Abstract
Colorectal cancer (CRC) is the third leading cause of cancer-related death around the world. In this study, we investigated the roles of LncRNA RP11-462C24.1 in CRC. The expressions of RP11-462C24.1 in CRC tissues and cells were measured. Then, the effects of RP11-462C24.1 on CRC proliferation, cell cycle, apoptosis, and invasion were evaluated both in vivo and in vitro; Last, the underlying mechanisms of concerning the signaling pathway regulated by RP11-462C24.1 was determined. From the results, we found that RP11-462C24.1 was significantly decreased in CRC tumor tissues and the CRC cell lines, which were most significant in SW480 and HT-29 cell lines; moreover, transient overexpression of RP11-462C24.1 suppressed the growth and migration while promoted apoptosis of SW480 and HT-29 cells, while knockdown of RP11-462C24.1 has shown the opposite effects; RP11-462C24.1 may also inhibit the growth of CRC tumors in xenograft mice models; additionally, 70 kD heat shock proteins (HSP70) has been identified as one of the most significantly deferentially expressed genes by RNA-seq, and we further confirmed that RP11-462C24.1 may affect the growth and metathesis of CRC cells via regulating HSP70 and PI3K/AKT signaling pathway. In summary, these results indicated that RP11-462C24 may function as a tumor suppressor in the development of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the most common gastrointestinal cancer (incidence number > 900,000/year), and it is the NO.1 cause of cancer-induced motility worldwide [1,2,3]. According to the results of previous reports, CRC may derive from a series of abnormal genetic and molecular alterations in colon epithelial cells, leading to the uncontrolled growth of the cancer cells [4, 5]. Currently, surgery, chemotherapy, and radiotherapy were the most commonly used methods for the treatment of CRC; however, none of the current anti-CRC methods have achieved the desired therapeutic efficacy, and the long-term prognosis of the CRC patients is still poor [6, 7]. Therefore, identifying novel diagnostic biomarkers and novel therapeutic targets for the early diagnosis and treatment of CRC is in high demand.
Long non-coding RNAs (LncRNAs) belongs to the non-coding RNA family. The length of LncRNAs are longer than 200 nucleotides, and some of the LncRNAs can reach to kilobases in length [8, 9]. Results of previous studies suggested that LncRNAs may participate in multiple biological activities, including the cell growth, migration and apoptosis, angiogenesis, embryo development and tumorigenesis via affecting the gene expression of their targets on post-transcriptional level [10,11,12]. Increasing evidence indicated that LncRNAs were aberrantly expressed in different types of cancers and may serve as diagnostic or prognostic markers, as well as potential therapeutic targets[13, 14]. In recent microarray analysis, Shi et al. have compared the LncRNA expression profile in CRC cancerous tissue samples and found that the expression of LncRNA RP11-462C24.1, a novel unnamed LncRNA, was significantly down-regulated in CRC tissues compared with the healthy tissues. Increased expression of RP11-462C24.1 in CRC tissues may indicate a better prognosis [15]. Nevertheless, the functions and mechanism of RP11-462C24.1 in the pathogenesis of CRC still requires further investigation.
In the present study, in vitro, in vivo and RNA-sequencing analysis have been performed to investigate the roles of RP11-462C24.1 in CRC. First, we compared the levels of RP11-462C24.1 in cancerous tissues and normal tissue samples obtained from the CRC patients, and the association between RP11-462C24.1 expression and the clinical features of the CRC patients have also been studied. Furthermore, the effects of RP11-462C24.1 on the growth and metathesis of CRC cells were also examined. Finally, the underlying mechanism of RP11-462C24.1 inducted anti-tumor effects were also investigated.
Materials and methods
Patients
We included 60 CRC patients hospitalized at Affiliated Fengxian Hospital, The Third School of Clinical Medicine, Southern Medical University between January 2016 and February 2017 in the present study. All participators were diagnosed as CRC pathologically, and patients with the history of radio and/or chemotherapies were excluded from this study. After surgery, the paired CRC tumor samples and the paired adjacent tissue samples were collected and immediately stored in liquid nitrogen. We have received the signed informed consent from all the participators of this study, and also obtained the ethical documents issued by the ethical committee of Affiliated Fengxian Hospital, The Third School of Clinical Medicine, Southern Medical University.
Cell culture
Human CRC cell lines LoVo, SW1116, HCT 116, SW480 and HT-29 were used in this study for the cell analysis. The CRC cells were obtained from Shanghai Institutes for Biological Sciences (Shanghai, China). The colonic mucosa cells NCM460 (INCELL, San Antonio, TX, USA) were used as the control group. RPMI-1640 medium (Invitrogen, Waltham, MA, USA) supplied with 10% of fetal bovine serum (FBS, Invitrogen) was used for cell culture, and cells were maintained at 37 ˚C and 5% CO2 in an incubator.
