Abstract
Drought stress has been known to adversely affect growth, development, and productivity of plants to varying extent. Being a multifaceted trait, drought tolerance involves interaction of an array of genes, pathways, and mechanisms. A unique regulatory scheme is adopted by different plants, which provides tolerance to drought stress in association with biochemical and physiological mechanisms. Transcriptome analysis of a drought tolerant [Nagina 22 (N-22)] and drought sensitive (IR-64) cultivars provides insights into the genes/pathways/mechanisms involved in terminal drought stress tolerance. In the present study, comparative physio-biochemical analyses of the rice cultivars under terminal drought stress substantiated their performance. Whole transcriptome analysis of leaf and root from the rice cultivars exposed to terminal drought stress revealed 6077 and 10,050 differentially expressed genes (DEGs) in leaf of N-22 and IR-64, respectively, under drought stress. A maximum of 2682 genes were up-regulated exclusively in N-22 while 7198 genes were down-regulated exclusively in leaf of IR-64. Interestingly, the highest number (2594) of genes was down-regulated exclusively in roots of IR-64, while only 1497 gene were up-regulated exclusively in root of N-22. Differential expression of OsNAC10, OsbZIP23, OsABA8ox1, OsCPK4, OsLEA3, and OsNCED4 along with the GO terms enriched with up-regulated genes for transcription factors (TFs), redox homeostasis, and ABA signaling in N-22 under terminal drought stress play crucial roles in stress tolerance. The stress-responsive genes for transcription factors, redox homeostasis, and ABA signaling up-regulated in N-22 were mainly responsible for terminal drought tolerance. These stress-associated genes can be utilized for genetic improvement of rice for drought tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.), one of the important cereal crops, serves as a staple food for more than half of the global population. Changes in climatic conditions, decreasing availability of fresh water, and erratic rainfall have significantly affected the productivity of crops. Drought stress is one of the critical factors that pose challenges to the cultivation of rice. Being a complex trait, drought tolerance requires complementary effects of biochemical, physiological, and molecular factors to cope with the abiotic stress. Hence, combining conventional breeding with genomics-assisted molecular breeding has been recommended to improve rice productivity in the changing global climate with frequent and severe drought stress events.
Abiotic stresses cause the increased generation of reactive oxygen species (ROS), which not only work as signaling molecules initially at lower concentrations but also cause oxidative damage to several biological molecules when accumulated in excess (Samota et al. 2017; Kumar et al. 2017). ROS-induced oxidation of biomolecules causes site-specific modifications, fragmentation, altered electric charge, and increased susceptibility to degradation. Such oxidative stress is mitigated through antioxidative/defensive enzymatic and non-enzymatic mechanisms involved in the free-radical scavenging process (Caverzan et al. 2016). Increased peroxidation/degradation of lipids has been reported to be common in plants under environmental stresses (Mishra et al. 2011). Malondialdehyde (MDA) level in plant tissue is considered a biochemical marker of lipid peroxidation/cellular membrane damage (Møller and Kristensen 2004) under abiotic stresses.
An array of metabolites like soluble sugar, proline, and phenolic compounds get accumulated in plant tissues upon exposure to stressful conditions. Proline, an amino acid (actually an imino acid), acts as an excellent osmolyte, serves as an antioxidative defense molecule, and a signaling molecule. Proline helps in maintaining membrane integrity, osmotic balance, and concentration of ROS within a normal range; thus, it minimizes oxidative burst (Sasi et al. 2021). Thus, proline enhances stress tolerance by protecting/stabilizing cellular membranes and enzymes (Kumar et al. 2017). Compatible solutes, such as soluble sugars and proline, play a significant role in osmotic adjustment as well as structural stability during abiotic stresses (Romero-Aranda et al. 2006). Increased concentrations of compatible solutes might alleviate the deleterious stress either via osmotic adjustment or by conferring desiccation tolerance. Synthesis/accumulation of polyphenolic compounds get stimulated in plant tissues in response to abiotic stresses. The accumulation of phytophenolics has been reported earlier, which help to maintain cellular homeostasis under abiotic stresses (Elhamid et al. 2014). A strong correlation between antioxidant activity and the accumulation of total phenolic content (TPC) was reported by Samota et al. (2017) in rice.
Drought stress at the reproductive stage has significant negative impact on panicle emergence and anther dehiscence, which might result in severe yield losses. Various defense mechanisms including enzymatic and non-enzymatic antioxidative defense machinery have been reported earlier to combat the harmful effects of drought stress (Kumar et al. 2017). Attempts are being made to analyze transcriptome data for different tissues grown under different conditions and collected at different developmental stages of rice to decipher the candidate genes/pathways associated with drought stress tolerance (Shankar et al. 2016). Previous studies on transcriptome profiling suggested that rice senses and responds rapidly to drought stress by modulating the expression of genes (Lenka et al. 2011; González-Schain et al. 2015; Sinha et al. 2018). Although much has been worked on drought stress responses in rice, lesser has been explored on cellular functions, signaling pathways, and molecular mechanisms for terminal drought stress (Sinha et al. 2018). The level of drought stress tolerance in several rice genotypes has been assessed based on the biochemical/physiological responses. Moreover, transcriptome profiling has been used to reveal the differentially expressed genes (DEGs) under stress (Shankar et al. 2016). Modulation in transcription factor activities, metabolic pathways, and carbon assimilation has been reported in drought tolerant rice genotypes (Lenka et al. 2011; Sinha et al. 2018; Prathap et al. 2019).
Efforts are still being made to decipher the genes/pathways involved in drought stress tolerance at different developmental stages in rice. Interestingly, redox homeostasis, antioxidant activity, ion transport, hormone signaling, etc. have been reported to be involved in drought stress tolerance in rice (Shankar et al. 2016; Mawlong et al. 2018). Dehydration-responsive element-binding (DREB) and late embryogenesis abundant (LEA) proteins were identified as the key modulators of abiotic stress tolerance (Cui et al. 2011; Liang et al. 2019). However, deciphering the genes/pathways/mechanisms involved in drought stress tolerance remains elusive, mainly because it is a multi-genic trait. Abscisic acid (ABA) has been reported to be involved in tolerance to several abiotic stresses (Zong et al. 2016; Liu et al. 2018; Chen et al. 2019), but its function/mechanism is still not deciphered. Induction of an intricate signaling network upon sensing stress and expression of the stress-responsive genes has been reported to be reprogramed by the synergistic action of transcription factors (TFs). Several TF families like NAC, bZIP, WRKY, AP2/ERF, homeodomain, MYB, bHLH, CAMTA, and NF-Y were demonstrated to be important players in abiotic stress tolerance through knockdown/knockout or overexpression studies (Lindemose et al. 2013; Castilhos et al. 2014; Ali et al. 2016).
Though some of the drought-responsive genes have been identified through genome-wide studies (Wang et al. 2011), the function of many of such genes is not known and needs to be investigated. Considerable genotypic variations do exist among the wild relatives and local rice cultivars; the desired level of improvement for drought stress tolerance in rice has not yet been achieved. RNA-seq analysis and the system biology approach are powerful tools to probe such complex traits. Several transcriptome studies on drought tolerance in rice published in the last decade have further improved our understanding at the systemic level (Lenka et al. 2011; Shankar et al. 2016; Zhang et al. 2016; Borah et al. 2017). Biochemical, physiological, genetic, genomic, and epigenomic approaches are being combined to analyze the complex nature of abiotic stress tolerance in crop plants (Kumar et al. 2017, 2022b). A large number of DEGs are obtained in comparative transcriptome studies, many of which are responsible for the necessary morphological and physiological changes (Borah et al. 2017; Kumar et al. 2022c).
Although transcriptome profiling of Nagina-22 [N-22, one of the most drought and heat tolerant cultivars (Vikram et al. 2011; Shanmugvadivel et al. 2017; Casartelli et al. 2018; Yadav et al. 2023)] and IR-64 rice cultivars under drought stress has been carried out by several researchers, the expression profiling was performed mostly at the seedling stage (Feng et al. 2009; Lenka et al. 2011; Shankar et al. 2016). Drought stress at the reproductive stage severely effects on the productivity and yield of rice; therefore, transcriptome analysis of contrasting rice genotypes for water-deficit stress at the reproductive stage might provide deeper insights into the regulatory networks, cellular and metabolic functions responsible for the abiotic stress tolerance. Hence, a comprehensive RNA-seq analysis of leaf and root tissues from a pair of contrasting rice cultivars (N-22, and IR-64) grown under control and terminal drought stress was carried out. Our comparative RNA-seq analysis of rice cultivars provides insights into the expression divergence of genes like OsNAC10, OsbZIP23, OsABA8ox1, OsLEA3, and CAMK/OsCPK4. The function of such genes encoding for the transcription factors, LEA/LTP proteins, involved in ABA biosynthetic process and redox homeostasis against drought stress might be responsible for better drought tolerance of N-22. These stress-responsive genes might be useful for screening the germplasm as well as in rice breeding programs towards improving drought tolerance and the yielding potential of rice under changing climatic conditions.
