Abstract
Global deregulation in miRNA expression is a hallmark of cancer cell. An estimated 2300 mature miRNAs are encoded by human genome; role of many of which in carcinogenesis and as cancer biomarkers remains unexplored. In this study, we investigated the utility of miR-3692-3p, miR-3195, and miR-1249-3p as biomarkers in non-small cell lung cancer (NSCLC). For this prospective study, 115 subjects, including 75 NSCLC patients and 40 controls, were recruited. The expression of miR-3692-3p, miR-3195, and miR-1249-3p was checked using qRT-PCR. The miRNA expression was correlated with survival outcome and therapeutic response. There were no significant differences in the mean age of NSCLC patients and controls (56.2 and 55.3 years, respectively; p = 0.3242). Majority of NSCLC patients (67%) were smokers. We observed a significant upregulation of miR-3692-3p expression (p < 0.0001), while the expression of miR-3195 (p = 0.0017) and miR-1249-3p was significantly downregulated (p < 0.0001) in the serum of NSCLC patients as compared to controls. The expression of miR-1249-3p was significantly upregulated in lung adenocarcinoma versus lung squamous cell carcinoma (p = 0.0178). Interestingly, patients who responded to chemotherapy had higher expression of miR-1249-3p than non-responders (p = 0.0107). Moreover, patients with higher expression of miR-3195 had significantly longer overall survival (p = 0.0298). In multivariate analysis, miR-3195 emerged as independent prognostic factor for overall survival. We conclude that the miR-3195 may have prognostic significance, while miR-1249-3p may predict therapeutic response in NSCLC. Further studies are warranted to elucidate the role of these miRNAs in lung carcinogenesis and their utility as candidate cancer biomarkers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, lung cancer is the most commonly diagnosed cancer accounting for 11.6% of all cancers in both the genders combined [1]. It is also the leading cause of cancer-related deaths accounting for 18.4% of all cancer-related deaths globally [1]. Non-small cell lung cancer (NSCLC), which constitutes 80–85% of all lung cancer, is classified into two histologically and molecularly distinct subtypes: lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). The therapeutic management of NSCLC, particularly LUAD, depends on the presence of specific driver mutations, including EGFR, ALK, ROS1, BRAF, and RET in tumor tissue, which are usually characterized by Sanger sequencing, next-generation sequencing, or PCR-based assays [2,3,4]. In the absence of targetable driver mutations, the role of immunotherapy becomes crucial specifically in a subset of NSCLC patients with high tumoral expression of programmed death-ligand 1 or high tumor mutation burden [5]. However, overall survival (OS) of advanced-stage NSCLC is still very poor despite the availability of several targeted agents and immunotherapeutic drugs. This is primarily due to the development of drug resistance and lack of more effective prognostic or predictive biomarkers which can be detected using minimal or non-invasive methods.
MicroRNAs (miRNAs), which are 18–25 nucleotide long, small, non-coding RNAs, are a class of epigenetic biomarkers involved in post-transcriptional gene silencing [6]. They are stable in various body fluids and can be detected using various molecular platforms, including quantitative PCR-based assays, microarray, and small RNA sequencing, making them an attractive target as cancer biomarkers. Numerous studies have observed deregulated miRNA expression in tumor tissue and body fluids of various human malignancies, including NSCLC [7,8,9,10,11]. There are approximately 2300 mature miRNAs known to be expressed in human system [12]; however, the role of many miRNAs as cancer biomarkers and in disease pathogenesis still remains unexplored. In our previous study, we identified several differentially expressed miRNAs in the serum of NSCLC patients compared to controls using small RNA sequencing [13]. However, in that study, we validated the expression of only ten differentially expressed miRNAs using reverse transcription quantitative polymerase chain reaction (qRT-PCR) [13]. The analysis of small RNA sequencing data revealed significant downregulation of miR-3195 {mean reads per kilobase of transcript per million mapped reads (RPKM) 42.404 in NSCLC vs. 230.251 in controls; p < 0.05} and miR-1249-3p {mean RPKM 16.675 in NSCLC vs. 69.548 in controls; p < 0.05} and a significant upregulation of miR-3692-3p {mean RPKM 7.089 in NSCLC vs. 0.692 in controls; p < 0.05} expression in the serum of NSCLC patients compared to controls [13]. Hence, in this study, we investigated the utility of miR-3692-3p, miR-3195 and miR-1249-3p as cancer biomarkers by profiling their expression in the serum of NSCLC patients and controls. Limited studies available so far indicate that aforementioned miRNAs may have some role in various human malignancies. However, their role as circulating cancer biomarkers has not been evaluated yet. To the best of our knowledge, this is the first report on profiling of these miRNAs in liquid-biopsy sample in NSCLC and their utility as diagnostic, prognostic, and predictive biomarkers.