Transfection
siRNA and overexpression plasmid of RP11-462C24.1 (GenePharma Co.,Ltd, Shanghai, China) were synthesized and the CRC cells were transfected by RP11-462C24.1 siRNA, RP11-462C24.1 siRNA negative control (NC), RP11-462C24.1 overexpression plasmid or empty plasmid vector (control) by using lipofectamine 3000 (Invitrogen) following the manufacturer’s protocols. Quantitative real-time PCR (RT-qPCR) was performed to determine transfection efficiency. To confirm the relationship between RP11-462C24.1 and HSP70, SW480 or HT-29 cells were co-transfected with RP11-462C24.1 over-expression and HSP70 shRNA (GenePharma Co.,Ltd) by using lipofectamine 3000. The sequences were as follows: RP11-462C24.1 siRNA forward, TAATTTGTTTCTAGATGTGTG, and reverse, CACATCTAGAAACAAATTAAT; HSP70 shRNA forward, TGCTGACACCAGGCTGGTTGTCAGAAGT TTTGGCCACTGACTGACTTCTGACACAGCCTGGTGT, and reverse, CCTGACACCAGGCTGTGTCAGAAGTCAGTCAGTGGCCAAAACTTCTGACAACCAGCCTGGTGTC; siRNA NC forward, UUCUCCGAACGUGUCACGUTT, and reverse, ACGUGACACGUUCGGAGAATT.
Reverse transcript and RT-PCR
The total RNAs in the cultured cells or tissue samples were isolated using TRIzol reagent (Invitrogen). cDNAs were then synthesized by PrimeScript RT Master Mix kit (Takara, Dalian, China). Next, RT-PCR has been conducted to detect the RP11-462C24.1 expression in different samples by SYBR premix Ex Taq kit (Takara). ABI 7500 Biosystems (Applied Biosystems Life Technologies, Foster City, USA) was used for the PCR reaction. The relative expression level of RP11-462C24.1 was normalized to GAPDH by 2−ΔΔCt method, and GAPDH has been applied for normalization. The thermo profiles were used: 95 ºC 30 s and 40 cycles of 95 ºC 5 s + 60 ºC 30 s. The primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China), and sequences were: RP11-462C24.1, F: 5′-GAACTTCCTCCCACTTATCCCTTAG-3′, R: 5′-GATGATTTTTGCTGTTCTAAGTGATGTAT-3′; GAPDH, F: CAACTCCCTCAAGATTGTCAGCAA, R: GGCATGGACTGTGGTCATGA.
Cell proliferation analysis
The proliferation of the cells of different treatments was detected using the cell counting kit-8 (CCK-8) kit (purchased from Beyotime, Shanghai, China). Briefly, SW480 or HT-29 cells were collected washed by PBS (pH 7.4), trypsinized and seeded onto 96-well plates. At 12, 24 or 48 h, cells were treated by CCK-8 solution (10 μl/well) and incubated at 37 ºC. Cell proliferation was determined by the OD value that was detected at 450 nm by a microplate reader (Thermo Fisher Scientific, Inc).
Cell apoptosis analysis
CRC cells of different treatments were collected at 48 h after transfection and stained by Annexin-V-fluorescein isothiocyanate (FITC, 5 μl) and propidium iodide (PI, 2.5 μl). FACSCalibur (BD Biosciences, San Jose, CA) flow cytometer was used to examine the cell apoptosis and cell cycles.
Cell invasion analysis
The invasion ability of the cells was determined by Transwell assay. The upper of the transwell chambers (Corning Inc., Corning, USA) covered with thick layer of Matrigel (Solarbio, Beijing, China) were seeded with CRC cells (5 × 104 cells/well) and incubated at 37 °C on 24-well cell cure plates. On day 2, the lower chambers were treated by methanol, and the CRC cells invaded into the lower chamber were then stained with crystal violet and imaged by a microscope.
RNA-seq analysis
Total RNAs were isolated by TRIzol (Invitrogen). The quality of the RNAs was evaluated by Nanodrop 2000 (Thermo Fisher Scientific, Inc). The samples were sent to Vazyme Biotech (Nanjing, China) for RNA-sequencing.