Materials and methods
Plant materials, growth conditions, and drought stress imposition
Mature and healthy seeds of contrasting rice cultivars Nagina-22 [N-22, early maturing (90–95 days) aus-type rice] and IR-64 (high-yielding, irrigated, terminal drought sensitive variety) were used in the present study. The selection of contrasting rice cultivars for this study was dependent on the earlier reports (Vikram et al. 2011; Shanmugvadivel et al. 2017; Casartelli et al. 2018) as well as that of our pilot study on comparative evaluation of the changes in stress-associated biochemical parameters under terminal drought stress (results not shown). The seeds were surface-sterilized using 0.1% mercuric chloride (HgCl2), followed by washing with distilled water. Twenty-five days old seedlings were transplanted in 12″ pots filled with puddled soil and grown under 70–75% relative humidity with a 35 °C/14 h day and 25 °C/10 h night cycle. While the control plants (3 plants in each of the nine pots) were maintained well-watered throughout the season, another set of plants was imposed with drought stress, at reproductive stage of plant, by withholding irrigation for 4–5 days just before panicle initiation (65 days after transplanting N-22). On a three-fourth reduction in soil moisture content and wilting of leaves [relative water content (RWC) of leaf ~ 58%], leaf (younger leaves, excluding the flag leaf) and root tissue samples were collected from the randomly selected pots in six replications (each sample comprised of the tissues from 3 plants in a pot) from the control (unstressed) as well as drought stressed plants for biochemical, physiological, and molecular analyses.
Estimation of soil moisture and relative water contents
Soil moisture content (SMC) was estimated by the gravimetric method using soil samples collected from pots at a depth of 5 cm. The soil sample was placed in a pre-weighed petri-plate, and the weight of the soil was recorded immediately. The soil was dried at 60 °C in an oven until the constant dry weight (DW) of the soil was achieved. The SMC was calculated using the formula: SMC = [(weight of wet soil) − (weight of dried soil)] ÷ (weight of dried soil).
RWC of leaf was measured by collecting leaf tissues (10 cm piece) at one-third distance from the tip, cutting it into 5 mm pieces in a pre-weighed petri-plate, covered with a lid, and fresh weight (FW) of the leaf was recorded. The petri-plate was filled with distilled water and stored at room temperature for 4 h till the leaf achieved turgidity. Turgid weight (TW) of the leaf was recorded, the leaf pieces were blot dried and dried in an oven at 60 °C until constant weight (DW) was achieved. The RWC was calculated using the formula:
RWC (%) = [(FW − DW) ÷ (TW − DW)] × 100.
Estimation of antioxidant activity
The antioxidant activity, in the tissues collected from control and drought treated plants at reproductive stage of growth, was measured using the stable DPPH radical method described earlier (Kumar et al. 2017). Fresh tissue (1.0 g) samples were ground into fine powder and extracted with 10 mL of ethanol (90%) by constant shaking for 48 h at room temperature. The extract was centrifuged at 13,000 rpm for 10 min and supernatant was used for the estimation of antioxidant activity. For this, 0.5 mL alcoholic solution of DPPH radical (0.2 mM) was added to 100 μL of the sample extract, mixed vigorously, and incubated in dark for 45 min. Then absorbance (A517) was measured, and the capacity to scavenge DPPH radicals was calculated using the formula:
Scavenging (%) = [(A0 − A1) ÷ A0)] × 100; where A0 is absorbance of the control reaction and A1 is absorbance of the sample at 517 nm. The inhibitory concentration at 50% (IC50, extract concentration that cause 50% scavenging of DPPH radical) was also determined.
Estimation of proline content
Proline content in the plant tissue samples was estimated following the method described earlier (Kumar et al. 2017), using the Ninhydrin reagent. The proline-ninhydrin chromophore was extracted with 4.0 mL toluene and absorbance was recorded at 520 nm. Proline content in plant tissue was calculated using the formula:
Proline (μM/g DW) = DMI × [(μg Proline/mL × mL Toluene) ÷ 115.5 μg/μM] ÷ [g sample) ÷ 5] where DMI = Dry matter index (Fresh weight ÷ Dry weight) of leaf tissues.
Estimation of total phenolic content
The tissue samples collected from control and drought treated plants of the contrasting rice cultivars were used to estimate TPC following the procedure described earlier (Singleton et al. 1999). In brief, 1.0 g fresh tissue was ground into a fine powder, and 20 mL of trichloro acetic acid (0.1%) was added. The content was centrifuged at 12,000 rpm for 10 min at room temperature, and 0.5 mL of the aqueous extract was added to 20.5 mL of Folin-Ciocalteu reagent (10% v/v) and 2 mL of 7.5% sodium carbonate. The reaction mixture was incubated for 40 min at 45 °C and absorbance (A765) was recorded. Gallic acid was used as a phenol standard to express TPC (mg) in terms of Gallic acid equivalent in per gram of the sample tissue.
Estimation of lipid peroxidation
Lipid peroxidation in terms of MDA level in the plant tissues was determined based on the thiobarbituric acid reaction as described previously (Kumar et al. 2017). The sample extract was prepared by grinding 1.0 g of fresh tissues in a 20 mL TCA (0.1%) solution, followed by centrifugation for 10 min at 12,000 rpm. One mL of supernatant was reacted with a 4 mL TCA solution containing 0.6% thibarbituric acid. The reaction mixture was heated at 95 °C for 30 min, cooled on ice, and then centrifuged for 10 min at 12,000 rpm. The absorbance of the reaction mix was recorded at 532 and 600 nm, and the MDA level in the tissue samples was calculated using the extinction coefficient of 155 mM−1 cm−1 using the following formula: MDA level (nmol) = ΔA(532 nm−600 nm) ÷ (1.56 × 105).
Estimation of chlorophyll content
Total chlorophyll content in leaf tissues (0.5 g) from the contrasting rice cultivars grown under control as well as drought stress, was extracted in 50 mL of dimethyl sulfoxide (DMSO) following the method described elsewhere (Kumar et al. 2017). The absorbance of the extract was recorded at 645 and 663 nm using DMSO as blank. The total chlorophyll content was calculated on a dry weight (DW) basis using the following formula: Total chlorophyll content (mg/g DW) = [{(20.2 × A645) + (8.02 × A663)} × (Vol ÷ Wt)] × DMI, where Vol = volume (mL) of DMSO used to extract tissue sample, Wt = weight (mg) of the sample tissue, DMI = Dry matter index (Fresh weight ÷ Dry weight) of leaf tissues.
Assessment of agronomic performance under drought stress
To assess the agronomic performance of the rice cultivars, the number of panicles as well as the number of well-filled and chaffy seeds was counted for the control and drought treated plants of both the cultivars. The number of well-filled pale-yellow seeds and whitish chaffy seeds was also determined to estimate the effects of terminal drought stress on grain formation/filling and thus the agronomic performance of the rice cultivars.
RNA isolation, cDNA library preparation, and Illumina sequencing
Total RNAs were isolated using the TRIzol method in three technical replications from leaf and root tissues collected in three biological replications by pooling tissues from three plants. The total RNAs from technical replicates were pooled in equal amounts and a total of 24 libraries (two tissues from two rice cultivars grown under two conditions in three replications) were prepared. Standard steps for mRNA enrichment, RNA fragmentation, first- and second-strand cDNA synthesis, purification, sequencing-adaptor ligation, and PCR amplification as per the TruSeq RNA Sample Preparation Kit (Illumina) were followed to prepare RNA-seq libraries. The libraries were sequenced using PE 150 bp chemistry at the Illumina platform. Raw sequence data were submitted to the NCBI under the BioProject IDs: PRJNA628020 and PRJNA833055.
Quality check and RNA-seq data analysis
Raw data for each sample was assessed for its quality using the Fast QC-toolkit. Adapter contamination and low-quality reads were removed using Trimmomatic software. The resultant high-quality reads were mapped to the rice genome annotation project (RGAP) data using the TopHat pipeline and assembled using Cufflinks software to construct a unique transcript sequence. The number of mapped reads for each gene was normalized to reads per kilobase per million (RPKM). DEGs were calculated for a > fourfold change at P < 0.05 using Cuffdiff software.
Gene ontology analysis
Gene ontology (GO) enrichment analysis for the DEGs was performed using AgriGO v2 software as detailed earlier Kumar et al. (2021), which revealed the enriched GO terms. The background list of genes and GO annotations were extracted from the RGAP database.
