Abstract
Mounting evidence indicates that the long non-coding RNA (lncRNA) LINC00460 plays an oncogenic role in tumor progression; however, the role of LINC00460 in cervical cancer (CC) remains unknown. In this study, we found that LINC00460 was frequently upregulated in CC tissues and cell lines. Knockdown of LINC00460 repressed CC cell growth and invasion in vitro and attenuated tumorigenesis in vivo. Mechanistically, miR-361-3p was predicted as a direct target of LINC00460 by bioinformatics analysis, which was further confirmed by qRT-PCR, dual-luciferase reporter assays, and rescue experiments. Furthermore, miR-361-3p targeted the 3′ untranslated region (UTR) of Gli1 mRNA and repressed its expression. Taken together, our study revealed that LINC00460 functions as an oncogenic lncRNA in CC, indicating the likely participation of the LINC00460/miR-361-3p/Gli1 pathway in the disease. Accordingly, our results provide new insight into CC tumorigenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer (CC) is one of the most common malignancies in women globally and a prominent cause of death in women in many developing nations [1]. Although standard vaccination against human papillomavirus, rapid surgical treatment, and periodic cancer screening have resulted in a substantial decline in the prevalence of CC, it remains among the most prolific lethal diseases in women. Therefore, it is critical to elucidate the mechanisms underlying the disease and determine novel biomarkers for the prevention and treatment of CC.
Modern integrative genomic research has shown that the vast majority of human genome transcripts are non-coding RNAs with no protein-coding capacity [2]. Long non-coding RNAs (lncRNAs), which are greater than 200 nt in length, are recently discovered vital members of the non-coding RNA family [3]. lncRNAs can function as competing endogenous RNAs (ceRNAs) and sponge microRNAs (miRNAs), thereby derepressing compound target genes. Mounting evidence indicates that aberrant lncRNA expression is involved in different human diseases, particularly cancers [4]. To date, many human lncRNAs have been shown to be dysregulated in CC, such as LINC00473, LNMICC, XLOC_006390, and SNHG12 [5,6,7,8], which contribute to the development and progression of CC. In particular, LINC00460, a newly identified lncRNA encoded on chromosome 13q33.2, has recently been described to function as an oncogene in several cancers, including lung cancer, colorectal cancer, papillary thyroid carcinoma, and hepatocellular carcinoma [9,10,11,12]. However, little is known concerning the role of LINC00460 in CC.
In this work, we investigated the potential involvement of LINC00460 in CC. We first examined the expression level of LINC00460 in human CC cells and tissues and evaluated its effects on cell growth, cell cycle distribution, and cell invasion in vitro and tumorigenesis in vivo. In addition, we explored the underlying mechanism of LINC00460 function in CC. Our results indicated that LINC00460 mediates miR-361-3p/Gli1 signaling to promote the growth and invasion of CC cells. Thus, this research provides a better understanding of CC pathogenesis.
Materials and methods
Patient tissue collection and cell culture
CC tumor tissues and paired adjacent normal tissues were collected from 20 patients at Shanghai the Eighth People’s Hospital (Shanghai, China). Among them, 19 patients were diagnosed with squamous cell carcinoma, and one patients diagnosed with adenocarcinoma. According to the International FIGO, eight cases were in stage IA1-IB1, and 12 cases were in stage IB2-IIA2. The average age of the patients was 58.4 years (range 38–75 years). When stratified by differentiation grade, 8 patients were poor and 12 patients were moderate. None of the patients had lymph node metastasis. This study was approved by the Ethics Committee of Shanghai the Eighth People’s Hospital (2020-008), and written informed consent for use of the specimens for research purposes was obtained from each patient. Tissues were collected during surgery and instantly frozen in liquid nitrogen. CC cell lines (HeLa and CaSki) were procured from the American Type Culture Collection (Manassas, VA, USA). These cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum.
RNA quantification
Intact RNA was extracted from tissues or cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and RNA was quantified using a NanoDrop spectrophotometer. Quantitative real-time PCR (qRT-PCR) was performed using SYBR Premix Ex Taq™ II (TaKaRa Bio, Tokyo, Japan). The mRNA or comparable miRNA expression levels were normalized to that of the internal control GAPDH or U6, respectively. Relative quantification was conducted using the 2−ΔΔCT method.