Western blot assay
Expressions of protein in the CRC cells with different treatments were evaluated by western blot assay. Briefly, cells were lysed by lysis buffer (Thermo Fisher Scientific, Inc) containing 1% protease inhibitor cocktail to isolate the proteins, and then protein concentration in each sample was measured by BCA kit (purchased from Beyotime). Next, appropriate amount of proteins (30 μg/μl) were separated by 10% SDS–PAGE gels, and when gel electrophoresis was accomplished, the protein on the gels were transferred onto PVDF membranes. Then the PVDF membranes were blocked by 5% non-fat milk solution containing 0.05% Tween 20 and incubated with the primary antibodies (anti-Hspa1a, anti-Hspa1b, anti-p-PI3K, anti-PI3K, anti-p-Akt, anti-Akt, anti-Caspase-3, anti-Bcl-2 and anti-Bax) at 4 °C overnight. GAPDH was served as the control. The following day, the protein bands were incubated by secondary antibodies (Beyotime) and chemiluminescent reagent BeyoECL Plus (Beyotime). Finally, the bands were photographed by Tanon 6100 Chemiluminescent Imaging System (Tanon, Shanghai, China).
Xenograft tumor model
SW480 cells were transfected with RP11-462C24.1 overexpression plasmid and SW480 cells treated with plasmid vector (empty vector) were served as the control group. Then, 3 × 106 transfected cells were injected into nude mice (6 week-old C57BL/6, purchased from Shanghai Institutes for Biological Sciences) subcutaneously. The formation of the tumors was examined weekly. At week 5 post-implantation, mice were sacrificed and the tumor volume and the tumor weight were measured. The animal study has been performed with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of Affiliated Fengxian Hospital, The Third School of Clinical Medicine, Southern Medical University (Grant No. 2017051287).
Statistics
The data were presented as mean ± standard deviation (SD), and the statistical analyses were performed using IBM SPSS Statistic v17.0 (IBM Co., Armonk, NY, USA). Comparison of the means between two groups was analyzed by student t-test and analysis of variance (ANOVA) was used to the comparison of the differences among multiple groups. Chi-square test was used to determine the association between the RP11-462C24.1 expression and clinical features. P < 0.05 was set as statistically significant.
Results
Down-regulation of RP11-462C24.1 in CRC tumor samples and CRC cell lines
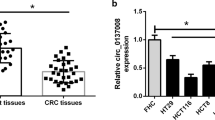
First, the levels of RP11-462C24.1 in CRC cancerous samples and the adjacent non-tumorous samples were compared using RT-qPCR methods. We found that LncRNA RP11-462C24.1 was significantly decreased in CRC tumors compared with the adjacent non-tumorous samples (Fig. 1a, p< 0.001); moreover, decreased the expression level of LncRNA RP11-462C24.1 was associated with the advanced TNM (tumor node metastasis) stage (Table 1, p < 0.05) and metathesis (Table 1, p < 0.01). Next, the expression levels of RP11-462C24.1 between 5 different colorectal cancer cell lines (LoVo, SW1116, HCT116, HT-29 and SW480) as well as healthy colonic mucosa cells (NCM460 cell) were also compared by RT-qPCR. We observed that the levels of RP11-462C24.1 were decreased in all colorectal cancer cell lines in comparison with the NCM460 cells. Among the 5 cell lines, the expression levels of LncRNA RP11-462C24.1 in SW480 and HT-29 cells were the lowest (Fig. 1B, p< 0.01). Based on these results, these two cell lines were used for the following in vitro and animal studies.
Down-regulation of LncRNA RP11-462C24.1 in CRC tumor samples and cell lines. a RP11-462C24.1 was significantly reduced in cancerous tissues samples compared with the adjacent tissues obtained from CRC patients (N = 60); b The expression of RP11-462C24.1 were significantly reduced in colorectal cancer cell lines compared with NCM460 normal colonic mucosa cells (N = 5). c Loss of RP11-462C24.1 decreases the expression of RP11-462C24.1 in SW480 and HT-29 colorectal cancer cell lines (N = 5). d Gain of RP11-462C24.1 increases the expression of RP11-462C24.1 in SW480 and HT-29 colorectal cancer cell lines (N = 5). ***p < 0.001, tumor tissues vs adjacent tissue; *p < 0.05, LoVo, SW1116 or HCT116 vs NCM 460; **p < 0.01, HT-29 or SW480 vs NCM460; **p < 0.01, siRNA vs NC; overexpression vs control
LncRNA RP11-462C24.1 inhibits the proliferation while promotes apoptosis of SW480 and HT-29 cells in vitro
To further investigate whether RP11-462C24.1 could affect the oncogenic behavior of CRC cells in vitro, the CRC cells were then transfected with RP11-462C24.1 overexpression plasmid or siRNA, and the transfection efficiency was shown in Fig. 1c, d. It was observed that RP11-462C24.1 siRNA significantly decreased and RP11-462C24.1 overexpression plasmid increased the levels of RP11-462C24.1 both in SW480 and HT-29 cells (p < 0.01), suggesting that transfection has been successfully performed. Next, CCK-8 cell proliferation assay, cell cycle assay and flow cytometry apoptosis analysis have been conducted. We found that transient overexpression of RP11-462C24.1 dramatically suppressed the proliferation ability of SW480 and HT-29 cells (Fig. 2a, b), while knockdown of RP11-462C24.1 promoted the proliferation of the CRC cells at 24 h and 48 h (Fig. 2c, d) after transfection; moreover, the overexpression of RP11-462C24.1 led to the arrest of the cell cycle at G1 stage for the CRC cells (Figs. 3a and 4a) while RP11-462C24.1 siRNA has shown the opposite effects (Figs. 3b and 4b). On the other hand, the transfection of RP11-462C24.1 overexpression plasmid significantly increased the apoptosis of CRC cells (Figs. 5a and 6a) while RP11-462C24.1 siRNA markedly decreased the apoptosis of the cells in vitro (Figs. 5b and 6b).