Validation of DEGs by RT-qPCR
To validate expression profile of the genes revealed by transcriptome analysis, the expression level of some of the selected genes was validated by RT-qPCR. Total RNAs isolated from root/leaf tissues of N-22/IR-64 grown under control/terminal drought were subjected to DNase I treatment, followed by reverse transcription using superscript II (Invitrogen). RT-qPCR validation of the genes was performed in triplicate using the SYBR Green PCR Master Mix kit (Applied Biosystems, CA, USA), following the manufacturer’s instructions. Relative gene expression level (fold-change) was determined by the 2−ΔΔCt method following the procedure described elsewhere (Kumar et al. 2022a) using actin and tubulin as internal reference genes.
Statistical analysis
The biochemical experiments were carried out with three technical and three biological replications. Statistical analysis was performed using the analysis of variance (ANOVA), Post-hoc Tukey test, and/or Duncan’s multiple range test (DMRT) at P ≤ 0.05 to compare the means (n = 3) of treatments for significance. The standard deviation (± SD) was calculated and represented as an error bar.
Results
Morpho-physiological responses of rice cultivars under drought stress
Terminal drought stress imposed by withholding irrigation just before panicle initiation by a three-fourth reduction in SMC caused ~ 14% decrease in RWC, which resulted in wilting/rolling-off the leaves (Supplementary Fig. S1). However, a greater reduction in RWC (57.5 ± 1%) was observed in the leaf of IR-64 compared to that (61.5 ± 1%) observed in the leaf of N-22. This also resulted in a delayed (5 − 7 days) emergence of panicle and adversely affected seed setting/grain filling, to varying degrees, in the rice cultivars due to the terminal drought stress.
Biochemical responses of rice under drought stress
Antioxidant potential, in terms of DPPH scavenging, was observed to increase significantly in leaf of N-22 under stress. A considerably higher level of DPPH scavenging, even under control condition, was observed in leaf of N-22 (drought tolerant) cultivar (Fig. 1A). Similarly, a significant increase in TPC in the leaves of N-22 was recorded under drought stress. However, the increase in TPC was not significant in the leaves of IR-64 (Fig. 1B). Under drought stress, endogenous proline content also increased significantly in both the rice cultivars, but it increased considerably (~ 2.5-fold) in the leaf of N-22 compared to that (~ 1.4-fold) in the leaf of IR-64) under stress (Fig. 1C).
Biochemical responses in leaf of contrasting rice cultivars under terminal drought stress. (A) DPPH free radical scavenging activity in N-22 and IR-64, B total phenolic content, (C) proline content in leaf of rice cultivars under terminal drought. Drought stress was imposed by withholding irrigation for 4 − 5 days just before panicle initiation. The data presented are for three technical replicates, each with three biological replications (n = 3). Means followed by different lower-case letters are significantly different (P < 0.05). Error bar represents the standard deviation (± SD)
Physio-biochemical changes in rice under drought stress
Under stress, a significant decrease in total chlorophyll content was observed in leaf of both rice cultivars (Fig. 2A). While ~ 9% decrease in total chlorophyll content was recorded in the case of N-22 (drought tolerant cultivar), the decrease was significantly higher (~ 25%) in the case of IR-64 (drought sensitive cultivar). Moreover, in leaf of N-22 only a minor increase in lipid peroxidation (in terms of MDA level) was recorded under stress but it was considerably (~ 2.5-fold) higher (even under control conditions) in the leaf of IR-64 (Fig.. 2B).
Physio-biochemical changes in leaf of contrasting rice cultivars under terminal drought. A Total chlorophyll content in leaf, B lipid peroxidation (in terms of MDA level) in leaf of N-22 and IR-64 in response to drought stress. Drought stress was imposed by withholding irrigation for 4 − 5 days just before panicle initiation. The data presented are for three technical replicates, each with three biological replications (n = 3). Means followed by different lower-case letters are significantly different (P < 0.05). Error bar represents the standard deviation (± SD)
Effect of drought stress on the agronomic performance of rice
Imposition of terminal drought stress caused a considerable (> 46%) reduction in the number of panicles per plant in the case of IR-64, while it was ~ 42% in the case of N-22 (Fig. 3A). Moreover, the drought stress had a considerable effect on seed setting/grain filling in both rice cultivars, resulting in a significant increase in the formation of chaffy seeds, which was higher (~ 17%) in the drought sensitive rice cultivar (IR-64) compared to that (~ 13%) in the case of N-22 (Fig. 3B). Such reduction in the number of panicles per plant and seed setting/grain filling resulted in a considerable reduction in the overall grain yield of the rice cultivars, particularly in the drought sensitive (> 60%) rice (IR-64), which was higher than that (> 33%) observed in case of drought tolerant (N-22) cultivar (data not shown).
Agronomic performance of rice cultivars under terminal drought stress. A Effect of drought stress on the number of panicle per plant, B effect of stress on seed setting/grain filling resulting in the formation of chaffy seeds. Drought stress was imposed by withholding irrigation for 4–5days just before panicle initiation. The means followed by different lower-case letters are significantly different (P < 0.05). Error bar represents the standard deviation (± SD)
Transcriptome sequencing and mapping on the reference genome
To reveal transcriptional variation in gene expression under terminal drought stress in the contrasting rice cultivars, eight RNA-seq libraries (leaf and root tissues from two rice cultivars grown under control and drought stress) in three replications were sequenced with an average of 23 million raw reads for each library (Table S1). Filtering of raw reads [to remove low-quality reads (Phred score ≥ 33)], trimming of reads, and reference-based mapping of the RNA-seq data on the rice reference genome (TIGR v7) showed an average mapping efficiency of ~ 85.61%, indicating a good quality of the RNA-seq data generated.
Differential expression of genes in contrasting rice cultivars
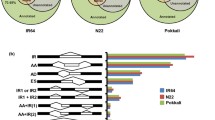
Comparative transcriptome analysis of the DEGs in leaf and root of N-22 and IR-64 under drought stress resulted in the identification of several up-regulated and down-regulated genes (based on Log2 fold-change > 2). A total of 6077 and 10,050 DEGs in leaf, while 4204 and 5715 DEGs in root, of N-22 and IR-64, respectively, were detected under terminal drought stress. Further comparative analysis of the RNA-seq data indicated a total of 2682 genes to be exclusively up-regulated in the leaf of N-22, while only 1447 genes that were exclusively up-regulated in leaf of IR-64 under drought stress (Fig. 4A). Likewise, a total of 7198 genes were exclusively down-regulated in leaf of IR-64 (compared to 1990 genes down-regulated in the leaf of N-22). In the root of N-22, 1497 genes were exclusively up-regulated, but 1677 genes were exclusively down-regulated; in contrast to 2091 that were genes exclusively up-regulated and 2594 genes exclusively down-regulated in the root of IR-64 under stress (Fig. 4B). Thus, the exclusively up-regulated (2091) and down-regulated (2594) genes were comparatively higher in root of IR-64.
Interestingly, the genes (802) up-regulated in the leaf of N-22 were down-regulated in the leaf of IR-64, and the genes (134) down-regulated in the leaf of N-22 were up-regulated in the leaf of IR-64 (Fig. 5A). Similarly, 193 genes up-regulated in the root of N-22 were down-regulated in the root of IR-64, while 168 genes down-regulated in the root of N-22 were up-regulated in the root of IR-64 (Fig. 5B).
Highly expressed genes in roots of contrasting rice cultivars
Analysis of the top 20 up-regulated genes in the root of N-22 indicated that genes for chitinase 10 (CHIT5) family protein (581-fold), thaumatin family domain-containing protein (78.5-fold), late embryogenesis abundant protein, group 3 (66.5-fold), LTPL157 for an LTP family protein (57.05 fold), dehydrin (45.21-fold), and FAD-binding/arabino-lactone oxidase domains containing protein 13 (23.54-fold) were highly up-regulated under stress (Table S2). Similarly, analysis of the top 20 down-regulated genes in the roots of N-22 included glutathione S-transferase (~ 27-fold), MYB 15 family TF (~ 25-fold), rho-GTPase-activating protein (~ 24-fold), zinc finger/C3HC4 type domain-containing protein (~ 24-fold), and DUF260 domain-containing protein (~ 21-fold) (Table S3).
In contrast, the top 20 up-regulated genes in roots of IR-64 under drought stress (Table S4) included LEA3 (~ 548-fold), dehydrin (~ 351-fold), transporters (~ 171-fold), and glutathione S-transferase (~ 157-fold). Besides, the top 20 down-regulated genes in roots of IR-64 under drought stress (Table S5) included the RNA-binding protein (~ 180-fold), glycosyl hydrolase (~ 143fold), WRKY21 (~ 117-fold), C2 domain-containing protein (~ 107-fold), and proline-rich protein (~ 74-fold).
