Abstract
Objective
This study evaluates Diabetology.co.in, an innovative algorithm-driven prescription system developed for personalized and precision treatment of treatment-naive patients with type 2 diabetes. It focuses on integrating computational medicine with clinical practice, leveraging artificial intelligence for optimized diabetes management.
Methods
A retrospective pilot study was conducted at a tertiary multispeciality hospital, assessing Diabetology.co.in’s alpha version. Data from the last fifty adult patients with type 2 diabetes from the outpatient Endocrinology OPD were analyzed. These patients were treatment-naive, excluding pregnant women and those with positive insulin antibodies or glucocorticoid use. Data, including clinical and laboratory parameters, were manually input into the system, which then generated evidence-based prescription recommendations using its algorithmic processing.
Results
The system processed data from 50 patients, with an average age of 41.9 years and a 40% female demographic. The application effectively utilized inputs like body mass index, glomerular filtration rate, and HbA1c levels to generate prescriptions. Metformin was universally recommended, with insulin prescribed for half of the patients, and SGLT2 inhibitors for 30%. The software’s suggestions showed a significant match with actual clinical prescriptions, indicating its accuracy and potential in aiding clinical decision-making. Notably, the software identified an overprescription tendency in clinician practices and provided insights into patient profiles through advanced data analysis capabilities, such as correlations between triglyceride levels and BMI.
Conclusion
Diabetology.co.in demonstrated high efficacy in generating precise and personalized treatment recommendations for newly diagnosed type 2 diabetes patients. It aligns closely with actual clinical prescriptions, showcasing its potential in reducing overprescription and contributing to evidence-based diabetes care.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Type 2 diabetes has traditionally been managed using a generalized approach, relying largely on clinicians’ judgments and mental assessments of clinical and laboratory data. However, as the medical landscape evolves, there is an increasing emphasis on precision medicine, personalized treatment, and evidence-based decision-making. While precision medicine seeks to offer individualized treatments based on unique patient factors, there is a notable absence of software applications in India that provide this level of tailored prescription support. Recognizing this gap and the potential of algorithm-driven decision-making, we introduced Diabetology.co.in. This is the world’s first algorithm driven prescription system dedicated to treatment naive patients with type 2 diabetes. By harnessing the power of artificial intelligence and cluster classification studies, which identify specific patient subgroups with diverse treatment needs, our system aims to revolutionize diabetes management by integrating computational medicine into daily outpatient practices. While the system’s development has recently been completed, it currently operates in an alpha version, intended exclusively for the internal use of the clinician-developer (author 1).
The primary objective of this study was to alpha-test the system, evaluating the accuracy and relevance of the outputs generated when applied to retrospective data from adult, non-pregnant patients treatment naive with type 2 diabetes who visited the outpatient Endocrinology OPD. The secondary objective was to analyze the clinical profile of newly diagnosed patients with type 2 diabetes visiting the OPD and generate some possible insights.
Materials and Methods
Study design
This research was designed as a retrospective, single-center pilot study to evaluate the functioning of Diabetology.co.in, an online Algorithm-based personalized and precision medicine system developed to generate evidence-based prescriptions for treatment naive patients with type 2 diabetes. The platform, currently in its alpha version, was assessed for its feasibility and the precision of its generated outputs.
Inclusion and exclusion criteria
The study included anonymous data from the last fifty treatment-naive adult patients with type 2 diabetes, sourced from the outpatient Endocrinology OPD of a tertiary multispeciality hospital in the last 6 months. Data of pregnant women those having insulin antibodies positive (Any of the following- GAD65, IAA, IA-2 and/or ZnT-8) and those on glucorticoids were excluded from the study.
Data collection
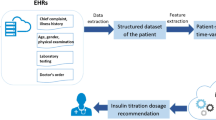
The data of the last 50 patients who matched the inclusion and exclusion criteria were extracted from the existing EMR and were manually entered in the Diabetology.co.in system. The system generated the final prescription from the data collected. Both the input variables entered and the output variables generated were captured by the system.
Software design
Figures 1, 2, 3, 4, 5, 6, and 7 illustrate how the application works. Figure 8 illustrates the application's usage procedure and the flow of its algorithm.