Gain of RP11-462C24.1 inhibits while loss of RP11-462C24.1 promotes SW480 and HT-29 colorectal cancer cell proliferation. a, b Gain of RP11-462C24.1 inhibits SW480 and HT-29 colorectal cancer cell proliferation determined by CCK-8 proliferation assay (N = 5). c, d Loss of RP11-462C24.1 promotes SW480 and HT-29 colorectal cancer cell proliferation determined by CCK-8 proliferation assay (N = 5). *p < 0.05, **p < 0.01, overexpression vs control; siRNA vs NC
Gain of RP11-462C24.1 promotes while loss of RP11-462C24.1 inhibits SW480 colorectal cancer cell cycle. a, b Gain of RP11-462C24.1 promotes SW480 colorectal cancer cell cycle determined by flow cytometry methods (N = 5). c, d Loss of RP11-462C24.1 inhibits SW480 colorectal cancer cell cycle determined by flow cytometry methods (N = 5). G1, S and G2 states were presented in Fig. 3. *p < 0.05, overexpression vs control, siRNA vs NC
Gain of RP11-462C24.1 promotes while loss of RP11-462C24.1 inhibits HT-29 colorectal cancer cell cycle. a, b Gain of RP11-462C24.1 promotes HT-29 colorectal cancer cell cycle determined by flow cytometry methods (N = 5). c, d Loss of RP11-462C24.1 inhibits HT-29 colorectal cancer cell cycle determined by flow cytometry methods (N = 5). *p < 0.05, overexpression vs control, siRNA vs NC
Gain of RP11-462C24.1 promotes while loss of RP11-462C24.1 inhibits SW480 colorectal cancer cell apoptosis. a, b Gain of RP11-462C24.1 promotes SW480 colorectal cancer cell apoptosis determined by flow cytometry apoptosis analysis (N = 5). c, d Loss of RP11-462C24.1 inhibits SW480 colorectal cancer cell apoptosis determined by flow cytometry apoptosis analysis (N = 5). **p < 0.01, overexpression vs control, siRNA vs NC
Gain of RP11-462C24.1 promotes while loss of RP11-462C24.1 inhibits HT-29 colorectal cancer cell apoptosis. a, b Gain of RP11-462C24.1 promotes HT-29 colorectal cancer cell apoptosis determined by flow cytometry apoptosis analysis (N = 5). c, d Loss of RP11-462C24.1 inhibits HT-29 colorectal cancer cell apoptosis determined by flow cytometry apoptosis analysis (N = 5). **p < 0.01, overexpression vs control, siRNA vs NC
The effect of RP11-462C24.1 on the invasion of SW480 and HT-29 CRC cells in vitro
Next, we determined whether RP11-462C24.1 could affect the invasion ability of CRC cells in vitro by Transwell assay. We found overexpression of RP11-462C24.1 markedly decreased the invasion ability of CRC cells in vitro (Fig. 7a, b), and RP11-462C24.1 siRNA increased the invasion (Fig. 7c, d) of the cells in vitro.
Gain of RP11-462C24.1 inhibits while loss of RP11-462C24.1 promotes the ability of invasion in SW480 and HT-29 colorectal cancer cell lines. a, b Gain of RP11-462C24.1 inhibits SW480 and HT-29 colorectal cancer cell invasion determined by Transwell analysis (N = 5). c, d Loss of RP11-462C24.1 promotes SW480 and HT-29 colorectal cancer cell INVASION determined by Transwell analysis (N = 5). **p < 0.01, overexpression vs control, siRNA vs NC; ***p < 0.001, overexpression vs control, siRNA vs NC
Effects of RP11-462C24.1 on the tumorigenesis of SW480 cells in vivo
Furthermore, the in vivo tumorigenic effects of RP11-462C24.1 were also investigated by nude mice xenograft tumor models. As shown in Fig. 8b, c, the average volume of the tumors was significantly smaller in RP11-462C24.1 overexpression group compared with the control mice; moreover, we also found the average tumor weight measured after the mice were sacrificed significantly lower in RP11-462C24.1 overexpression group in comparison with the control mice (Fig. 8c, d, p < 0.01).