Highly expressed genes in leaf of contrasting rice cultivars
The top 20 up-regulated genes in the leaf of N-22 showed considerably (> 50-fold) increased expression under the terminal drought stress (Table S6), including those for LEA protein (~ 422fold), LTPL102 (~ 290-fold), asparagine synthetase (~ 170-fold), glyoxalase family protein (~ eightfold), transferase (~ 85-fold), and glycine-rich cell wall structural protein (~ 83-fold). While a different set of genes were highly up-regulated in leaf of IR-64 (Table S8). The top 20 down-regulated genes in the leaf of N-22 included those for Type-A response regulator (~ 88-fold), CPuORF26 conserved peptide (~ 59-fold), OsFTL2 (~ 39-fold), respiratory burst oxidase (~ 21-fold), LSD1 zinc-finger domain-containing protein (~ 20-fold), 9-cisepoxycarotenoid dioxygenase 1 (~ 20-fold), and HAD-superfamily hydrolase (~ 20-fold) (Table S7).
Similarly, the top 20 up-regulated genes in the leaf of IR-64 included those for the DNA directed RNA polymerase (~ 60-fold), ribulose bisphosphate carboxylase (~ 51-fold), photosystem I P700 (~ 44-fold), ATP synthase (~ 42-fold), NADPH-dependent oxido reductase (~ 39-fold), and photosystem II D2 protein (~ 37-fold) (Table S8). The top 20 highly down-regulated genes in the leaf of IR-64 included the genes for LTPL124 (~ 161-fold), glycine-rich cell wall protein (83.5-fold), SCP-like extracellular protein (~ 71-fold), aquaporin (~ 55-fold), and peroxi-redoxin (~ 48-fold) (Table S9).
Gene ontology of DEGs under stress
To gain a better insight into the functional role of DEGs, gene ontology (GO) analysis was performed. This indicated that the up-regulated DEGs in N-22 were associated with 46 GO terms enriched in leaf, while the down-regulated DEGs were associated with 66 GO terms. The up-regulated and down-regulated DEGs in roots of N-22 were associated with 51 and 30 GO terms, respectively. Similarly, the up-regulated and down-regulated DEGs under drought stress in the roots of IR-64 were associated with 24 and 39 GO terms, respectively. Further investigations on the GO terms revealed that GO terms like transport, response to abiotic stimulus, catabolic process, intracellular membrane-bounded organelle, nucleolus, plasma membrane, etc. were enriched with the significantly up-regulated genes in the leaf of N-22 under stress (Table S10). On the other hand, GO terms like gene expression, signaling, nucleotide binding, hydrolase activity, chromatin binding, transporter activity, etc. were enriched with the down-regulated genes in leaf of N-22 (Fig. 7, Table S11). Similarly, GO terms like regulation of metabolic process, regulation of gene expression, cellular development process, nucleotide/nucleic acid metabolism, signaling, transporter activity, hydrolase activity, kinase activity, transferase activity, etc. were enriched with up-regulated genes in the leaf of IR-64 under terminal drought stress (Table S12). The GO terms like cellular macromolecule biosynthesis, gene expression/translation, structural molecule activity, translation factor activity, ribonucleoprotein complex, etc. were enriched with down-regulated genes in the leaf of IR-64 under stress (Table S13).
In roots of N-22, the up-regulated DEGs were associated with GO terms like biosynthetic and metabolic processes, gene expression, translation, response to abiotic stimulus, catalytic and hydrolase activities, RNA binding, mitochondrion, nucleolus, vacuole, ribonucleoprotein complex, macromolecular complex, etc. (Table S14). Whereas, the down- regulated DEGs in the root of N-22 were associated with the GO terms like transport, localization, protein modification, signal transduction, regulation of cellular and biological processes, nitrogen compound metabolic processes, nucleobase and nucleic acid metabolic process, transcription regulator activity, kinase and transferase activities, and plasma membrane binding (Table S15). Similarly, GO terms like transport and localization, protein binding, cytoplasm, plastid, intracellular organelle, membrane-bounded organelle, cytoskeleton, endosome, intracellular non-membrane-bounded organelle, etc. were enriched with the up-regulated DEGs in the roots of IR-64 (Table S16). Likewise, the down-regulated DEGs were associated with the GO terms like response to abiotic stimulus, signal transduction, protein modification, regulation of the cellular process, macromolecule modification, nitrogenous compound metabolic process, nucleobase and nucleic acid metabolic process, protein metabolic process, transcription regulator activity, kinase and transferase activity, transporter activity, plasma membrane, etc. in the roots of IR-64 (Table S17).
Cultivar-specific GO terms/gene expression under drought stress
The GO terms for the biological process enriched with the down-regulated genes in leaf of N-22 included epigenetic regulation of gene expressions, and multicellular development (embryonic and post-embryonic development) (Fig. 6A). Interestingly, these GO terms were enriched with the up-regulated genes in the leaf of IR-64 (Fig. 6B). Likewise, the GO terms for molecular function like hydrolase and RNA binding activities were enriched with down-regulated genes in the leaf of N-22 (Fig. 7A), whereas such genes were up-regulated in IR-64 leaf (Fig. 7B). GO terms for cellular components, particularly nucleolus activity, were enriched with up-regulated genes in the leaf of N-22 (Fig. 8A); whereas those were enriched with the down-regulated genes in the leaf of IR-64 (Fig. 8B).
Gene ontology (GO) analysis of biological processes in contrasting rice cultivars under drought stress. A GO terms under-represented (with down-regulated genes) in the leaf of N-22 (drought tolerant), B GO terms enriched (with up-regulated genes) in the leaf of IR-64 (drought sensitive) rice cultivar
Gene ontology (GO) analysis of cellular components in contrasting rice cultivars under drought stress. A GO terms enriched (with up-regulated genes) in the leaf of N-22 (drought tolerant), B GO terms under-represented (with down-regulated genes) in the leaf of IR-64 (drought sensitive) rice cultivar
Dynamics of ABA metabolism under drought stress
Analysis of the genes involved in ABA metabolism in the leaf and root of N-22 and IR-64 under terminal drought stress suggests that the gene (ZEP, LOC_Os04g37619) coding for zeaxanthin epoxidase (which catalyzes the conversion of zeaxanthin to violaxanthin) was down-regulated in both N-22 and IR-64 under drought stress (Table S18). Expression of the gene for five different isoforms of 9-cis-epoxycarotenoid dioxygenase (NCED1, 2, 3, 4, and 5), a key rate-limiting enzyme, was checked and it was observed that the expression of NCED1 (LOC_Os02g47510) was considerably down-regulated in both the leaf and root of N-22, while it was up-regulated in the root of IR-64. NCED2 (LOC_Os12g24800) was considerably (~ 20-fold) down-regulated in the leaf (no change in the root) of N-22, but it was slightly (~ onefold) down-regulated in the leaf and considerably (~ 17-fold) up-regulated in the root of IR-64.
Expression of NCED3 (LOC_Os03g44380) was ~ fourfold down-regulated in roots of N-22 but it was ~ onefold up-regulated in roots of IR-64.In contrast, NCED4 (LOC_Os07g05940) was considerably up-regulated (~ 24-fold) in the leaf of N-22, while it was only ~ 1.5-fold up-regulated in the leaf of IR-64. Expression of NCED5 (LOC_Os12g42280) was also ~ 2.8-fold down-regulated in roots of N-22 but ~ 1.8-fold up-regulated in roots of IR-64 under drought stress (Table S18). Expression of the gene for short-chain dehydrogenase/reductase (SDR, LOC_Os03g45194) was ~ 5.4-fold up-regulated in the leaf of N-22, while it was ~ 3.1-fold up-regulated in the leaf of IR-64. Aldehyde oxidase gene (AO1, LOC_Os10g04860), which converts abscisic aldehyde to abscisic acid, was up-regulated (1.3-fold) in root of N-22, but down-regulated (~ twofold) in root of R-64. The gene (ABA8Ox3, LOC_Os09g28390) coding for the first enzyme of the ABA catabolic pathway (catalyzing hydroxylation of ABA to 8′-hydroxy ABA) was considerably (~ 18-fold and 14-fold) down-regulated in leaf and root of N-22, respectively, compared to that (~ twofold and 1.4-fold down-regulated) in leaf and root of IR-64 (Table S18).
Differential expression of TFs under drought stress
Variation in the expression of TFs was examined in the leaf and root of N-22 and IR-64 under drought stress. In the leaf of N-22, a total of 483 (281 up-regulated and 202 down-regulated) TFs, while 931 (376 up-regulated and 555 down-regulated) TFs in leaf of IR-64 were differentially expressed. Likewise, in the root of N-22, a total of 518 (232 up-regulated and 286 down-regulated) TFs, while 720 (288 up-regulated and 432 down-regulated) TFs in root of IR-64 were differentially expressed. Among these, some of the well-known TFs for abiotic stress tolerance including NAC, AP2/EREBP, WRKY, bHLH, MYB, MYB-related, C2H2, and bZIP were differentially expressed (up-regulated in leaf), in N-22 under drought stress (Table S19).