Materials and methods
Patients
For this prospective study, we enrolled 115 subjects, including 75 newly diagnosed and treatment-naive NSCLC patients and 40 controls from the Dept. of Pulmonary Critical Care and Sleep Medicine and Dept. of Medical Oncology, All India Institute of Medical Sciences, New Delhi, India, between years 2014–2018. The diagnosis of NSCLC was confirmed by histopathology examination of biopsy or cytology specimens. For staging, CT scans of the chest and upper abdomen and if needed, CT or MRI scan of the brain and radionuclide bone scan were performed. Staging was done as per the recommendations of International Association for the Study of Lung Cancer Staging Committee for NSCLC [14]. Demographic and epidemiological details of all patients were also collected. The response to therapy was assessed in those patients who underwent chemotherapy for at least three cycles using RECIST v1.1 criteria [15]. The controls included patients with non-malignant pulmonary disorders, including chronic obstructive pulmonary disease, pulmonary tuberculosis, sarcoidosis, and bronchiectasis. The study was approved by institutional ethics committee (IEC-155/07.04.2017, RP-13/2017) and written informed consent was obtained from all the subjects, including NSCLC patients and controls.
Collection and processing of samples for the isolation of serum RNA
We collected 5 mL of peripheral blood in sterile vacutainers (BD), which was kept at room temperature (RT) for 15–30 min for clotting. The clotted blood was centrifuged at 1500×g for 15 min at RT for serum separation. The serum was again centrifuged at 10,000×g for 15 min to remove any remaining cellular debris. Serum was aliquoted and transferred in a fresh sterile nuclease-free microcentrifuge tube and was stored at − 80 °C until further use.
Quantification of miRNA expression using qRT-PCR
Quantification of the expression of all miRNAs was performed using miScript SYBRGreen PCR system (QIAGEN, Germany) as per the manufacturer’s instructions. Briefly, an equal amount of synthetic spike-in control, C. elegans miR-39 miRNA mimic, was added to each serum sample before RNA isolation for normalization. RNA was isolated from 200 μL of serum using miRNeasy Serum/Plasma kit (QIAGEN, Germany) as per the manufacturer’s instructions. RNA was eluted in 14 μL of RNase-free water and stored at − 80 °C until further use. 5 μL of RNA was used for reverse transcription (RT) using miScript II RT kit (QIAGEN, Germany) as per recommended protocol. After RT, cDNA was diluted in 200 μL of RNase-free water, aliquoted, and stored at − 80 °C. The quantification of expression of miR-3692-3p, miR-3195, and miR-1249-3p was performed using miScript SYBRGreen PCR kit (QIAGEN, Germany) as per the manufacturer’s instructions. C. elegans miR-39 was used as an exogenous control (Ce_miR-39_1 miScript® Primer Assay, QIAGEN, Germany) for normalizing the expression of miRNA expression. The relative quantification of miRNA expression was determined using ∆Ct method, where ∆Ct = Ct (miRNA of interest) − Ct (reference miR-39).
Statistical analysis
The descriptive data were expressed as mean ± SD, mean ± SE or median (range). Median values of quantitative variables were used as cut-off for high and low values. Student’s t test was used to compare the mean of two groups while the linear relationship between two continuous variables was established using Pearson correlation coefficient. Kaplan–Meier curve was applied for the survival analysis followed by log rang test to compare the statistical significance between the groups. We performed Cox proportional hazard analysis for those miRNAs which correlated with OS in univariate analysis. OS was calculated from the date of diagnosis to death or last follow-up, while progression-free survival (PFS) was calculated date of diagnosis to date of progression, relapse, or death. The end point of the study was 10th October 2019. Receiver operating characteristic (ROC) curves were plotted to calculate the area under the curve (AUC), sensitivity, and specificity of each miRNA. All statistical analysis was done using STATA 11.2. p < 0.05 was considered as statistically significant.