The in vivo effects of RP11-462C24.1 on the tumorigenic ability of SW480 cells. a The expression of RP11-462C24.1 was significantly increased after transfection with RP11-462C24.1 over-expression plasmid (N = 5). b The average volume of the tumors was significantly increased in RP11-462C24.1 over-expression group during the 5 weeks of tumor formation (N = 5). c Photos of the dissected tumors obtained from the nude mice (N = 5). d The average weight of the tumors in RP11-462C24.1 over-expression group was significantly decreased after the mice were sacrificed (N = 5). **p < 0.01, ***p < 0.001, overexpression vs control
Genome-wide RNA sequencing for the analysis of the deferentially expressed genes for SW480 cells transfected by RP11-462C24.1 siRNA
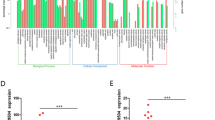
Furthermore, we performed sequencing for the analysis of changes in the expression of genes in SW480 cells treated with RP11-462C24.1 siRNA or without treatment. As shown in Fig. 9, we found that the expression of 1254 genes significantly altered in RP11-462C24.1 siRNA treated SW480 cells compared with the control. Among them, 684 genes were upregulated and 570 genes were downregulated (Fig. 9). Furthermore, the gene ontology (GO) and pathway analysis were performed to determine the influence of RP11-462C24.1 siRNA on the genome-wide expression of the genes, and results were shown in Figs. 10 and Fig. 11. According to Fig. 10, the main biological process, cellular component and molecular function were primary metabolic process, binding and intracellular, respectively. From Fig. 11, it demonstrated 20 top enriched pathways.
RP11-462C24.1 may affect the proliferation and migration of CRC cells via regulating HSP70 and PI3K/AKT in vitro
Based on the sequencing results, the transient knockdown of RP11-462C24.1 siRNA significantly increased the expressions of two paralogs of 70 kilodalton heat shock protein (HSP70), Hspa1a and Hspa1b (21.21 fold and 12.89 fold, respectively). Therefore, to explore the association between RP11-462C24.1 and HSP70 in CRC, SW480 and HT-29 cells of different treatments were collected, and the expressions of Hspa1a and Hspa1b were determined by western blot. As shown in Fig. 12, RP11-462C24.1 overexpression plasmid significantly suppressed in the expression levels of Hspa1a and Hspa1b in CRC cells and transient knockdown of RP11-462C24.1 using siRNA has shown the opposite effects. Moreover, the overexpression of RP11-462C24.1 also suppressed the activation of PI3K/AKT signaling pathway in SW480 or HT-29 cells by decreasing the expression levels of p-PI3K and p-AKT, as well as the downstream protein Bcl 2 (anti-apoptotic factor), and promoting the expression of Bax and Caspase 3 (pro-apoptotic factors, Fig. 12a, b). Meanwhile, the transient knockdown of RP11-462C24.1 active the PI3K/AKT signaling in CRC cells in vitro (Fig. 12c, d).
Gain of RP11-462C24.1 suppresses while loss of RP11-462C24.1 promotes HSP70/PIAK/AKT signaling pathway in CRC in vitro. a, b Gain of RP11-462C24.1 decreases Hspa1a, Hspa1b, p-PI3K, p-AKT and Bax while increases Bax and Caspase 3 expressions in SW480 and HT-29 colorectal cancer cell lines (N = 5). c, d Loss of RP11-462C24.1 increases Hspa1a, Hspa1b, p-PI3K, p-AKT and Bax while decreases Bax and Caspase 3 expressions in SW480 and HT-29 colorectal cancer cell lines (N = 5)
HSP70 shRNA may partially abrogate RP11-462C24.1 overexpression plasmid induced anti-tumor effects on CRC cells
Finally, the association between RP11-462C24.1, HSP70, and PI3K/AKT signaling in the pathogenesis of CRC were further explored. SW480 and HT-29 cells were co-transfected with RP11-462C24.1 overexpression plasmid and HSP70 shRNA, and the growth, as well as invasion of CRC cells with different treatment, were examined. As shown in Figs. 13 and 14, HSP70 shRNA partially abrogated RP11-462C24.1 over-expression plasmid induced anti-tumor effects by promoting cell growth and invasion and suppressing the cell apoptosis of both SW480 and HT-29 in vitro(p < 0.05).