Oligonucleotides and transfection
Small interfering RNAs (siRNAs) for LINC00460 (si-LINC00460 1#, 5′-GACTGAGCGTGGGAAAGAAGA-3′; si-LINC00460 2#, 5′-GAAAGACTGAGCGTGGGAAAG-3′), miR-361-3p mimic (5′-TCCCCCAGGTGTGATTCTGATTT-3′) and inhibitor (5′-AAATCAGAATCACACCTGGGGA-3′), and their corresponding negative controls were purchased from GenePharma (Shanghai, China). Lentivirus expressing LINC00460 shRNA was purchased from Hanbio (Shanghai, China). Cell transfection was performed using Lipofectamine 2000. After 48 h, the cells were collected for subsequent experiments.
Cell proliferation assay
To measure cell proliferation, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay and 5-ethynyl-2′-deoxyuridine (EdU) assay were performed. For MTT assay, transfected cells seeded in 96-well plates were stained with 100 μl sterile MTT dye (0.5 mg/ml, Sigma, St. Louis, MO, USA) at designated times for 4 h at 37 °C, followed by the exchange of culture medium with 150 μl dimethyl sulfoxide (Sigma). After 10 min of shaking at the same temperature, the absorbance of the stained cells was measured at 490 nm. An EdU assay kit (Beyotime Biotechnology, Jiangsu, China) was used to determine cell proliferation rates according to the manufacturer’s instructions.
Cell cycle analysis
The cell cycle profile was evaluated according to a standard method [13]. Briefly, transfected cells (1 × 106) were collected and then fixed with 70% ethanol. After washing, the cells were incubated with both DNase-free RNase and propidium iodide (Sigma) for 30 min at 37 °C. A FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) with CellQuest software was used to evaluate the cell cycle.
In vitro Matrigel invasion assay
An invasion assay was performed in a 24-well Transwell chamber containing polyethylene membranes (8-μm pore size), which were coated with cold Matrigel [14]. After the cancer cells were harvested by trypsinization, they were suspended in serum-free DMEM and plated in the upper chamber at a density of 1 × 105 cells. The lower chamber was filled with DMEM containing 10% fetal bovine serum. After 24 h of incubation at 37 °C, the invaded cells adhering to the lower surface were fixed, stained with crystal violet, and counted under a microscope (Olympus Corp., Tokyo, Japan) to determine their relative numbers.
Western blotting
Cells were lysed with RIPA buffer supplemented with phosphatase and protease inhibitors. The concentration of protein was measured with the BCA Protein Assay Kit (Beyotime Biotechnology). From each sample, 30–50 μg of total protein was separated via SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were then blocked and incubated with primary antibodies against Gli1 (Proteintech, Wuhan, China) and GAPDH (Proteintech), followed by incubation with an HRP-conjugated secondary antibody and development with ECL reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Luciferase reporter assay
A fragment containing the miR-361-3p binding site of LINC00460 or the Gli1 3′ untranslated region (UTR) was cloned into the pmirGLO luciferase vector. HeLa and CaSki cells were co-transfected with a proper reporter and miRNA using Lipofectamine 2000. Cells were harvested and lysed after 48 h. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to measure luciferase activity.
In vivo tumorigenesis assays
The animal study was performed in accordance with the Guidelines for the Animal Care and Use (Shanghai Eighth People Hospital). Four-to-six-week-old female BALB/c nude mice were obtained from the Shanghai Laboratory Animal Center (Shanghai, China). HeLa (5 × 106 cells each) and CaSki cells (2 × 106 cells each) infected with LINC00460 shRNA lentivirus were injected subcutaneously into the nude mice (n = 4 per group). Every 5 days, tumor volume (V) was measured using the following formula: V = 0.5 × length × width2. After 30 days, the mice were sacrificed, and the tumors were excised, followed by IHC analysis of Ki67.
Bioinformatical analyses
Comparison of LINC00460 expression levels in CC and normal tissue were analyzed with Gene Expression Profiling Interactive Analysis (GEPIA; https://gepia.cancer-pku.cn/). Prognosis based on LINC00460 expression was also analyzed using GEPIA. The potential LINC00460 and miR-361-3p binding sites was predicted using miRanda (https://www.miranda.org/) and MiRDB (https://mirdb.org/), and miR-361-3p and Gli1 mRNA binding sites was predicted using Targetscan (https://www.targetscan.org/vert_72/).