Differential expression of key drought-responsive genes
Differential expression of some of the well-known drought-associated genes was checked in N-22 and IR-64 (Table 1). Most of the NAC domain-containing TFs were up-regulated in leaf of N22, whereas they were down-regulated in leaf of IR-64. LEA genes (OsLEA3-1 and OsLEA3-2) were highly (10 − 70 times more) up-regulated in the leaf of N-22, compared to that in leaf of IR-64. OsDREB1A and OsDREB1B were up-regulated in the leaf and root of N-22, while they were down-regulated in the root of IR-64. OsDREB2A was considerably up-regulated in the leaf of N-22 compared to that in IR-64. Similarly, OsNCED4 was considerably (~ 16 times) up-regulated in the leaf of N-22 compared to that in IR-64. In contrast, OsbZIP12 7 showed tissue-specific differential expression in the rice cultivars. It was threefold up-regulated in the leaf but onefold down-regulated in the root of N-22, whereas it was 1.3-fold down-regulated in the leaf but 39 fold up-regulated in the root of IR-64.
Redox homeostasis under drought stress
Differential expression of AOX genes [OsAOX1a (LOC_Os04g51150), OsAOX1d (LOC_Os04g51160), OsAOX1e (LOC_Os02g21300), and OsAOX1c (LOC_Os02g47200)] was observed in the contrasting rice cultivars. While OsAOX1a and OsAOX1d were up-regulated in the leaf (but down-regulated in root) of N-22, they were down-regulated in the leaf (but up-regulated in root) of IR-64 under drought stress. OsAOX1e showed ~ 2 times more up-regulated expression in leaf of N-22 compared to that in IR-64, and ~ 4 times more down-regulated expression in the root of IR-64. Up-regulated expression of glutathione S-transferase (GST, LOC_Os01g27210) in the leaf (twofold) and root (eightfold) of N-22, while down-regulated (~ 2.2-fold) expression in leaf of IR-64, indicate its role in terminal drought tolerance (Table S20).
Differential expression of genes of signaling pathway under stress
Some of the signaling pathway genes showed differential expression in the contrasting rice cultivars under terminal drought stress. Calcium/calmodulin-dependent protein kinase (CAMK, LOC_Os02g03410) was up-regulated in N-22 but down-regulated in IR-64 (Table S21). However, another CAMK (LOC_Os02g34600) gene was up-regulated in the leaf, but down-regulated in the root of both the rice cultivars. Other two CAMK genes (LOC_Os07g42940 and LOC_Os03g17700) were up-regulated in the leaf of N-22, but down-regulated in the leaf of IR-64. Receptor-like kinases (which play a vital role in signal transduction) were differentially expressed under drought stress. A protein kinase domain-containing protein (LOC_Os03g06410) was up-regulated in N-22, but down-regulated in leaf as well as the root of IR-64 (Table S21).
Up-regulated expression of genes in Nagina-22 under stress
Certain genes involved in various metabolic pathways/defense responses were significantly/exclusively up-regulated in leaf and root of N-22 (Table S22). Isocitrate lyase (a key enzyme in glyoxylate cycle that allows to uses lipids as a source of energy) was significantly up-regulated in leaf as well as root of N-22 under terminal drought stress. Moreover, cytochrome P450 (the enzyme involved in NADPH- and/or O2-dependent hydroxylation reactions) playing a crucial role in biosynthesis of secondary metabolites, antioxidants, and phytohormones was up-regulated in leaf and root of N-22 under drought stress. Gene (LOC_Os04g57880) for a heat shock protein DnaJ (heat-shock protein 40, HSP40) that function as molecular chaperone (independently or as co-chaperone of HSP70) was exclusively up-regulated in N-22 (both in leaf and root). Furthermore, genes for peptide transporters (LOC_Os01g65130 and LOC_Os01g65140, low-affinity nitrate transporters contributing in nitrogen allocation) were exclusively up-regulated in both root and leaf of N-22 (but down-regulated in IR-64) under terminal drought stress. Most of these genes, except for isocitrate lyase (LOC_Os07g34520), were considerably down-regulated in leaf as well as root of IR-64 under terminal drought stress (Table S22).
RT-qPCR validation of transcriptome data
Differential expression of some of the stress-responsive genes (observed on RNA-seq analysis) was validated by RT-qPCR (Fig. 9). The genes showed a similar pattern of expression, which confirmed trustworthiness of the RNA-seq data.
Discussion
Abiotic stresses adversely affect plant growth, development, productivity, and pose threats to global food security. Several reports on transcriptome analysis of rice under drought stress have been published in the last two decades, which contribute a lot to understanding drought tolerance at the systematic level (Hu et al. 2006; Feng et al. 2009; Lenka et al. 2011; Shankar et al. 2016; Zhang et al. 2016; Borah et al. 2017; Kumar et al. 2022c). Effects of drought stress on the performance of plants and stress-responsive genes have been studied mainly at the seedling stage (Feng et al. 2009; Lenka et al. 2011; Shankar et al. 2016; Zhang et al. 2016); however, only a few studies focused on comparative RNA-seq analysis of contrasting rice genotypes under drought stress at the reproductive stage (Ereful et al. 2020). Comparative/comprehensive studies on molecular, biochemical, and physiological parameters responsible for stress tolerance and better performance of plant provide valuable insights on the mechanisms of interest (Zhang et al. 2016; Li et al. 2018). Many of the high-yielding rice cultivars (e.g., IR-64) are sensitive to drought stress, particularly in the reproductive stage. Hence, to have better insights on the mechanisms adopted by rice plant against terminal drought stress, we used a pair of contrasting rice cultivars (N-22 and IR-64) exposed to reproductive stage drought for comparative whole transcriptome analysis.
A considerable (three-fourth) decrease in SMC, resulting in ~ 14% reduction in RWC and rolling-off/wilting of leaves, indicated the successful imposition of drought stress on the plants. Moreover, delayed (5 − 7 day) emergence of panicle, reduced seed setting/grain filling, and lesser grains (Fig. 3) indicated the differential agronomic performance of the cultivars under terminal drought stress. Increased antioxidant potential, total phenolics, and proline contents in the leaf of a rice cultivar under stress, with a considerably higher level in N-22 (Fig. 1), indicate the involvement of biochemical mechanisms in providing stress tolerance to N-22. Moreover, a significant (~ 25%) decrease in total chlorophyll content in the leaf of IR-64, compared to only ~ 9% decrease in leaf of the N-22, and a considerable increase in lipid peroxidation in the leaf of IR-64 (compared to that in the leaf of N-22) under stress (Fig. 2) indicate the reasons for poor performance of IR-64 (Fig. 3) under terminal drought stress, as evidenced by its yield potential. On the contrary, a comparatively lesser increase in lipid peroxidation and a lesser reduction in chlorophyll content in the leaf of N-22 under terminal drought stress must be responsible for the better physio-biochemical responses of N-22 under stress.
In addition to the above mentioned physio-biochemical changes, the comparative molecular analysis indicates important roles of a number of differentially expressed genes in the leaf and root of N-22 for its better performance under terminal drought stress. Interestingly, the up-regulated expression of genes in the leaf of N-22, compared to that in root, was more closely correlated with stress tolerance. Our comparative analysis revealed a greater number of drought-responsive genes to be differentially expressed in the leaf of the drought tolerant (N-22) cultivar compared to that in the root under stress. This suggests an important role of leaf in managing with drought stress. Such a finding corroborates with the earlier report by Zhang et al. (2017) showed a larger number (11,231) of transcripts to differentially express in the leaf of drought tolerant Oryza rufipogon compared to that (7025) in the root. A similar finding was also reported for alkalinity stress tolerance in rice (Li et al. 2018). The up-regulated expression of genes in the leaf of N-22 playing a more important role under direct-sown (stressful) conditions has also been reported (Kumar et al. 2022c). This tissue- and cultivar-specific differential expression of genes might be responsible for the terminal drought tolerance of N-22.
A gene (LOC_Os05g46480) for late embryogenesis abundant (LEA) protein was observed to be highly (~ 422-fold) up-regulated in the leaf as well as root (~ 66-fold) of the drought tolerant (N-22) cultivar (Table S6), which must be responsible for protecting the plant under the drought stress. LEA proteins have been reported earlier to protect cell membranes, stabilize cellular macromolecules, and act as water_binding molecules (Liang et al. 2019). Similarly, a gene (LOC_Os04g33920) for protease inhibitor/seed storage/LTP family protein was highly up-regulated in leaf (~ 290-fold) and root (~ 66-fold) of N-22 which help in protecting the plant under abiotic/biotic stresses. Moreover, LTPL124 (LOC_Os04g52260) showed down-regulated (~ 161fold) expression in leaf of (IR-64) under stress. Such non-specific lipid transfer proteins (nsLTPs/LTPs) have been reported to be induced in response to abiotic/biotic stresses to protect the plant by transferring fatty acid metabolism-related proteins and phospholipids across the membrane (Liu et al. 2015). Higher expression of nsLTP genes was reported under drought and salinity stress in wheat (Fang et al. 2020).