HSP70 shRNA may partially abrogate RP11-462C24.1 over-expression plasmid induced anti-tumor effects on SW480 cells. a HSP70 shRNA may partially counteracted the suppressive proliferation induced by RP11-462C24.1 over-expression in SW480 colorectal cancer cells (N = 5). b HSP70 shRNA may partially counteracted the promotive apoptosis induced by RP11-462C24.1 over-expression in SW480 colorectal cancer cells (N = 5). c HSP70 shRNA may partially counteracted the suppressive invasion induced by RP11-462C24.1 over-expression in SW480 colorectal cancer cells (N = 5). *p < 0.05, overexpression vs control, overexpression + HSP70 shRNA vs overexpression; **p < 0.01, overexpression vs control
HSP70 shRNA may partially abrogate RP11-462C24.1 over-expression plasmid induced anti-tumor effects on HT-29 cells. a HSP70 shRNA may partially counteracted the suppressive proliferation induced by RP11-462C24.1 over-expression in HT-29 colorectal cancer cells. b HSP70 shRNA may partially counteracted the promotive apoptosis induced by RP11-462C24.1 over-expression in HT-29 colorectal cancer cells. c HSP70 shRNA may partially counteracted the suppressive invasion induced by RP11-462C24.1 over-expression in HT-29 colorectal cancer cells. *p < 0.05, overexpression vs control, overexpression + HSP70 shRNA vs overexpression; **p < 0.01, overexpression vs control
Discussion
In this study, we found that the expression of LncRNA RP11-462C24.1 was down-regulated in colorectal cancer tumor samples and cell lines, which was consistent with previous observations, and more importantly, we reported for the first time that RP11-462C24.1 may function as a tumor suppressor in CRC via affecting HSP70 and PI3K/AKT signaling pathway.
Dysregulation of LncRNAs in different types of cancers has been observed in many previous studies [16, 17]. In CRC, the regulatory roles of LncRNAs, either as tumor suppressors or oncogenes, have also been reported [18,19,20]. RP11-462C24.1 is a recently identified LncRNA by Shi et al. in 2014. RP11-462C24.1 locates in chr4q25, and it consists of four exons. The length of RP11-462C24.1 is 1,136 bp. The patterns of the expressions of RP11-462C24.1 in the tissue samples and the serum of patients are not uniform. Shi et al. reported that RP11-462C24.1 was markedly decreased in the CRC tumor samples, and the expression levels of RP11-462C24.1 may be negatively associated with the tumor metastasis and positively correlated with the prognosis of the CRC patients [15]. On the other hand, Wang et al. suggested that the circulating RP11-462C24.1 was overexpressed in the serum samples of CRC patients and the combination of three LncRNAs, LOC285194, Nbla12061, and RP11-462C24.1 and may serve as diagnostic biomarkers [21]. In the present work, results of RT-PCR analysis showed that RP11-462C24.1 was decreased in CRC cancerous tissues in comparison the adjacent tissues. Our finding was consistent with the results of Shi et al. Moreover, we also discovered the expression of RP11-462C24.1 in CRC tumor was negatively correlated with the TNM stage and tumor metathesis of the patients; furthermore, LncRNA RP11-462C24.1 was also decreased in different CRC cell lines, particularly the SW480 cells and HT-29 cells. Taken together, results of the present study indicated that LncRNA RP11-462C24.1 was abnormally decreased in CRC and may function as a tumor suppressor.
Tumor cells were characterized by uncontrolled cell growth, increased invasion ability and resistance to apoptosis [22,23,24]. Our data indicated that the transient overexpression of RP11-462C24.1 suppressed the proliferation and invasion and on the other hand, increased the apoptosis of CRC cells, while transfection of RP11-462C24.1 siRNA has shown the opposite effects. Moreover, the tumor-suppressive effects of RP11-462C24.1 in CRC were also confirmed by nude mice xenograft models in vivo. Taken together, these results indicated that RP11-462C24.1 could regulate the growth and invasion of the colorectal cancer cells in vitro and also in vivo.
Since reports on the roles of RP11-462C24.1 in cancers were limited, we performed genome-wide RNA sequencing to further explore the possible underlying mechanism of how RP11-462C24.1 could regulate the growth and invasion of the colorectal cancer cells. Results of sequencing results suggested that compared with un-treated SW480 cells, the expression 684 genes were significantly increased while 570 genes were decreased in RP11-462C24.1 siRNA transfected cells. Furthermore, a gene ontology (GO) analysis was performed and revealed that RP11-462C24.1 may participate in the metabolic process, cellular response to chemical stimulus, positive regulation of cellular process, proliferation, and biological process SW480 cells. Finally, the mRNAs deferentially expressed in cells treated with or without RP11-462C24.1 siRNA were further subjected to pathway analyses. Twenty pathways were significantly enriched (p < 0.05), including p53 signaling pathway, cell cycle pathways and MAPK pathways, all of which have been proved to function as important signaling pathways in the development of CRC.