This is the main screen of the application in which the clinician enters all the patient data. The variables we have collected are mentioned in Table 1
Variables
The input variables collected are enlisted in Table 1.
Experimental procedure
The Body Mass Index (BMI) of patients was determined using their weight (in kilograms) and height (in meters) with the formula: BMI} = weight (kg) divided by height (m) [2]. The obesity criteria were determined based on the published cut-off for Asians [1].
The estimated glomerular filtration rate (eGFR) was computed using the CKD-EPI equation [2]. Cardiovascular risk stratification was done using American College of Cardiology ASCVD risk score [3]. The cluster classification was done using Anjana et al. data for diabetes clusters in Indian population [4]. Diabetes remission criteria were generated using proprietary diabetes remission risk score which is developed based on existing data set on the topic [5]. The mixed-meal stimulation score is based on our unpublished mixed-meal calculator developed from existing data-set and validated in small internal study. The starting dose of basal insulin was determined using Hollman-Turner formula formula based on Treat-to-target study [6]. Fib-4 score was calculated using the formula given by Sterling et al. [7]. The standard treatment recommendation as based on American diabetes association standards of care 2023 [8]. The chronic kidney status was determined as per the KDIGO guidelines [9].
Statistical analysis
The primary objective of this study was to alpha-test the diabetes medication treatment generated by the application and its algorithm. The results produced were then compared to the medications prescribed by author 1 to the same patient.
The secondary aim was to examine the profiles of newly diagnosed type 2 diabetes patients visiting the endocrinology outpatient clinic. Parametric data is presented as mean ± standard deviation, while nonparametric data is shown as median with interquartile range.
Data analysis was conducted using the OpenAI ChatGPT Advanced Data Analysis 2023 (Beta version).
Results
We entered the data of the last 50 newly diagnosed patients with type 2 diabetes who attended the OPD. The average age of the patients was 41.9 years ± 14.2 and 40% of them were female. Just one of the 50 patients presented with osmotic symptoms at the time of presentation. Thirty percent of the patient had painful neuropathy at the time of presentation. None of them had retinopathy at baseline. There were no patients having a prior history of pancreatititis, cardiovascular disease, or gastroparesis. None of the patients had a personal or family history of medullary thyroid carcinoma. Two of the patients were smokers.
The application has a personalization that adjusts the treatment based on income of the patient, the preference for oral versus injectable, pill burden etc. Forty percent of the patient was in the high income group, 50% from the mid-income, and 10% from the low income group. (No specific data was used for inferring the income and it was based on the clinician's assessment).
The mean weight of the patient was 87 kg ± 21.5 and the mean BMI was 31.5 kg/m2 ± 7.28. Forty percent were classified as having class III Obesity and three had class II obesity. Twenty percent were overweight. We failed to capture the waist circumference in our patients; hence, the data was not added to the software.
We had failed to capture some key data at the time of the patient initial evaluation. For example, the HDL-C level was not recorded for a large number of the patients. While the ALT values were recorded, AST and platelet counts were not recorded in many of the patients and hence the Fib-4 score could not be calculated. Table 2 shows the analysis of the laboratory data of the patients.
The mean HBA1c at the time of presentation in our patients was 9.29% ± 2.21. The highest HBA1c was 13% at the time of first presentation.
One of the patient had a recent dengue infection and had an abnormally high ALT level. Since this was an outlier, the data was removed and the ALT mean values were calculated from the remaining patients.
Twenty percent of the patients were perceived to have high cardiovascular risk. However, statin was indicated in all patients. Fifty percent of the patients were suggested to require high intensity statin and the other needing moderate intensity statin. None of the patient has retinopathy at the time of presentation. Ten percent patient had painful diabetic neuropathy. Ten percent of the patients has diabetic kidney disease status in G2A1 category and 10% patient had diabetic kidney disease in G1A2 category. The rest has DKD status of G1A1.