Statistical analysis
Data were analyzed in GraphPad Prism using Student’s t-test for comparison between groups. The correlation between factors was determined by Pearson’s correlation analysis. Data are presented as means ± SD. P values < 0.05 were considered significant.
Results
LINC00460 is upregulated in CC
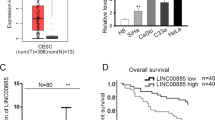
To evaluate the role of LINC00460 in CC, the Gene Expression Profiling Interactive Analysis (GEPIA) database was used to retrieve data corresponding to LINC00460 expression in both CC and normal tissues. Figure 1a shows that LINC00460 expression was lower in the normal tissues than in the CC tissues. Remarkably, survival analysis indicated that higher LINC00460 expression was associated with a lower overall survival rate of CC patients (Fig. 1b). Subsequently, we detected LINC00460 expression in CC and adjacent normal tissues from 20 CC patients. As expected, LINC00460 expression levels were elevated in CC tissues compared with the adjacent normal tissues (Fig. 1c). Consistently, LINC00460 upregulation was also observed in the CC cell lines (HeLa and CaSki) compared with the normal cervical epithelia (Fig. 1d). These observations revealed that LINC00460 may play an oncogenic role in CC progression.
Expression of LINC00460 in CC tissues and cell lines. a LINC00460 expression data corresponding to CC tissues (n = 306) and normal tissues (n = 13) retrieved from the GEPIA database. b Correlation analyses between LINC00460 expression and overall survival in 288 CC patients determined by Kaplan–Meier analysis. c LINC00460 expression determined by qRT‑PCR in 20 paired CC tissues and normal adjacent tissues. d qRT‑PCR analysis of LINC00460 expression in HeLa and CaSki (human CC cell lines) cells and normal cervical tissues. e qRT-PCR analysis of LINC00460 expression in HeLa and CaSki cells after transfection with si-LINC00460 1#, 2# or si-NC. Data are presented as means ± SD of three independent experiments. **P < 0.01 compared with the normal or si-NC group
LINC00460 knockdown inhibits CC cell growth and invasion in vitro
To define the biological roles of LINC00460 in CC cells, its expression was first knocked down in HeLa and CaSki cells by transfection with siRNAs (Fig. 1e), and then loss-of-function tests were performed. MTT assay revealed that the proliferation of HeLa and CaSki cells transfected with si-LINC00460 1# or 2# was repressed compared with that of the control cells (Fig. 2a, b). Meanwhile, we chose si-LINC00460 1# for the following experiments. The EdU assay further verified the suppressive effect of si-LINC00460 1# on the proliferation of HeLa and CaSki cells (Fig. 2c, d). Further, the flow cytometry results confirmed cell cycle arrest in the G0 phase by si-LINC00460 1# (Fig. 2e, f). Thereafter, we assessed the influence of si-LINC00460 on cell invasion, which revealed that LINC00460 knockdown drastically weakened the invasive abilities of HeLa and CaSki cells (Fig. 2g, h). These observations suggested that LINC00460 knockdown suppresses several malignancy parameters in human CC cells.
LINC00460 knockdown impaired CC cell proliferation and invasion in vitro. a, b MTT assays of HeLa and CaSki cells transfected with si-LINC00460 1#, 2# or si-NC. c, d EdU immunofluorescence staining of transfected CC cells. e, f Flow cytometric analysis of the effect of si-LINC00460 1# on the cell cycle. g, h Transwell invasion assays for the determination of the effect of si-LINC00460 1# on cell invasion. Data are presented as means ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with the si-NC group
LINC00460 knockdown decreases tumorigenicity of CC cells in vivo
To determine if LINC00460 affects tumorigenesis, HeLa and CaSki cells infected with sh-LINC00460 or sh-NC lentivirus were inoculated into nude mice. Our results showed that the tumors formed by LINC00460-knockdown cells were smaller and of lower weight than those formed from control cells (Fig. 3a–d). Immunohistochemical analysis revealed that the LINC00460-knockdown tumors exhibited fewer Ki67-positive cells (Fig. 3e, f). These observations suggested that LINC00460 positively regulates the tumorigenicity of CC cells in vivo.