Results
Demographic characteristics
The mean age of 75 NSCLC patients and 40 controls was almost similar (56.2 and 55.3 years, respectively; p = 0.3242). Majority of the patients (82.7%) and controls (67.5%) were males (p = 0.054). Histologically, 42 patients were of LUAD (mean age 53.6 years; 31 males and 11 females) while 33 patients were of LUSC (mean age 59.6 years; 31 males and 2 females). Majority of NSCLC patients were smokers (66.7%) with more prevalence in patients with LUSC (87.9%) than with LUAD (50%). The demographic characteristics of lung cancer patients as well as controls are summarized in Table 1.
Differential expression of miR-1249-3p, miR-3195, and miR-3692-3p in NSCLC
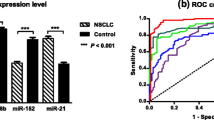
We compared the expression of miR-1249-3p, miR-3195, and miR-3692-3p in the serum of 75 NSCLC patients with 40 controls. The expression of miR-1249-3p (p < 0.0001; Fig. 1a) and miR-3195 (p = 0.0017; Fig. 1b) was significantly downregulated, while that of miR-3692-3p was significantly upregulated (p < 0.0001; Fig. 1c) in NSCLC as compared to controls (Table 2).
Circulating serum miRNAs as diagnostic biomarker for NSCLC. The box plot shows the differential expression of a miR-1249-3p, b miR-3195, and c miR-3692-3p in the serum of NSCLC patients (n = 75) as compared to controls (n = 40) as measure by qRT-PCR. ROC curves were generated to assess the diagnostic potential of d miR-1249-3p, e miR-3195, and f miR-3692-3p for NSCLC
We also assessed the diagnostic performance of all 3 miRNAs using ROC curves. Amongst 3 miRNAs, only miR-1249-3p exhibited reasonable sensitivity of 73.3% at 95% specificity and an area under the curve (AUC) of 0.879 (Table 2, Fig. 1d). The miR-3195 and miR-3692-3p had very low AUC and sensitivity at 95% specificity indicating that they may not be good diagnostic biomarkers for NSCLC (Table 2, Fig. 1e, f).
Correlation of miRNA expression with survival and therapeutic response
For our study population, median OS and PFS was 6.4 (range 0.2–30.7) and 5.7 (range 0.2–30.7) months, respectively. We correlated the expression of miRNAs with OS and PFS using median ∆Ct of individual miRNAs as cut-off (Table 3). The expression of miR-1249-3p and miR-3692-3p was not correlated with OS and PFS in univariate analysis (Table 3). However, NSCLC patients with higher expression of miR-3195 had significantly longer OS as compared to patients with lower expression of miR-3195 (p = 0.0298; Fig. 2a). On multivariate analysis, the expression of miR-3195 emerged as an independent prognostic factor for OS (HR 2.54; p = 0.024; Table 4).
Circulating serum miRNAs as prognostic and predictive biomarker for NSCLC. a Kaplan–Meier curve for correlation between miR-3195 expression and OS in NSCLC patients. NSCLC patients were categorized into two groups based on median ∆Ct of miR-3195. Those with < median ∆Ct levels of miR-3195 were classified as patients with higher expression while those with ≥ median ∆Ct levels of miR-3195 were classified as patients with lower expression of miR-3195. The curve indicates that patients with higher expression of miR-3195 have significantly longer OS as compared to patients with lower expression. b The box plot shows significantly higher expression of miR-1249-3p in responders as compared to non-responders to chemotherapy
We also correlated the expression of miRNAs with response to therapy. NSCLC patients who achieved complete response (CR), partial response (PR), or stable disease (SD) after 3 cycles of chemotherapy were classified as responders (n = 23), while those with progressive disease were classified as non-responders (n = 10). Interestingly, responders had higher expression of miR-1249-3p than non-responders (mean ∆Ct of 8.71 vs. 10.51, respectively; p = 0.0107; Fig. 2b). None of the other miRNAs correlated with therapeutic response (Table 5).
We also correlated various clinicopathological parameters with OS and PFS. The median PFS was significantly higher for responders than non-responders (p = 0.0027; Table 6). However, other parameters, including age, gender, smoking status, histology, stage, and ECOG PS were not correlated with OS or PFS (Table 6).
Correlation of miRNA expression with clinico-pathological parameters
We looked into correlation between miRNA expression and aforementioned clinico-pathological parameters. We observed a significant correlation between the expression of miR-1249-3p and age, gender, smoking status, T factor, ECOG PS and histology (Table 7). The expression of miR-1249-3p was significantly upregulated in lung adenocarcinoma as compared to lung squamous cell carcinoma (p = 0.0178, Table 7). The expression of miR-3195 was significantly correlated with age, gender, T factor, and ECOG PS while that of miR-3692-3p was only correlated with gender (Table 7).