Output from the software
Metformin was the most recommended medicine by the software and was recommended to all patients. Insulin was recommended to 50% patients. Interestingly, all were recommended to start premixed insulin by the software. SGLT2 inhibitors was recommended to 30% of the patients while acarbose was recommended to thirty. Ten percent of the patients was recommended GLP-1 Receptor agonist. It is interestingly to note that none of the patients were recommended DPP-IV as the initial treatment by the software.
We compared the output from the software with the actual medication prescribed by us. The average number of medicines that matched between the software suggestion and the actual prescription is approximately 2.1. On average, there are 0.3 medicines suggested by the software but not prescribed, and 1.2 medicines prescribed but not suggested by the software.
In 20% of the treatments, all medicines suggested by the software matched those that were actually prescribed. In 80% of the treatments, there was a partial match between the medicines suggested and those prescribed. There were no treatments where none of the suggested medicines matched the prescribed ones.
We used the Open AI ChatGPT tool to perform some deep analysis which could be missed by us on preliminary statistical analysis. The software found that there was a positive correlation between triglyceride levels and BMI, implying that as BMI increases, triglyceride levels might increase. The urine albumin/creatinine ratio (UACR) positively correlated with height. This was an interesting finding that needs to be looked at further with larger dataset and analysis.
Discussion
Alpha testing is an early phase of the software testing process where the software is tested in-house by the development team or by a specialized testing team within the organization. The primary objective of alpha testing is to identify all possible issues and bugs before releasing the software to beta testing or to the public [10]. This study was an alpha-test to check for functionality and accuracy of Diabetology.co.in which is a novel personalized and precision medicine system developed by us.
The application takes input from the user in form of clinical, laboratory, and personalized data and generates a prescription. We used the application restrospectively in fifty patients with newly diagnosed type 2 diabetes. The application worked as expected without any glitches.
On entering the data, the first thing we realized was that there are many data points that are generally missed by us during routine collection which may be useful for making the right decision and making calculations based on established formulae in patients with type 2 diabetes. For example, we failed to collect the platelet count values and SGOT values because of which we were unable to calculate the Fib-4 score and get treatment optimized from a NASH/NAFLD perspective. Similarly, we failed to collect waist circumference of our patients and hence could not get the correct cluster classification of our patients based on the study by Anjana et al.
Despite this, the application is able to give a treatment recommendation based on entering just two essential parameter, HBA1c and Serum creatinine. However, the accuracy of the treatment decision improves with addition of more data. In almost 100% of cases, the treatment for diabetes matched the treatment recommended by us. There was an exact match in 20% cases and partial match in 80% of cases.
Interestingly, in most cases in which there was a partial match, we prescribed more medication than that recommended by the software. Hence, it can be concluded that the software could potential check overprescription from our side.
In all patients that we prescribed insulin, the use of insulin was recommended by the software application as well. In all cases recommended insulin therapy in newly diagnosed type 2 diabetes patients, premixed/co-formulation insulin was the first choice which aligns with the frequent practice of insulin prescription based in India [11].
The most common medicines that were suggested by the software but not prescribed by us included SGLT2 inhibitor. The most common medicines that were prescribed by us but not suggested by the software included DPP-IV inhibitor and acarbose. This could suggest a potential bias in our prescription methodology.
Although we had a small dataset, we did perform a profile analysis of the fifty newly diagnosed patients whose data we entered into the platform. The analysis was made particular easy by the inbuilt OpenAI-based ChatBot system. The use of this feature could help physicians gain deeper insights into their own prescription practices, biases, and potential data correlation which could be used for future prescriptions. The mean BMI of our patient (31.5 kg/m2 ± 7.28) was higher than that reported in other studies from India. Additionally, the mean HbA1c at the time of presentation in our dataset was higher than that reported by other studies (9.29% ± 2.21) [12]. Our practice is in a more urban affluent setting and this could be the reason for the difference in these parameters.
The AI-based data-analysis found correlation between BMI and triglyceride which has been previously well established. However, it found a positive correlation between the urine albumin/creat ratio (UACR) and the height of the patient. This an interesting finding which needs to be looked at closer in larger studies in future.