Highly up-regulated (~ 170-fold) expression of asparagine synthetase.
(LOC_Os03g18130) and glyoxalase family protein (~ 89-fold) genes in the leaf of N-22 corroborate with earlier reports on drought tolerance in wheat (Curtis et al. 2018) and detoxifying methylglyoxal generated under stress in rice (Schmitz et al. 2018). A glycine-rich cell wall structural protein (LOC_Os01g06310), up-regulated (~ 84-fold) in the leaf of N-22 but down-regulated in IR-64, was reported to play an important role in abiotic stress tolerance (Czolpinska and Rurek 2018). To our surprise, some of the genes (e.g. LOC_Os03g61150, > 431-fold up-regulated; LOC_Os03g61160, > 261-fold up-regulated) coding for expressed proteins were considerably up-regulated in leaf of N-22. Such genes need to be annotated for their function /role in terminal drought stress tolerance (Table S6). A network module-based analysis by Lv et al. (2019) suggests zinc finger domain TF (LOC_Os08g06280) to be > 20-fold down-regulated in N-22 (Table S7) that interacts with three different gene products, including axing efflux carrier family protein (LOC_Os01g69070), OsRR2 type-A response regulator (LOC_Os02g35180, 88-fold down-regulated), and Ca2+-activated RelA/spot homolog (LOC_Os05g06920, 23-fold down-regulated in the leaf of N-22) to play an important role in drought tolerance in rice. The down-regulated (~ 60-fold) expression of CPuORF26 (LOC_Os05g47540) in the leaf of N-22 under stress (Table S7) corroborates with the findings of Shaik and Ramakrishna (2013). Significantly up-regulated expression of glutathione S-transferase (GST, LOC_Os01g27210) in the N-22 that protects plant from oxidative stress corroborates with the findings of Chen et al. (2012) in Arabidopsis.
GO enrichment analysis clearly differentiated the responses of N-22 from IR-64 based on their molecular functions, biological processes, and cellular components involved in stress tolerance. GO terms like gene expression, transporter and hydrolase activities, embryonic development, signaling processes, RNA binding, chromatin, and epigenetic regulation, etc. were enriched with the down-regulated genes in the leaf of N-22. Similarly, the GO terms like response to abiotic stimulus, catabolic process, intracellular membrane-bound organelles, etc. were enriched with the up-regulated genes under stress. Such enrichment of the GO terms with up-regulated or down-regulated genes might be responsible for drought stress tolerance observed in N-22. These findings corroborate the observations of Martin et al. (2020) in Brachipodium. In the roots of N-22, GO terms like response to abiotic stimulus, translation, biosynthetic and metabolic processes, catalytic and hydrolytic activities, RNA binding, etc. were enriched with up-regulated DEGs under stress. Similarly, the GO terms signal transduction, transcriptional regulation, kinase and transferase activities, regulation of cellular and biological processes, protein modification, nitrogen metabolic process, transporter activity, etc. were enriched with the down-regulated genes in the roots of N-22. Such a variety-specific schema of drought tolerance in N-22 corroborates with the findings of Hassan et al. (2019) reported in cotton under drought stress.
Our observations on the up-regulated expression of NAC domain-containing TFs in the leaf of N-22, and the down-regulated expression in the leaf of IR-64 (Table 1, Table S19) are in agreement with the drought-inducible up-regulated expression of OsNACs (OsNAC1, OsNAC6, OsNAC10, OsNAC14, etc.) reported earlier by Nuruzzaman et al. (2013) and Shim et al. (2018). Previous studies reported NAC, bZIP, AP2/ERF, homeodomain, MYB, WRKY, and bHLH to play crucial roles in abiotic stress tolerance in plants (Nuruzzaman et al. 2013; Castilhos et al. 2014; Shao et al. 2015). We observed 6.2-fold up-regulated expression of SNAC1 in the leaf of N-22 under drought stress, while it was ~ 1.3-fold down-regulated in the leaf of IR-64 (Table 1). Overexpression of SNAC1 (LOC_Os03g60080) was reported to improve drought, salt, and cold tolerance in transgenic rice (Hu et al. 2006). Moreover, overexpression of OsNAC10 (LOC_Os11g03300) in the roots of transgenic rice was reported to significantly improve drought tolerance (Jeong et al. 2010). In the present study, we observed up-regulated expression of OsNAC10 and OsNAC14 in N-22, but down-regulated expression in IR-64 under stress (Table 1).
Overexpression of OsNAC14 (LOC_Os01g48446) in transgenic rice showed enhanced drought stress tolerance at vegetative as well as reproductive stages. However, induction of OsNAC14 expression under stress was reported to be predominant in the leaf than that in the root (Shim et al. 2018). This finding corroborates with our observation of 3.15-fold up-regulation of OsNAC14 in the leaf of N-22 compared to 1.39-fold up-regulation in root under stress (Table 1). Enhanced expression of NAC in drought tolerant cultivar (N-22) compared to that in the sensitive cultivar (IR-64), to drought stress at reproductive stage, substantiates that NAC could be utilized as a candidate gene to improve drought tolerance in rice. Some of the NAC TFs have recently been reported to play an important role in abiotic stress tolerance in rice (Yuan et al. 2019). The up-regulated expression of OsRAD51A1, which we also observed in the leaf of N-22 under drought stress (but 1.67-fold down-regulated in the leaf of IR-64), was reported by Tripathi et al. (2016) that increase DNA repair efficiency and alleviate cell death to confer stress tolerance.
Our observations on the up-regulated expression of mitogen-activated protein kinase (MAPK) signalling pathway genes (LOC_Os02g03410, LOC_Os07g42940, and LOC_Os03g17700) in the leaf of N-22, but down-regulated in IR-64, are in agreement with the earlier report of Campo et al. (2014). Other protein kinases playing vital roles in downstream signaling, including LOC_Os03g06410 (a protein kinase), were up-regulated in the leaf and root of N-22, but down-regulated in IR-64, which is in agreement with the earlier findings (Kim et al. 2003). Moreover, a cysteine-containing protein kinase (CRK, LOC_Os12g41490) was 9.5-fold up-regulated in the leaf of N-22, compared to only 2.83-fold up-regulated in the leaf of IR-64 during drought stress. Several of the receptor-like kinases, including CRK, were reported to be involved in ABA signaling and stress tolerance in plants (Lu et al. 2016).
ABA has been reported to play important roles in drought stress tolerance in plants. Among the ABA biosynthesis pathway enzymes, NCED catalyzes the rate-limiting reaction (Fig. 10), and five NCED genes are known in rice (Saika et al. 2007). We observed down-regulated expression of NCED1 under drought stress, which corroborates with the findings of Ye et al. (2011). However, OsNCED4 (LOC_Os07g05940) was considerably (~ 24-fold) up-regulated in leaf of N-22 under stress, which might help to increase the ABA concentration required for better stress tolerance (Table S18). OsNCED4 was reported to be positively regulated by OsbZIP23 during drought stress (Zong et al. 2016), which corroborates with our findings of up-regulated (2.87-fold) expression of bZIP23 (LOC_Os02g52780) in the leaf of N-22 under stress, but down-regulated (1.26-fold) in the leaf of IR-64.
Thus, our findings indicate that N-22 quickly senses stress and enhances the expression of drought-responsive genes. Considerably down-regulated (~ 14 − 18-fold) expression of the gene involved in ABA catabolism (OsABA8Ox3, LOC_Os09g28390) in N-22 must be responsible for higher ABA concentration in cells for improved stress tolerance. Moreover, the up-regulated expression of SAPK2 (LOC_Os07g42940, calmodulin-dependent protein kinaseslike7) in the leaf of N-22, but down-regulation in IR-64 under stress (Table 1) and its role in drought stress tolerance is in agreement with Lin et al. (2020). Our findings also indicate that the ABA-independent pathway is equally involved in protecting N-22 plants under drought stress. While expression of ABA-dependent genes was reported to be regulated through bZIP, the genes of the ABA-independent pathway are regulated through DRE and C-repeat (CRT) cis-acting elements with the help of DREB or CRT-binding factor (CBF) (Cui et al. 2011). TFs like MYB/MYC and WRKY also work in an ABA-independent manner (Liu et al. 2018). The considerably up-regulated expression of the isoforms of stress-associated genes like DREB, MYB, MYC, and WRKY was observed in the leaf of N-22 under stress (Table 1, Table S19). All of these indicate that both ABA-dependent and ABA-independent pathways, are operational in N-22 in making it a drought tolerant cultivar.