We also tried to establish a correlation between the expressions of various miRNAs in NSCLC patients. The expression of miR-3195 significantly correlated with the expression of miR-1249-3p (r = 0.4966; p < 0.0001), while a weak but significant correlation was observed between the expression of miR-3692-3p and miR-3195 (r = 0.2108; p = 0.0237). We could not find any significant correlation between the expression of miR-3692-3p and miR-1249-3p (r = − 0.0825; p = 0.3806).
Discussion
An estimated 2300 mature miRNAs are encoded by the human genome [12], the function and clinical utility of many of which remain unknown. In the present study, we profiled the expression of miR-3692-3p, miR-3195, and miR-1249-3p in the serum of NSCLC patients and controls since there is very limited data available for their clinical utility as cancer biomarkers. We found an upregulation of miR-3692-3p and a downregulation of miR-3195 and miR-1249-3p expression in the serum on NSCLC patients suggesting that differential expression of these miRNAs might contribute to the development of NSCLC. However, we did not check the expression of these miRNAs in paired tumor tissue, which limits our understanding of the tumor origin of these miRNAs or their direct involvement in carcinogenesis. Since few studies have reported their expression in tumor tissues of various malignancies, they might be released in the circulation due to tumor cell lysis or death making their detection feasible using minimally or less-invasive methods.
The expression of miR-3692 has been found to be upregulated in osteosarcoma cells [16], in esophageal squamous cell carcinoma tissue [17] and in breast cancer (BC) cells subjected to compression [18]. In contrast, few studies have reported downregulation of miR-3692-5p expression in the plasma of BC patients [19], in the extracellular vesicles derived from urine samples of pancreatic cancer patients [20] and in hepatocellular carcinoma (HCC) tissue [21]. In another study, microarray analysis revealed a weak expression of miR-3692 in prostate cancer (PC) tissue sample [22]. The expression of miR-3692 was found to be upregulated in biochemical recurrence (BCR) positive versus BCR-negative and pT3 stage tumors versus pT2 stage tumors of PC patients [22]. Inducible T-cell co-stimulator (ICOS) gene is considered to be a target of miR-3692-3p since 3′ untranslated region (3′ UTR) of ICOC mRNA contains a potential binding site for miR-3692-3p [23]. Considering the documented role of ICOS in immune modulation, it is emerging as an attractive immunotherapeutic target [24, 25]. Hence, understanding the role of miR-3692-3p in the regulation of ICOS expression is highly desirable.
A downregulation of miR-3195 expression has been reported in lung tumor tissue as compared to paired normal lung tissue using miRNA microarray [26], which is in line with our observation in serum samples. However, further validation using qRT-PCR did not reveal any significant differences in its expression between paired tumor and normal lung tissue [26]. The expression of miR-3195 was also found to be significantly downregulated in colorectal cancer (CRC) tissue [27] and in oral squamous cell carcinoma tissue [28]. A moderate expression of miR-3195 is also reported in PC tissue sample [22]. The expression of miR-3195 was also found to be downregulated in poorly differentiated as compared with well differentiated HCC while it was upregulated in hepatitis virus B (HBV)-positive as compared to HBV-negative HCC cell lines [29]. In contrast, the expression of miR-3195 was upregulated in tumor mass as compared to border region (an area between tumor mass and brain where tumor and normal cells co-exist) of glioblastoma patients [30].
A number of biological factors are known to induce the expression of miR-3195 in various human malignancies. Melatonin in PC [31] and gastric cancer (GC) [32] while p53 in osteosarcoma [33] were found to induce miR-3195 expression. Both p53 and melatonin are known to have tumor-suppressive activities. Overexpression of miR-3195 along with miR-374b-suppressed angiogenesis by inhibiting the expression of angiogenesis-related genes, including HIF-1α, HIF-2α and VEGF, and inhibited motility of PC cells under hypoxic condition [31]. HIF-2α was identified as one of the important targets of miR-3195 [31]. The expression of miR-3195 was found to be downregulated in GC cells when cultured as non-adherent spheroid body as compared to parental cell [34] further supporting the role of this miRNA in maintaining stem cell properties and as a tumor suppressor. Its expression was, however, upregulated in the side population (SP) of PC cells as compared to non-SP [35]. SP are known to have features of cancer stem cells, including self-renewal, multipotent differentiation, a slow cell cycle, and chemo-/radio-resistance. Supporting this, a significant overexpression of miR-3195 was found in the exosomes and cells of cisplatin-resistant A549 lung adenocarcinoma cells [36]. Few other studies have reported higher expression of miR-3195 in melanoma cells when grown as monolayer in serum containing media versus cells grown as spheroids in stem cell media [37], in serum-starved osteosarcoma cells in comparison to asynchronously growing cells [38] and also in neuroendocrine-differentiated PC cells [39].