The 70 kilodalton heat shock proteins (HSP70) is a member of the heat shock proteins. Results of previous studies indicated that HSP70 was significantly up-regulated in different types of cancers, including oral squamous cell carcinoma[25], esophageal cancer [26], gastric cancer [27], lung cancer [28] and CRC [29]. Our sequencing results indicated that RP11-462C24.1 siRNA significantly increased the expression of two paralogs of HSP70, Hspa1a, and Hspa1b; however, the relationship between RP11-462C24.1 and HSP70 in CRC has not been discussed. We report for the first time in the present work that RP11-462C24.1 over-expression significantly decreased the expression of HSP70 in CRC cells, and knockdown of RP11-462C24.1 have shown the opposite effects, suggesting that RP11-462C24.1 can exert anti-tumor function, at least partially, through affecting the expression of HSP70 family. Moreover, PI3K/Akt signaling is known as one of the most important signaling pathways that involved in the development of different types of cancers [30,31,32], and the relationship between PI3K/AKT and HSP70 signaling has been discussed previously [33, 34]; however, whether RP11-462C24.1 can regulate the expression of HSP70 through PI3K/AKT signaling pathway is still unclear. We found that transient over-expression of RP11-462C24.1 significantly suppressed the activation of the PI3K/AKT signaling, while the RP11-462C24.1 siRNA led to the increased expression of phosphorylated of PI3K and AKT. Interestingly, we also found that the transfection of HSP70 shRNA can partially abrogate RP11-462C24.1 over-expression plasmid induced anti-tumor effects on colorectal cancer cells. Taken together, our results suggested that RP11-462C24.1 may regulate the growth and metathesis of CRC cells through affecting HSP70 expression via regulating PI3K/AKT signaling pathway.
However, there were still some limitations in our study. First, the CRC samples selected in our study are too few, larger samples need to be recruited for clinical data comparison in the future. Then, whether RP11-462C24.1 regulates cell migration should be further verified by wound healing or Boyden chamber assays. Last, the effect of HSP70 depletion by itself (without overexpressing RP11-462C24.1) should be thoroughly illustrated in the future.
To sum up, we reported that LncRNA RP11-462C24.1 may function as a tumor suppressor in CRC through regulating the expression of HSP70 via PI3K/AKT signaling pathway. Our study has provided novel evidence for the potential use of LncRNA RP11-462C24.1 as a diagnostic biomarker or the therapeutic target for the early diagnosis and management of the disease.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Rusiecki J, Cifu AS. Colonoscopy surveillance after colorectal cancer resection. JAMA. 2017;318:2346–7.
Sun X, Liu S, Chen P, et al. miR-449a inhibits colorectal cancer progression by targeting SATB2. Oncotarget. 2017;8:100975–88.
Zhang S, Jin J, Tian X, Wu L. hsa-miR-29c-3p regulates biological function of colorectal cancer by targeting SPARC. Oncotarget. 2017;8:104508–244.
Costa V, Lo Dico A, Rizzo A, et al. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget. 2017;8:24292–302.
Pan Q, Meng L, Ye J, et al. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT). Cancer Lett. 2017;392:26–38.
Pang L, Wang DW, Zhang N, Xu DH, Meng XW. Elevated serum levels of MMP-11 correlate with poor prognosis in colon cancer patients. Cancer Biomark. 2016;16:599–607.
Wu J, Long Z, Cai H, et al. High expression of WISP1 in colon cancer is associated with apoptosis, invasion and poor prognosis. Oncotarget. 2016;7:49834–47.
Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233:6750–7.
Ouyang S, Zheng X, Zhou X, Chen Z, Yang X, Xie M. LncRNA BCAR4 promotes colon cancer progression via activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:92815–26.
Tong W, Yang L, Yu Q, Yao J, He A. A new tumor suppressor lncRNA RP11-190D6.2 inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells. Onco Targets Ther. 2017;10:1227–355.
Liu L, Yue H, Liu Q, et al. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7:15787–800.
Zhang S, Zhong G, He W, Yu H, Huang J, Lin T. lncRNA up-regulated in nonmuscle invasive bladder cancer facilitates tumor growth and acts as a negative prognostic factor of recurrence. J Urol. 2016;196:1270–8.