LINC00460 knockdown decreased the tumorigenicity of CC cells in vivo. HeLa and CaSki cells infected with sh-LINC00460 or sh-NC lentivirus were utilized for tumorigenesis in vivo. a, b Tumor growth curves after subcutaneous injection of the infected cells into nude mice. The tumor volumes were measured every 5 days after inoculation. c, d Tumor weights measured after the experiment. e, f The proliferation index (right) determined based on the percentage of Ki67-positive cells (left). Data are presented as means ± SD of three independent experiments. **P < 0.01 compared with the sh-NC group
LINC00460 interacts with miR-361-3p in CC
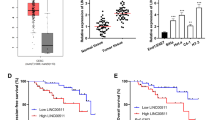
Emerging evidence indicates a close link between lncRNAs and miRNAs in the regulation of biological processes [15]. To identify miRNAs that may interact with LINC00460, we performed bioinformatics analyses using miRDB, miRanda, and lncBase. We found that LINC00460 harbors a putative target site for miR-361-3p, whose expression was shown to be a free prognostic indicator of favorable survival in CC [16]. To determine whether LINC00460 regulates miR-361-3p expression in CC cell progression, the association between LINC00460 and miR-361-3p was investigated. When LINC00460 was knocked down, miR-361-3p expression was upregulated based on qRT-PCR (Fig. 4a). Furthermore, LINC00460 expression decreased after miR-361-3p was overexpressed but recovered after miR-361-3p was inhibited (Fig. 4b, c). In addition, miR-361-3p expression was suppressed in CC tissues compared with adjacent normal tissues (Fig. 4d), revealing an inverse correlation between miR-361-3p and LINC00460 expression (Fig. 4e). A dual-luciferase reporter assay validated the binding site between LINC00460 and miR-361-3p (Fig. 4f). Furthermore, MTT assay indicated that the suppression of CC cell growth and invasion triggered by suppression of LINC00460 was reversed by miR-361-3p inhibitor (Fig. 4g, h). Therefore, LINC00460 may promote CC cell growth and invasion by repressing the activity of miR-361-3p.
LINC00460 regulated miR-361-3p expression in CC cells. a After LINC00460 knockdown, miR-361-3p was significantly upregulated. b, c In response to miR-361-3p overexpression, LINC00460 mRNA expression was significantly decreased but increased following miR-361-3p inhibition. d In human CC tissues, miR-361-3p expression levels were significantly decreased compared with those in normal tissues. e An inverse correlation was observed between mRNA expression of miR-361-3p and LINC00460 in human CC tissues. f LINC00460 directly interacted with miR-361-3p. g MTT assay showed that LINC00460 knockdown suppressed CC cell growth, which was reversed by miR-361-3p inhibitor (miR-361-3p-in). h Transwell invasion assay showed that LINC00460 knockdown repressed CC cell invasion, which was reversed by miR-361-3p inhibitor (miR-361-3p-in). Data are presented as means ± SD of three independent experiments. **P < 0.01 compared with the si-NC, NC mimic, NC inhibitor, or normal group; ##P < 0.01 compared with the si-LINC00460 group
LINC00460 regulates Gli1 through miR-361-3p
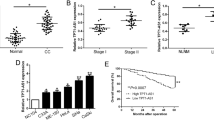
A previous study revealed that miR-361-3p interacts with the 3′UTR of Gli1 mRNA, and overexpression of miR-361-3p downregulates Gli1 mRNA and protein levels [17], thus identifying Gli1 as a direct target of miR-361-3p. Similarly, our results showed that Gli1 expression was downregulated when miR-361-3p was overexpressed; however, Gli1 expression recovered when miR-361-3p was repressed in HeLa and CaSki cells (Fig. 5a, b). Luciferase reporter gene assays further verified the binding site between miR-361-3p and the Gli1 3′UTR (Fig. 5c). These results indicated that miR-361-3p directly targets the 3′UTR of Gli1 and represses its expression. Next, we evaluated whether LINC00460 regulates Gli1 through miR-361-3p in CC cells. Western blotting assay revealed that Gli1 protein expression was decreased in response to si-LINC00460 transfection but significantly increased following miR-361-3p suppression (Fig. 5d, e), suggesting that the inhibitory effect on Gli1 expression induced by LINC00460 knockdown was reversed by miR-361-3p inhibition. We finally measured the mRNA levels of Gli1 in CC and corresponding normal tissues. The results showed that the expression of Gli1 was greater in CC tissues than in paired adjacent tissues (Fig. 5f). There was an inverse correlation between miR-361-3p and Gli1 expression in CC tissues (Fig. 5g). In addition, LINC00460 expression was correlated with Gli1 expression (Fig. 5h), which was consistent with the results from the GEPIA database (Fig. 5i). These results demonstrated that LINC00460 interacts with miR-361-3p to decrease its suppressive effect on Gli1, subsequently enhancing the expression of Gli1.