Like many other miRNAs, miR-1249 is also reported to have dual role as tumor suppressor and oncogenic miRNA in various human malignancies. In our study, we found a significant downregulation of miR-1249-3p expression in the serum of NSCLC patients, which is in line with other studies in BC tissue [40] and CRC tissue and cell lines [41]. The miR-1249 acts like a tumor suppressor miRNA in BC and CRC since its overexpression resulted in reduced cell proliferation and migration and reversed EMT under in vitro condition [40, 41]. In BC, Homeobox 8 (HOXB8), which is an oncogene, was found to be its direct target [40], while in CRC, vascular endothelial growth factor A (VEGFA) and high-mobility group AT-hook 2 (HMGA2) were directly targeted by miR-1249 [41]. In CRC tissue, the expression of miR-1249 was negatively correlated with pN stage, pM stage, TNM stage, OS, and VEGFA or HMGA2 expression [41], while in BC, it was negatively correlated with the expression of migration inhibitory factor antisense RNA 1, a long non-coding RNA, which acts as a sponge of miR-1249-3p [40]. Interestingly, p53 was found to be the regulator of miR-1249 in CRC and p53-induced miR-1249 expression inhibited tumor growth, EMT, and angiogenesis in vitro and vivo [41]. The expression of miR-1249 was also downregulated in tumor tissue of HCC patients as compared to normal liver tissue [42].
Some studies have also reported oncogenic functions of miR-1249. In two recent studies, the expression of miR-1249 was markedly upregulated in glioma [43] and HCC [44] tissues and cell lines. Silencing of miR-1249 resulted in reduced cell proliferation, colony formation, and EMT in glioma and HCC [43, 44]. The miR-1249 was found to directly target adenomatous polyposis coli 2 (APC2), a tumor suppressor gene of Wnt/β-catenin signaling, in glioma [43] and heterogeneous nuclear ribonucleoprotein K in HCC [44]. High miR-1249-3p expression was found to correlate with poor OS of HCC patients [44], while an inverse correlation was observed between miR-1249 and APC2 expression in glioma [43]. As an oncomiR, the activation of miR-1249-5p has also been reported in non-alcoholic fatty liver disease-related HCC, where it was found to regulate the expression of 112 genes and act as a master regulator [45]. Indeed, in one study, the expression of miR-1249 was highly upregulated and associated with poor prognosis in HCC [46]. Further, overexpression of miR-1249 resulted in constitutive activation of hedgehog signaling [46]. Glioma-associated oncogene 1 (Gli1), which is a transcription factor of hedgehog signaling, was found to regulate the expression of miR-1249 in HCC by binding to its promoter [46]. Gli1-induced miR-1249 inhibited the expression of tumor suppressor patched-1 (PTCH-1) by directly interacting with 3′ UTR of PTCH-1 mRNA [46]. Overexpression of miR-1249 resulted in silencing of PTCH-1 expression and induction of Gli1 activity. In addition, overexpression of miR-1249 expression resulted in increased cell proliferation, colony formation, enhanced nuclear localization of Gli1 protein, and induced invasion and migration [46]. In HCC tumor tissue samples, a positive correlation was observed between Gli1 and miR-1249 expression [46]. Hence, Gli1–miR-1249–PTCH-1 feedback loop seems to play an important role in HCC progression.
The expression of miR-1249 is also reported in K562 leukemia cells and microvesicles secreted by K562 cells [47]. The expression of miR-1249-5p was also found to be upregulated in p53-deficient cancer cell-derived exosomes of CRC patients [48]. Further, overexpression of miR-1249-5p significantly downregulated p53 mRNA expression in fibroblasts since p53 is directly targeted by miR-1249-5p [48]. Overexpression of miR-1249-5p also resulted in enhanced proliferation of fibroblasts due to lack of p53 [48]. On the contrary, inhibition of miR-1249-5p expression in fibroblasts resulted in the upregulation of p53 and p21 expression [48]. This study supports the crucial role of exosomal miRNAs in modulating tumor microenvironment by engaging in cell-to-cell communication. Since there is an existing loss of tumor suppressive signals in cancer cells due to numerous genetic aberrations, this study emphasizes the role of exosomal miRNAs in silencing tumor suppressor machinery of cells present in tumor microenvironment, which might further enhance the oncogenic signals leading to more aggressive tumor phenotype. The higher expression of miR-1249 has also been found to correlate with postoperative tumor relapse in small cell carcinoma of the esophagus [49]. However, in one study, there was no significant difference in the expression of miR-1249 between PC tissue and benign prostate hyperplasia [50].