Alternative oxidase (AOX), being one of the terminal oxidases, plays an important role in generation of reactive oxygen species (ROS) by minimizing the generation of superoxide (Saha et al. 2016), which affects the metabolic process of the plant during abiotic and biotic stress. Genotype-specific expression of OsAOX1a and OsAOX1d (5 − tenfold up-regulated in N-22, but 1 − threefold down-regulated in IR-64), and tissue-specific differential expression of OsAOX1e (3 − fivefold up-regulated in leaf, and 2 − sixfold down-regulated in root) might help in minimizing the oxidative stress in N-22. This corroborates with the earlier reports by Feng et al. (2009) and Sasi et al. (2021). Another antioxidant enzyme glutathione S-transferase (GST, LOC_Os01g27210) was up-regulated in the leaf (twofold) and root (eightfold) of N-22. GST has been reported to quench the ROS with the help of glutathione (GSH), and thus, protect the cell from oxidative damage (Ding et al. 2017). Moreover, we found that the expression of glutathione (GRCP2, LOC_Os10g28000) is induced by stress, sowing higher expression in N-22.
Significantly up-regulated expression of isocitrate lyase (key enzyme in glyoxylate cycle) in leaf and root of N-22 under terminal drought stress might be involved in converting lipids to organic acids, which helps mobilization of amino acids from leaves. Our findings corroborate with those of Yuenyong et al. (2019) who suggested that isocitrate lyase plays multifunctional roles in salt tolerance by modulating energy metabolism under abiotic stress. Cytochrome P450 (LOC_Os03g55230, up-regulated in leaf and root of N-22 but down-regulated in IR-64 under terminal drought stress) has been reported to play important roles in mitigating abiotic stresses by getting involved (directly or indirectly) in biosynthesis of several antioxidants (see Pandian et al. 2020). Similarly, exclusively up-regulated expression of a heat shock protein DnaJ (LOC_Os04g57880) in leaf and root of N-22 (Table S22) functions as molecular chaperone to stimulate Hsp70 ATPase activity in alleviating ROS accumulation under heat stress; thereby reducing photoinhibition of PSII. These molecular actions complement the physio-biochemical changes in mitigating the oxidative stress created due to terminal drought. The exclusively up-regulated expression of peptide transporters (low-affinity nitrate transporters) in root and leaf of N-22 supports their important roles in transport of nitrate and other substrates (peptides, amino acids, auxin, etc.) under drought stress, as suggested by Fan et al. (2017). Our findings also corroborates with a recent study by Fang et al. (2017) wherein they demonstrated a peptide transporter to contribute in nitrogen allocation for increased grain yield in rice. Up-regulated expression of these genes (isocitrate lyase, cytochrome P450, heat shock protein DnaJ, and peptide transporters) was reported in panicle of N-22 under drought stress (Kaur et al. 2023).
Conclusion
Our findings reveal differential expression of OsNAC10, OsbZIP23, OsABA8ox1, OsCPK4, OsLEA3, and OsNCED4 in the contrasting rice cultivars under terminal drought stress. Gene ontology analysis indicated some of the GO terms to be enriched with up-regulated genes for transcription factors, redox homeostasis, and ABA signaling, while other GO terms (transcription, signaling process, nucleotide binding, hydrolase activity, chromatin binding) were represented with down-regulated genes in drought tolerant rice cultivar N-22. In addition, exclusively up-regulated expression of peptide transporters, heat shock protein DnaJ, and cytochrome P450 in N-22 tissues under drought stress must have complementary roles in making this a tolerant cultivar. This investigation on comparative RNA-seq analysis in leaf and root tissues from a pair of contrasting Indica rice cultivars imposed with terminal (reproductive stage) drought stress presents a combination of rice cultivars, tissues, and the stage of drought stress imposition. The study also indicates more important role of leaves (through differential expression of genes) in managing with terminal drought stress in N-22. The information might be utilized in the genetic improvement of rice for terminal drought tolerance and the development of climate-resilient crops.
Availability of data and materials
RNA-seq raw data are available at NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/bioproject) under the BioProject IDs: PRJNA628020 and PRJNA833055.
Abbreviations
- ABA:
-
Abscisic acid
- AO:
-
Aldehyde oxidase
- AOX:
-
Alternative oxidase
- AP2:
-
Apetala 2
- CAMK:
-
Calcium/calmodulin-dependent protein kinases
- DEGs:
-
Differentially expressed genes
- DMSO:
-
Dimethyl sulfoxide
- DPPH:
-
2,2-Diphenyl-1-picrylhydrazyl
- DREB:
-
Dehydration-responsive element-binding
- DW:
-
Dry weight
- FW:
-
Fresh weight
- GO:
-
Gene ontology
- GST:
-
Glutathione S-transferase
- ICL:
-
Isocitrate lyase
- LEA:
-
Late embryogenesis abundant
- LP:
-
Lipid peroxidation
- LTPL:
-
Lipid transfer protein-like
- MDA:
-
Malondialdehyde
- N-22:
-
Nagina 22
- NADP:
-
Nicotinamide adenine dinucleotide phosphate
- NCED:
-
9-Cis-epoxycarotenoid dioxygenase
- PTR:
-
Peptide transporter
- RGAP:
-
Rice genome annotation project
- ROS:
-
Reactive oxygen species
- RPKM:
-
Reads per kilobase per million
- RWC:
-
Relative water content
- SDR:
-
Short-chain dehydrogenase/reductase
- SMC:
-
Soil moisture content
- TF:
-
Transcription factor
- TIGR:
-
The institute for genome research
- TPC:
-
Total phenolics content
- TW:
-
Turgid weight
References
Ali K, Rai RD, Tyagi A (2016) Expression of bZIP transcription factor encoding gene in response to WDS in rice. Indian J Exp Biol 54:332–337
Borah P, Sharma E, Kaur A, Chandel G, Mohapatra T, Kapoor S, Khurana JP (2017) Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci Rep 7:42131
Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, Segundo BS (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165:688–704
Caverzan A, Casassola A, Brammer SP (2016) Antioxidant responses of wheat plants under stress. Genet Mol Biol 39:1–6
Casartelli A, Riewe D, Hubberten HM, Altmann T, Hoefgen R, Heuer S (2018) Exploring traditional aus-type rice for metabolites conferring drought tolerance. Rice 11:9
Castilhos G, Lazzarotto F, Spagnolo-Fonini L, Bodanese-Zanettini MH, Margis-Pinheiro M (2014) Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci 223:1–7
Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158:340–351
Chen Y, Chen Y, Shi Z, Jin Y, Sun H, Xie F, Zhang L (2019) Biosynthesis and signal transduction of ABA, JA, and BRs in response to drought stress of Kentucky Bluegrass. Int J Mol Sci 20:1289
Cui M, Zhang W, Zhang Q, Xu Z, Zhu Z, Duan F, Wu R (2011) Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol Biochem 49:1384–1391
Curtis TY, Bo V, Tucker A, Halford NG (2018) Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress. Food Energy Secur 7:e00126
Czolpinska M, Rurek M (2018) Plant glycine-rich proteins in stress response: an emerging, still prospective story. Front Plant Sci 9:302
Ding N, Wang A, Zhang X, Wu Y, Wang R, Cui H, Huang R, Luo Y (2017) Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol 17:e225
Elhamid EMA, Sadak MS, Tawfik MM (2014) Alleviation of adverse effects of salt stress in wheat cultivars by foliar treatment with antioxidant: changes in some biochemical aspects, lipid peroxidation, antioxidant enzymes and amino acid contents. Agric Sci 5:1269–1280
Ereful NC, Liu L, Greenland A, Powell W, Mackay I, Leung H (2020) RNA-seq reveals differentially expressed genes between two indica inbred rice genotypes associated with drought-yield QTLs. Agronomy 10:621
Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G (2017) Plant nitrate transporters: from gene function to application. J Exp Bot 68:2463–2475
Fang Z, Bai G, Huang W, Wang Z, Wang X, Zhang M (2017) The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front Plant Sci 8:1338
Fang Z-W, He Y-Q, Liu Y-K, Jiang W-Q, Song J-H, Wang S-P, Ma DF, Yin JL (2020) Bioinformatic identification and analyses of the non-specific lipid transfer proteins in wheat. J Integr Agric 19:1170–1185
Feng HQ, Li HY, Sun K (2009) Enhanced expression of alternative oxidase genes is involved in the tolerance of rice (Oryza sativa L.) seedlings to drought stress. Zeitschrift Für Naturforschung C 64:704–710
González-Schain N, Dreni L, Lawas LM, Galbiati M, Colombo L, Heuer S, Jagadish KS, Kater MM (2015) Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol 57:57–68
Hassan MM, Ma F, Islam F, Sajid M, Prodhan ZH, Li F, Shen H, Chen Y, Wang X (2019) Comparative transcriptomic analysis of biological process and key pathway in three cotton (Gossypium spp.) species under drought stress. Int J Mol Sci 20:2076