Botti G, Marra L, Malzone MG, et al. LncRNA HOTAIR as prognostic circulating marker and potential therapeutic target in patients with tumor diseases. Curr Drug Targets. 2017;18:27–34.
Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17:1750–7.
Shi D, Zheng H, Zhuo C, et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31.
Shi J, Zhang W, Tian H, Zhang Q, Men T. lncRNA ROR promotes the proliferation of renal cancer and is negatively associated with favorable prognosis. Mol Med Rep. 2017;16:9561–6.
Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8:84086–101.
Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892.
Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8:5286–97.
Zheng Y, Song D, Xiao K, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727–34.
Wang C, Yu J, Han Y, et al. Long non-coding RNAs LOC285194, RP11-462C241 and Nbla12061 in serum provide a new approach for distinguishing patients with colorectal cancer from healthy controls. Oncotarget. 2016;7:70769–78.
Cho HD, Lee JH, Moon KD, Park KH, Lee MK, Seo KI. Auriculasin-induced ROS causes prostate cancer cell death via induction of apoptosis. Food Chem Toxicol. 2018;111:660–9.
Park WH. MAPK inhibitors, particularly the JNK inhibitor, increase cell death effects in H2O2-treated lung cancer cells via increased superoxide anion and glutathione depletion. Oncol Rep. 2018;39:860–70.
Cui S, Su X, Dong L, et al. Programmed cell death ligand 1 protein levels predicted survival of non-small cell lung cancer. J Cancer. 2017;8:4075–82.
Nair S, Kotrashetti VS, Nayak R, Bhat K, Somannavar P, Hosmani J. HSP70 induces TLR4 signaling in oral squamous cell carcinoma: an immunohistochemical study. J Cancer Res Ther. 2013;9:624–9.
Wang XW, Shi XH, Tong YS, Cao XF. The prognostic impact of heat shock proteins expression in patients with esophageal cancer: a meta-analysis. Yonsei Med J. 2015;56:1497–502.
Saini J, Sharma PK. Clinical, prognostic and therapeutic significance of heat shock proteins in cancer. Curr Drug Targets. 2018;19:1478–90.
Ostheimer C, Gunther S, Bache M, Vordermark D, Multhoff G. Dynamics of heat shock protein 70 serum levels as a predictor of clinical response in non-small-cell lung cancer and correlation with the hypoxia-related marker osteopontin. Front Immunol. 2017;8:1305.
Gunaldi M, Kocoglu H, Okuturlar Y, et al. Heat shock protein 70 is a useful marker for predicting colorectal cancer. J BUON. 2015;20:1464–70.
Slattery ML, Mullany LE, Sakoda LC, et al. The PI3K/AKT signaling pathway: associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol Carcinog. 2018;57:243–61.
Chen C, Cai Q, He W, et al. AP4 modulated by the PI3K/AKT pathway promotes prostate cancer proliferation and metastasis of prostate cancer via upregulating L-plastin. Cell Death Dis. 2017;8:e3060.
Chen R, Li Y, Buttyan R, Dong X. Implications of PI3K/AKT inhibition on REST protein stability and neuroendocrine phenotype acquisition in prostate cancer cells. Oncotarget. 2017;8:84863–76.
Wang Y, Jia C, Li QS, Xie CY, Zhang N, Qu Y. BAG-1L protects SH-SY5Y neuroblastoma cells against hypoxia/re-oxygenation through up-regulating HSP70 and activating PI3K/AKT signaling pathway. Neurochem Res. 2017;42:2861–8.
Kong Q, Dai L, Wang Y, et al. HSPA12B attenuated acute myocardial ischemia/reperfusion injury via maintaining endothelial integrity in a PI3K/Akt/mTOR-dependent mechanism. Sci Rep. 2016;6:33636.
Author information
Authors and Affiliations
Contributions
Haiqing Zhang performed most of the experiments and wrote the manuscript, Guangjun Zhang and Haijun Liu performed some of the experiments, Yuanzhou Shan performed some of the statistical analysis, Xueli Zhang designed the study and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Informed consent
We have received the signed informed consent from all the participators of this study, and also obtained the ethical documents issued by the ethical committee of Affiliated Fengxian Hospital, The Third School of Clinical Medicine, Southern Medical University.
Research involving animal participants
The animal study has been performed with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of Affiliated Fengxian Hospital, The Third School of Clinical Medicine, Southern Medical University (Grant No. 2017051287.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, H., Zhang, G., Liu, H. et al. RP11-462C24.1 suppresses proliferation and invasion of colorectal carcinoma cells by regulating HSP70 through PI3K/AKT signaling pathway. Human Cell 34, 132–151 (2021). https://doi.org/10.1007/s13577-020-00426-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00426-7