6-methyladenosine
- METTL3:
-
Methyltransferase-like 3
- ALKBH5:
-
AlkB homolog 5
- FTO:
-
Fat mass and obesity–associated protein
- MeRIP:
-
Methylated RNA immunoprecipitation
- qRT-PCR:
-
Quantitative Real-time PCR
References
Gupta B, Kumar N. Worldwide incidence, mortality and time trends for cancer of the oesophagus. Eur J Cancer Prev. 2017;26(2):107–18. https://doi.org/10.1097/CEJ.0000000000000249.
Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–5. https://doi.org/10.1016/j.asjsur.2016.10.005.
Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. https://doi.org/10.1038/nrg2843.
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9. https://doi.org/10.1016/j.molmed.2014.06.005.
Eichelmann AK, Matuszcak C, Lindner K, Haier J, Hussey DJ, Hummel R. Complex role of miR-130a-3p and miR-148a-3p balance on drug resistance and tumor biology in esophageal squamous cell carcinoma. Sci Rep. 2018;8(1):17553. https://doi.org/10.1038/s41598-018-35799-1.
Huang H, Xu Y, Guo Z, Chen X, Ji S, Xu Z. MicroRNA-133b inhibits cell proliferation and promotes apoptosis by targeting cullin 4B in esophageal squamous cell carcinoma. Exp Ther Med. 2018;15(4):3743–50. https://doi.org/10.3892/etm.2018.5906.
Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. MiR-338-5p enhances the radiosensitivity of esophageal squamous cell carcinoma by inducing apoptosis through targeting survivin. Sci Rep. 2017;7(1):10932. https://doi.org/10.1038/s41598-017-10977-9.
Wang SS, Huang ZG, Wu HY, He RQ, Yang LH, Feng ZB, et al. Downregulation of miR-193a-3p is involved in the pathogenesis of hepatocellular carcinoma by targeting CCND1. PeerJ. 2020;8:e8409. https://doi.org/10.7717/peerj.8409.
Kong L, Wei Q, Hu X, Chen L, Li J. miR-193a-3p promotes radio-resistance of nasopharyngeal cancer cells by targeting SRSF2 gene and hypoxia signaling pathway. Med Sci Monit Basic Res. 2019;25:53–62. https://doi.org/10.12659/MSMBR.914572.
Yi Y, Chen J, Jiao C, Zhong J, Song Z, Yu X, et al. Upregulated miR-193a-3p as an oncogene in esophageal squamous cell carcinoma regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12(6):4779–84. https://doi.org/10.3892/ol.2016.5229.
Meng F, Qian L, Lv L, Ding B, Zhou G, Cheng X, et al. miR-193a-3p regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene. 2016;579(2):139–45. https://doi.org/10.1016/j.gene.2015.12.060.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–200. https://doi.org/10.1016/j.cell.2017.05.045.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–43. https://doi.org/10.1002/hep.28885.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. https://doi.org/10.1186/s12943-019-1036-9.
Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19(1):123. https://doi.org/10.1186/s12943-020-01239-w.
Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19(1):152. https://doi.org/10.1186/s12943-020-01267-6.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46. https://doi.org/10.1186/s12943-020-1146-4.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. https://doi.org/10.1186/s12943-019-1109-9.
Lin M, Zhang Z, Gao M, Yu H, Sheng H, Huang J. MicroRNA-193a-3p suppresses the colorectal cancer cell proliferation and progression through downregulating the PLAU expression. Cancer Manag Res. 2019;11:5353–63. https://doi.org/10.2147/CMAR.S208233.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. https://doi.org/10.1016/j.molcel.2012.10.015.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606. https://doi.org/10.1016/j.ccell.2017.02.013.
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19(1):40. https://doi.org/10.1186/s12943-020-01161-1.
Nagaki Y, Motoyama S, Yamaguchi T, Hoshizaki M, Sato Y, Sato T, et al. m(6) A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells. 2020. https://doi.org/10.1111/gtc.12792.
Acknowledgements
The study was supported by Natural Science Fund of Luoyang Central Hospital Affiliated to Zhengzhou University.
Funding
The study was supported by Natural Science Fund of Luoyang Central Hospital Affiliated to Zhengzhou University.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no competing financial interests.
Ethical approval
The Ethics Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University approved the study protocols and the use of these tissue samples (approval number: 201908–06) in accordance with the Declaration of Helsinki. The animal studies were approved by the Institutional Animal Care and Use Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University.
Informed consent
The informed consent was obtained from all patients in this research. The protocol of this study was approved by the Ethics Committee of Luoyang Central Hospital Affiliated to Zhengzhou University and carried out in accordance with the World Medical Association Declaration of Helsinki.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13577_2020_458_MOESM1_ESM.tif
Supplemental Figure 1 FTO does not affect miR-193a-3p expression. A. KYSE-150 and ECA109 cells were transfected with negative control or FTO siRNAs, and the FTO mRNA expression was detected by qRT-PCR. B. KYSE-150 and ECA109 cells were transfected with negative control or FTO siRNAs, and the miR-193a-3p expression was detected by qRT-PCR. **p<0.01. (TIF 1102 KB)
13577_2020_458_MOESM2_ESM.tif
Supplemental Figure The knockdown efficacy of siRNA against METTL3, METTL14 or WTAP in ESCC cells was determined by qRT-PCR (TIF 1198 KB)
13577_2020_458_MOESM3_ESM.jpg
Supplemental Figure 3 The correlation between ALKBH5 and prognosis of Esophageal carcinoma patients from TCGA database was analyzed GEPIA online tool (JPG 55 KB)
Rights and permissions
About this article
Cite this article
Xue, J., Xiao, P., Yu, X. et al. A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Human Cell 34, 502–514 (2021). https://doi.org/10.1007/s13577-020-00458-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00458-z