To our knowledge, no other software application is capable of generating detailed treatment recommendations while considering multiple parameters, compelling indications, and patients’ personal preferences. The application we are developing has the potential to revolutionize diabetes treatment based on an evidence-based precision medicine approach.
Conclusion
To conclude, the Diabetology.co.in was accurate in suggesting treatment recommendation for newly diagnosed patients with type 2 diabetes mellitus. In all patients, there was a partial or complete match with the prescription given by us vis-a-vis as suggested by the application. The application has potential for reducing over prescription of medications and also reveal any intrinsic bias a clinician may have towards treatment. The output and tables generated from the application have potential to be used in clinical research for deeper insights into our patient data.
References
Misra A. Ethnic-specific criteria for classification of body mass index: a perspective for Asian Indians and American Diabetes Association position statement. Diabetes Technol Ther. 2015;17(9):667–71.
Levey AS, Stevens LA, Schmid CH, Zhang Y, Castro AF III, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–12.
Wong ND, Budoff MJ, Ferdinand K, Graham IM, Michos ED, Reddy T, Shapiro MD, Toth PP. Atherosclerotic cardiovascular disease risk assessment: an American Society for Preventive Cardiology clinical practice statement. Am J Prev Cardiol. 2022;10:100335.
Anjana RM, Baskar V, Nair AT, Jebarani S, Siddiqui MK, Pradeepa R, Unnikrishnan R, Palmer C, Pearson E, Mohan V. Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Res Care. 2020;8(1):e001506.
Singla R, Gupta G, Dutta D, Raizada N, Aggarwal S. Diabetes reversal: Update on current knowledge and proposal of prediction score parameters for diabetes remission. Diabetes Metab Syndr. 2022;16(4):102452. https://doi.org/10.1016/j.dsx.2022.102452.
Wangnoo SK, Sethi B, Sahay RK, John M, Ghosal S, Sharma SK. Treat-to-target trials in diabetes. Indian J Endocrinol Metab. 2014;18(2):166.
Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, Sulkowski MS, Torriani FJ, Dieterich DT, Thomas DL, Messinger D. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–25.
ElSayed NA, Aleppo G, Aroda VR, Bannuru RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D, Johnson EL, Kahan S. 9. Pharmacologic approaches to glycemic treatment: standards of care in diabetes—2023. Diabetes Care. 2023;46(Supplement1):S140-57.
de Boer IH, Caramori ML, Chan JC, Heerspink HJ, Hurst C, Khunti K, Liew A, Michos ED, Navaneethan SD, Olowu WA, Sadusky T. KDIGO 2020 clinical practice guideline for diabetes management in chronic kidney disease. Kidney Int. 2020;98(4):S1-15.
Hulyalkar M, Gleich SJ, Kashyap R, Barwise A, Kaur H, Dong Y, Fan L, Murthy S, Arteaga GM, Tripathi S. Design and α-testing of an electronic rounding tool (CERTAINp) to improve process of care in pediatric intensive care unit. J Clin Monit Comput. 2017;31:1313–20.
Kalra S, Czupryniak L, Kilov G, Lamptey R, Kumar A, Unnikrishnan AG, Boudiba A, Abid M, Akanov ZA, Latheef A, Araz M. Expert opinion: patient selection for premixed insulin formulations in diabetes care. Diabetes Ther. 2018;9:2185–99.
Anjana RM, Unnikrishnan R, Deepa M, Pradeepa R, Tandon N, Das AK, Joshi S, Bajaj S, Jabbar PK, Das HK, Kumar A. Metabolic non-communicable disease health report of India: the ICMR-INDIAB national cross-sectional study (ICMR-INDIAB-17). Lancet Diabetes Endocrinol. 2023;11:474–89.
Funding
Grant from RSSDI Research Retreat in 2023.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical considerations
Approval for this study was granted by the Institutional Ethics Committee in 2023.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lakhani, O.J., Gupta, A., Tripathi, P. et al. An alpha-test of Diabetology.co.in—an algorithm-driven personalized and precision medicine prescription system for treatment-naive patients with type 2 diabetes. Int J Diabetes Dev Ctries (2024). https://doi.org/10.1007/s13410-024-01368-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13410-024-01368-8