Like most of other studies, our study has its own strengths and limitations. This is the first study to investigate the role of miR-3692-3p, miR-3195, and miR-1249-3p as circulating biomarkers in NSCLC. We checked their efficacy as liquid-biopsy-based diagnostic, prognostic, and predictive biomarkers in NSCLC, for which the literature is not available. However, small sample size limited to only locally advanced or metastatic NSCLC patients and shorter follow-up time limits the clinical impact of our study. In addition, correlation of expression of these miRNAs in lung tumor tissue and serum would have been ideal to find out whether these miRNAs are secreted from lung tumor cells and are not of blood cell origin. Detection of these miRNAs in tumor tissue of other malignancies, however, makes these claims futile. Finally, the mechanistic insights for exploring the role of these miRNAs in lung carcinogenesis were also not attempted in the present study.
Conclusions
In summary, the expression of miR-3195 and miR-1249-3p was downregulated, while miR-3692-3p was upregulated in the serum of NSCLC patients as compared to controls. The expression of miR-1249-3p correlated with response to therapy while miR-3195 emerged as independent prognostic factor for OS. The role of all these miRNAs in lung carcinogenesis is still unclear. Hence, more comprehensive mechanistic studies are needed to decipher their role in lung cancer and as candidate biomarkers for lung cancer.
Abbreviations
- NSCLC:
-
Non-small cell lung cancer
- LUSC:
-
Lung squamous cell carcinoma
- LUAD:
-
Lung adenocarcinoma
- miRNA:
-
MicroRNA
- RT:
-
Room temperature
- qRT-PCR:
-
Reverse transcription quantitative polymerase chain reaction
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- ECOG PS:
-
Eastern Cooperative Oncology Group performance status
- BC:
-
Breast cancer
- HCC:
-
Hepatocellular cancer
- PC:
-
Prostate cancer
- GC:
-
Gastric cancer
- BCR:
-
Biochemical recurrence
- ICOS:
-
Inducible T-cell co-stimulator
- UTR:
-
Untranslated region
- CRC:
-
Colorectal cancer
- HBV:
-
Hepatitis virus B
- SP:
-
Side population
- HOXB8:
-
Homeobox 8
- VEGFA:
-
Vascular endothelial growth factor A
- HMGA2:
-
High-mobility group AT-hook 2
- APC2:
-
Adenomatous polyposis coli 2
- Gli1:
-
Glioma-associated oncogene 1
- PTCH-1:
-
Patched-1
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. https://doi.org/10.3322/caac.20102.
Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. https://doi.org/10.1016/S1470-2045(10)70087-5.
Korpanty GJ, Graham DM, Vincent MD, et al. Biomarkers that currently affect clinical practices in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol. 2014;4:204. https://doi.org/10.3389/fonc.2014.00204.
Brahmer JR, Govindan R, Anders RA, et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J Immunother Cancer. 2018;6(1):75. https://doi.org/10.1186/s40425-018-0382-2.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. https://doi.org/10.1016/s0092-8674(04)00045-5.
Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. https://doi.org/10.1038/nrc3166.
Singh DK, Bose S, Kumar S. Role of microRNA in regulating cell signaling pathways, cell cycle, and apoptosis in non-small cell lung cancer. Curr Mol Med. 2016;16:474–86. https://doi.org/10.2174/1566524016666160429120702.
Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. https://doi.org/10.1002/emmm.201100209.
Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–46. https://doi.org/10.1158/2159-8290.CD-15-0893.
He Y, Lin J, Kong D, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61:1138–55. https://doi.org/10.1373/clinchem.2015.241190.
Alles J, Fehlmann T, Fischer U, et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47(7):3353–64. https://doi.org/10.1093/nar/gkz097.
Kumar S, Sharawat SK, Ali A, et al. Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients. Curr Probl Cancer. 2020. https://doi.org/10.1016/j.currproblcancer.2020.100540.