LINC00460 regulated Gli1 expression through miR-361-3p in CC cells. a Western blot and qRT-PCR indicated that the mRNA and protein expression levels of Gli1 were downregulated following miR-361-3p overexpression in HeLa and CaSki cells. b Western blot and qRT-PCR showed that the protein and mRNA expression levels of Gli1 were upregulated following miR-361-3p inhibition in HeLa and CaSki cells. c The miR-361-3p target site in the Gli1 3′UTR sequence; luciferase reporter assay confirmed the binding of miR-361-3p and Gli1. d, e The inhibitory effect of LINC00460 knockdown on Gli1 protein expression was partially restored by a miR-361-3p inhibitor (miR-361-3p-in) in HeLa and CaSki cells. f The mRNA expression levels of Gli1 were significantly elevated in CC tissues compared with normal adjacent tissues. g A negative correlation was observed between miR-361-3p and Gli1 mRNA expression in human CC tissues. h A serial correlation was observed between LINC00460 and Gli1 mRNA expression in human CC tissues. i Correlation analysis of LINC00460 and Gli1 mRNA expression in CC tissues and normal tissues from the GEPIA database. Data are presented as means ± SD of three independent experiments. **P < 0.01
Discussion
In this study, we demonstrated that LINC00460 is significantly upregulated in CC tissues and cell lines. Using loss-of-function assays, we found that knockdown of LINC00460 decreased in vitro cell growth and invasion and attenuated xenograft growth. Mechanistically, LINC00460 could function as a ceRNA that concealing miR-361-3p to eliminate its inhibitory effect on the target gene Gli1. Hence, LINC00460 may play a crucial role in the development and pathogenesis of CC.
Recently, lncRNAs have emerged as unique molecular players in human diseases, particularly cancers. Previous studies indicated that LINC00460 is upregulated and possibly promotes tumor growth in diverse types of cancer. For example, upregulated LINC00460 is correlated with poor prognosis in colorectal cancer and leads to tumorigenesis [10]; LINC00460 stimulates the gefitinib resistance of non-small cell lung cancer cells by regulating EGFR via sponging of miR-769-5p [18]. In addition, LINC00460 was shown to enhance head and neck squamous cell carcinoma growth and metastasis by enabling PRDX1 entry into the nucleus [19]. Nevertheless, the expression and role of LINC00460 in CC have not yet been clarified. Here, we established that the average LINC00460 level in CC tissues was higher than that in adjacent tissues. Functional assays revealed that inhibition of LINC00460 suppressed CC cell growth and invasion in vitro and tumorigenesis in vivo. Moreover, its knockdown induced G0/G1 arrest in CC cells. These results indicated that LINC00460 functions as an oncogene in CC.
Storing lncRNAs have been proven as endogenous ceRNAs for particular miRNAs and normalize their functions. Many miRNAs have been reported to potentially interact with LINC00460, such as miR-149-5p, miR-613, miR-485-5p, and miR-769-5p [10,11,12, 18]. By bioinformatics analysis and subsequent confirmatory experiments, we demonstrated that LINC00460 acts as a ceRNA to competitively sponge miR-361-3p, which has been shown to be a tumor suppressor in retinoblastoma and non-small cell lung cancer [17, 20] as well as a self-governing prognostic indicator of survival in CC [16]. Further, we found that knockdown of LINC00460 enhanced miR-361-3p expression, leading to destruction of its target gene Gli1.
Consequently, the impact of LINC00460 on CC cell growth and invasion could be attributed, at least in part, to its function as an antagonistic ceRNA that sponges miR-361-3p. Nonetheless, there were some limitations in this work. In particular, further in vivo investigation is needed, such as metastasis assays. In future research, we plan to increase the sample size and further examine the characteristics of LINC00460 in CC metastasis in vivo.