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do-Choi Y, Kim M, Reuzeau C, Kim J-K (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Kaur S, Seem K, Duhan N, Kumar S, Kaundal R, Mohapatra T (2023) Transcriptome and physio-biochemical profiling reveals differential responses of rice cultivars at reproductive-stage drought stress. Int J Mol Sci 24(2):1002
Kim JA, Agrawal GK, Rakwal R, Han KS, Kim KN, Yun CH, Heu S, Park SY, Lee YH, Jwa NS (2003) Molecular cloning and mRNA expression analysis of a novel rice (Oryza sativa L.) MAPK kinase, OsEDR1, an ortholog of Arabidopsis AtEDR1, reveal its role in defense/stress signalling pathways and development. Biochem Biophysics Res Commun 300:868–876
Kumar S, Beena AS, Awana M, Singh A (2017) Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front Plant Sci 8:1151
Kumar S, Pallavi, Chugh C, Seem K, Kumar S, Vinod KK, Mohapatra T (2021) Characterization of contrasting rice (Oryza sativa L.) genotypes reveals the Pi-efficient schema for phosphate starvation tolerance. BMC Plant Biol 21:282
Kumar S, Kumar S, Krishnan SG, Mohapatra T (2022a) Molecular basis of genetic plasticity to varying environmental conditions on growing rice by dry/direct-sowing and exposure to drought stress: Insights for DSR varietal development. Front Plant Sci 13:1013207
Kumar S, Seem K, Kumar S, Mohapatra T (2022b) RNA-seq analysis reveals the genes/pathways responsible for genetic plasticity of rice to varying environmental conditions on direct-sowing and transplanting. Sci Rep 12:2241
Kumar S, Seem K, Kumar S, Vinod KK, Chinnusamy V, Mohapatra T (2022c) Pup1 QTL regulates gene expression through epigenetic modification of DNA under phosphate starvation stress in rice. Front Plant Sci 13:871890
Lenka SK, Katiyar A, Chinnusamy V, Bansal KC (2011) Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J 9:315–327
Li N, Liu H, Sun J, Zheng H, Wang J, Yang L, Zhao H, Zou D (2018) Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci Rep 8:9586
Liang Y, Kang K, Gan L, Ning S, Xiong J, Song S, Xi L, Lai S, Yin Y, Gu J, Xiang J, Li S, Wang B, Li M (2019) Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol J 17:2123–2142
Lin Z, Li Y, Zhang Z, Liu X, Hsu C-C, Du Y, Sang T, Zhu C, Wang Y, Satheesh V, Pratibha P, Zhao Y, Song CP, Tao WA, Zhu JK, Wang P (2020) A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat Commun 11:613
Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878
Liu F, Zhang X, Lu C, Zeng X, Li Y, Fu D, Wu GJ (2015) Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J Exp Bot 66:5663–5681
Liu S, Lv Z, Liu Y, Li L, Zhang L (2018) Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet Mol Biol 41:624–637
Lu K, Liang S, Wu Z, Bi C, Yu Y-T, Wang X-F, Zhang DP (2016) Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5 enhances abscisic acid sensitivity and confers drought tolerance. J Exp Bot 67:5009–5027
Lv Y, Xu L, Dossa K, Zhou K, Zhu M, Xie H, Tang S, Yu Y, Guo X, Zhou B (2019) Identification of putative drought-responsive genes in rice using gene co-expression analysis. Bioinformation 15:480–489
Martin RC, Kronmiller BA, Dombrowski JE (2020) Transcriptome analysis of responses in Brachypodium distachyon overexpressing the BdbZIP26 transcription factor. BMC Plant Biol 20:174
Mawlong I, Ali K, Tyagi A (2018) Functional validation of a water deficit stress responsive AP2/ERF family transcription factor-encoding gene in Oryza sativa. Indian J Biochem Biophys 55:17–25
Mishra S, Jha AB, Dubey RS (2011) Arsenite treatment induces oxidative stress, upregulates antioxidant system, and causes phytochelatin synthesis in rice seedlings. Protoplasma 248:565–577
Møller IM, Kristensen BK (2004) Protein oxidation in plant mitochondria as a stress indicator. Photochem Photobiol Sci 3:730–735
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PVV, Jugulam M (2020) Role of cytochrome P450 enzymes in plant stress response. Antioxidants (basel) 9(5):454
Prathap V, Ali K, Singh A, Vishwakarma C, Krishnan V, Chinnusamy V, Tyagi A (2019) Starch accumulation in rice grains subjected to drought during grain filling stage. Plant Physiol Biochem 142:440–451
Romero-Aranda MR, Jurado O, Cuartero J (2006) Silicon alleviates the deleterious salt effect on tomato plant growth by improving plant water status. J Plant Physiol 63:847–855
Saha B, Borovskii G, Panda SK (2016) Alternative oxidase and plant stress tolerance. Plant Signal Behav 11:e1256530
Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, Arimura S, Miyao A, Hirochika H, Kamiya Y, Tsutsumi N, Nambara E, Nakazono M (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48:287–298
Samota MK, Sasi M, Awana M, Yadav OP, Amitha Mithra SV, Tyagi A, Kumar S, Singh A (2017) Elicitor-induced biochemical and molecular manifestations to improve drought tolerance in Rice (Oryza sativa L.) through seed-priming. Front Plant Sci 8:934
Sasi M, Awana M, Samota MK, Tyagi A, Kumar S, Sathee L, Krishnan V, Praveen S, Singh A (2021) Plant growth regulator induced mitigation of oxidative burst helps in the management of drought stress in rice (Oryza sativa L.). Environ Exp Bot 185:104413
Schmitz J, Rossoni AW, Maurin VG (2018) Dissecting the physiological function of plant Glyoxalase I and Glyoxalase I-like proteins. Front Plant Sci 9:1618
Shaik R, Ramakrishna W (2013) Genes and co-expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS ONE 8:e77261
Shankar R, Bhattacharjee A, Jain M (2016) Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci Rep 6:23719
Shanmugavadivel PS, AmithaMithra SV, Prakash C, Ramkumar MK, Tiwari R, Mohapatra T, Singh NK (2017) High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP Array. Rice 10:28
Shao HB, Wang HY, Tang XL (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6:902
Shim JS, Oh N, Chung PJ, Kim YS, Choi YD, Kim J-K (2018) Overexpression of OsNAC14 improves drought tolerance in rice. Front Plant Sci 9:310
Singleton VL, Orthofer R, Lamuela-Raventos RM (1999) Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin–Ciocalteu reagent. Methods Enzymol 299:152–178
Sinha SK, Sevanthi V, Chaudhary S, Tyagi P, Venkadesan S, Rani M, Mandal P (2018) Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic N starvation reveals differences in chloroplast and starch metabolism-related genes. Genes 9:206
Tripathi AK, Pareek A, Singla-Pareek SL (2016) A NAP-family histone chaperone functions in abiotic stress response and adaptation. Plant Physiol 171:2854–2868
Vikram P, Swamy BM, Dixit S, Ahmed HU, Cruz MTS, Singh AK, Singh NK (2011) qDTY1.1 a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 12:89
Wang D, Pan YJ, Zhao XQ, Zhu LH, Fu BY, Li ZK (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12:149
Yadav N, Amitha Mithra SV, Pandey R, Chinnusamy V, Singh AK, Singh NK (2023) Physiological response and agronomic performance of drought tolerance mutants of Aus rice cultivar Nagina 22 (Oryza sativa L). Field Crops Res 290:108760
Ye N, Zhu G, Liu Y, Li Y, Zhang J (2011) ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol 52:689–698
Yuan X, Wang H, Cai J, Bi Y, Li D, Song F (2019) Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol 19:278
Yuenyong W, Sirikantaramas S, Qu LJ, Buaboocha T (2019) Isocitrate lyase plays important roles in plant salt tolerance. BMC Plant Biol 19:472
Zhang ZF, Li YY, Xiao BZ (2016) Comparative transcriptome analysis highlights the crucial roles of photosynthetic system in drought stress adaptation in upland rice. Sci Rep 6:19349
Zhang F, Zhou Y, Zhang M, Luo X, Xie J (2017) Effects of drought stress on global gene expression profile in leaf and root samples of Dongxiang wild rice (Oryza rufipogon). Biosci Rep 37:BSR20160509
Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought resistance related genes. Plant Physiol 171:2810–2825
Acknowledgements
Financial support to Suresh Kumar from National Agricultural Science Fund (NASF/ABP70161/2018-19), Indian Council of Agricultural Research, Government of India, New Delhi, is gratefully acknowledged. Assistance in bioinformatics analyses rendered by Dr. Santosh Kumar is thankfully acknowledged.
Author information
Authors and Affiliations
Contributions
AT and TM conceived the experiment. AT and SK designed and supervised the experiments; SK and AT conducted the experiments, collected data, and analyzed them. SK prepared the manuscript, SK and TM revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tyagi, A., Kumar, S. & Mohapatra, T. Biochemical, physiological and molecular responses of rice to terminal drought stress: transcriptome profiling of leaf and root reveals the key stress-responsive genes. J. Plant Biochem. Biotechnol. (2023). https://doi.org/10.1007/s13562-023-00865-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13562-023-00865-x