Detterbeck FC, Boffa DJ, Kim AW, et al. The eighth edition lung cancer stage classification. Chest. 2017;151:193–203. https://doi.org/10.1016/j.chest.2016.10.010.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. https://doi.org/10.1016/j.ejca.2008.10.026.
Gao P, Teng Z, Ji L, et al. Interactions of ABLIMI and CXCL5 with miRNAs as a prognostic indicator for clinical outcome of osteosarcoma. Int J Clin Exp Med. 2016;9(8):15345–53.
He Z, Yi J, Liu X, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15(1):51. https://doi.org/10.1186/s12943-016-0533-3.
Kim BG, Kang S, Han HH, et al. Transcriptome-wide analysis of compression-induced microRNA expression alteration in breast cancer for mining therapeutic targets. Oncotarget. 2016;7(19):27468–78. https://doi.org/10.18632/oncotarget.8322.
Stückrath I, Rack B, Janni W, et al. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6(15):13387–40101. https://doi.org/10.18632/oncotarget.3874.
Yasui T, Yanagida T, Ito S, et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv. 2017;3(12):e1701133. https://doi.org/10.1126/sciadv.1701133.
Liu S, Pan H, Cao J, et al. MicroRNA-134 inhibits HCC cell growth and migration through the AKT/GSK3β/SNAIL signaling pathway. Int J Clin Exp Pathol. 2016;9(7):6877–86.
Stuopelytė K, Daniūnaitė K, Jankevičius F, et al. Detection of miRNAs in urine of prostate cancer patients. Medicina (Kaunas). 2016;52(2):116–24. https://doi.org/10.1016/j.medici.2016.02.007.
Wu D, Tang R, Qi Q, et al. Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cell Immunol. 2015;293(1):41–8. https://doi.org/10.1016/j.cellimm.2014.11.006.
Marinelli O, Nabissi M, Morelli MB, et al. ICOS-L as a potential therapeutic target for cancer immunotherapy. Curr Protein Pept Sci. 2018;19(11):1107–13. https://doi.org/10.2174/1389203719666180608093913.
Soldevilla MM, Villanueva H, Meraviglia-Crivelli D, et al. ICOS costimulation at the tumor site in combination with CTLA-4 blockade therapy elicits strong tumor immunity. Mol Ther. 2019;27(11):1878–91. https://doi.org/10.1016/j.ymthe.2019.07.013.
Pan HL, Wen ZS, Huang YC, et al. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. https://doi.org/10.1038/srep14331.
Huang L, Cai JL, Huang PZ, et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996–2008.
Schneider A, Victoria B, Lopez YN, et al. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci Rep. 2018;8(1):675. https://doi.org/10.1038/s41598-017-18945-z.
Morishita A, Iwama H, Fujihara S, et al. MicroRNA profiles in various hepatocellular carcinoma cell lines. Oncol Lett. 2016;12(3):1687–92. https://doi.org/10.3892/ol.2016.4853.
Hide T, Komohara Y, Miyasato Y, et al. Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine. 2018;30:94–104. https://doi.org/10.1016/j.ebiom.2018.02.024.
Sohn EJ, Won G, Lee J, et al. Upregulation of miRNA3195 and miRNA374b mediates the anti-angiogenic properties of melatonin in hypoxic PC-3 prostate cancer cells. J Cancer. 2015;6:19–28. https://doi.org/10.7150/jca.9591.
Zhu C, Huang Q, Zhu H. Melatonin inhibits the proliferation of gastric cancer cells through regulating the miR-16-5p-Smad3 pathway. DNA Cell Biol. 2018;37(3):244–52. https://doi.org/10.1089/dna.2017.4040.
Jiang J, Ma B, Li X, et al. MiR-1281, a p53-responsive microRNA, impairs the survival of human osteosarcoma cells upon ER stress via targeting USP39. Am J Cancer Res. 2018;8(9):1764–74.
Liu J, Ma L, Wang Z, et al. MicroRNA expression profile of gastric cancer stem cells in the MKN-45 cancer cell line. Acta Biochim Biophys Sin (Shanghai). 2014;46(2):92–9. https://doi.org/10.1093/abbs/gmt135.
Chen Y, Zhao J, Luo Y, et al. Downregulated expression of miRNA-149 promotes apoptosis in side population cells sorted from the TSU prostate cancer cell line. Oncol Rep. 2016;36(5):2587–600. https://doi.org/10.3892/or.2016.5047.
Qin X, Yu S, Xu X, et al. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. 2017;8(26):42125–35. https://doi.org/10.18632/oncotarget.15009.