In conclusion, this study demonstrated that LINC00460 expression is upregulated in CC tissues and may have an adverse prognostic influence in CC patients. The impact of LINC00460 on CC cell proliferation and invasion indicates that LINC00460 exhibits oncogenic properties in CC progression and tumorigenesis. Our results provide further insight into CC pathogenesis and may facilitate development of lncRNA-related therapeutics and diagnostics for CC.
Data availability
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Change history
18 May 2022
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s13577-022-00723-3
References
de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141(4):664–70. https://doi.org/10.1002/ijc.30716.
Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459. https://doi.org/10.1371/journal.pgen.1000459.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. https://doi.org/10.1016/j.cell.2009.02.006(S0092-8674(09)00142-1[pii]).
Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16(1):39. https://doi.org/10.1186/s12943-017-0588-9.
Shi C, Yang Y, Yu J, Meng F, Zhang T, Gao Y. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res. 2017;7(11):2157–68.
Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res. 2018;78(4):877–90. https://doi.org/10.1158/0008-5472.CAN-17-2356.
Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29(6):e95. https://doi.org/10.3802/jgo.2018.29.e95.
Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY, Li S, et al. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. 2019;234(5):6624–32. https://doi.org/10.1002/jcp.27403.
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. https://doi.org/10.1016/j.canlet.2018.01.060.
Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, et al. A Novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–97. https://doi.org/10.1016/j.omtn.2018.06.012.
Feng L, Yang B, Tang XD. Long noncoding RNA LINC00460 promotes carcinogenesis via sponging miR-613 in papillary thyroid carcinoma. J Cell Physiol. 2019;234(7):11431–9. https://doi.org/10.1002/jcp.27799.
Tu J, Zhao Z, Xu M, Chen M, Weng Q, Ji J. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485-5p to up-regulate PAK1. Biomed Pharmacother. 2019;118:109213. https://doi.org/10.1016/j.biopha.2019.109213.
Li F, Wang Z, Lu G. TRIM28 promotes cervical cancer growth through the mTOR signaling pathway. Oncol Rep. 2018;39(4):1860–6. https://doi.org/10.3892/or.2018.6235.
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77(23):6704–16. https://doi.org/10.1158/0008-5472.CAN-17-1915.
Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen L, et al. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38(1):60. https://doi.org/10.1186/s13046-019-1055-9.
Liu S, Song L, Yao H, Zhang L, Xu D, Li Q, et al. Preserved miR-361-3p expression is an independent prognostic indicator of favorable survival in cervical cancer. Dis Markers. 2018;2018:8949606. https://doi.org/10.1155/2018/8949606.
Zhao D, Cui Z. MicroRNA-361-3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp Ther Med. 2019;17(2):1154–62. https://doi.org/10.3892/etm.2018.7062ETM-0-0-7062.
Ma G, Zhu J, Liu F, Yang Y. Long noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through epidermal growth factor receptor by sponging miR-769-5p. DNA Cell Biol. 2019;38(2):176–83. https://doi.org/10.1089/dna.2018.4462.
Jiang Y, Cao W, Wu K, Qin X, Wang X, Li Y, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. https://doi.org/10.1186/s13046-019-1364-z.
Chen W, Wang J, Liu S, Wang S, Cheng Y, Zhou W, et al. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res. 2016;35:76. https://doi.org/10.1186/s13046-016-0357-4.
Acknowledgements
The study was supported by the Special Fund for Gynecology and Obstetrics of Medical Association Disease Diagnosis and Freatment Center of Shanghai Sixth People’s Hospital (02.LY01.03.23).
Author information
Authors and Affiliations
Contributions
FL, WZ and ZW conceived and designed this study. FL, WZ and ZW collected the samples and performed the experiment. FL wrote the paper. WZ and ZW revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shanghai the Eighth People’s Hospital, and written informed consent for use of the specimens for research purposes was obtained from each patient.
Consent for publication
All authors are responsible for the submission of this article and accept the conditions of submission.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s13577-022-00723-3
About this article
Cite this article
Li, F., Zhu, W. & Wang, Z. RETRACTED ARTICLE: Long noncoding RNA LINC00460 promotes the progression of cervical cancer via regulation of the miR-361-3p/Gli1 axis. Human Cell 34, 229–237 (2021). https://doi.org/10.1007/s13577-020-00447-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00447-2