Wozniak M, Sztiller-Sikorska M, Czyz M. Expression of miRNAs as important element of melanoma cell plasticity in response to microenvironmental stimuli. Anticancer Res. 2015;35(5):2747–58.
Ghosh T, Varshney A, Kumar P, et al. MicroRNA-874-mediated inhibition of the major G1/S phase cyclin, CCNE1, is lost in osteosarcomas. J Biol Chem. 2017;292(52):21264–81. https://doi.org/10.1074/jbc.M117.808287.
Dankert JT, Wiesehöfer M, Czyrnik ED, et al. The deregulation of miR-17/CCND1 axis during neuroendocrine transdifferentiation of LNCaP prostate cancer cells. PLoS ONE. 2018;13(7):e0200472. https://doi.org/10.1371/journal.pone.0200472.
Ding J, Wu W, Yang J, et al. Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol Res Pract. 2019;215(7):152376. https://doi.org/10.1016/j.prp.2019.03.005.
Chen X, Zeng K, Xu M, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131. https://doi.org/10.1038/s41419-018-1188-3.
Katayama Y, Maeda M, Miyaguchi K, et al. Identification of pathogenesis-related microRNAs in hepatocellular carcinoma by expression profiling. Oncol Lett. 2012;4(4):817–23. https://doi.org/10.3892/ol.2012.810.
Fang B, Li G, Xu C, et al. MicroRNA miR-1249 downregulates adenomatous polyposis coli 2 expression and promotes glioma cells proliferation. Am J Transl Res. 2018;10(5):1324–36.
Shu H, Hu J, Deng H. miR-1249-3p accelerates the malignancy phenotype of hepatocellular carcinoma by directly targeting HNRNPK. Mol Genet Genom Med. 2019;7(10):e00867. https://doi.org/10.1002/mgg3.867.
Seshachalam VP, Sekar K, Hui KM. Insights into the etiology-associated gene regulatory networks in hepatocellular carcinoma from The Cancer Genome Atlas. J Gastroenterol Hepatol. 2018;33(12):2037–47. https://doi.org/10.1111/jgh.14262.
Ye Y, Wei Y, Xu Y, et al. Induced MiR-1249 expression by aberrant activation of Hedegehog signaling pathway in hepatocellular carcinoma. Exp Cell Res. 2017;355(1):9–17. https://doi.org/10.1016/j.yexcr.2017.03.010.
Chen X, Xiong W, Li H. Comparison of microRNA expression profiles in K562-cells-derived microvesicles and parental cells, and analysis of their roles in leukemia. Oncol Lett. 2016;12(6):4937–48. https://doi.org/10.3892/ol.2016.5308.
Yoshii S, Hayashi Y, Iijima H, et al. Exosomal microRNAs derived from colon cancer cells promote tumor progression by suppressing fibroblast TP53 expression. Cancer Sci. 2019;110(8):2396–407. https://doi.org/10.1111/cas.14084.
Okumura T, Shimada Y, Omura T, et al. MicroRNA profiles to predict postoperative prognosis in patients with small cell carcinoma of the esophagus. Anticancer Res. 2015;35(2):719–27.
Scaravilli M, Porkka KP, Brofeldt A, et al. MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate. 2015;75(8):798–805. https://doi.org/10.1002/pros.22961.
Acknowledgements
The authors gratefully acknowledge the financial support from the Science and Engineering Research Board (SERB), Govt. of India, New Delhi (Grant no. SB/YS/LS-348/2013) and All India Institute of Medical Sciences, New Delhi (Grant no. A-516).
Funding
The authors gratefully acknowledge the financial support from the Science & Engineering Research Board (SERB), Govt. of India, New Delhi (Grant no. SB/YS/LS-348/2013) and All India Institute of Medical Sciences, New Delhi (Grant no. A-516).
Author information
Authors and Affiliations
Contributions
SK, VG, and MP performed all the laboratory work. SK and SKS analyzed and interpreted the experimental data. AS, PSM, SK, AM and RG collected, analyzed, and interpreted the clinical data. SK designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by ethics committee of All India Institute of Medical Sciences, New Delhi (Ref. No. IEC-155/07.04.2017, RP-13/2017). This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, S., Sharawat, S.K., Ali, A. et al. Differential expression of circulating serum miR-1249-3p, miR-3195, and miR-3692-3p in non-small cell lung cancer. Human Cell 33, 839–849 (2020). https://doi.org/10.1007/s13577-020-00351-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00351-9