Abstract
Background
Chemotherapeutic agents such as cisplatin are commonly used in patients with clinically unresectable or recurrent esophageal cancer (ESCA). However, patients often develop resistance to cisplatin, which in turn leads to a poor prognosis. Studies have shown that FAM111B may be involved in the development of tumors as an oncogene or tumor suppressor gene. However, the pathological role and corresponding mechanism of FAM111B in ESCA are still unclear.
Methods
The GEPIA web tool, ENCORI Pan-Cancer Analysis Platform and UALCAN-TCGA database were used to study the expression of FAM111B in ESCA. CCK-8, angiogenesis, Transwell and xenograft assays were applied to explore the biological function of FAM111B in ESCA. Western blot, RT-qPCR, and RNA-seq analyses were applied to study the FAM111B/GSDMA axis in the progression of ESCA cells. CCK-8 and xenograft assays were used to study the role of the FAM111B/GSDMA axis in determining the sensitivity of ESCA to cisplatin.
Results
Our results demonstrated that FAM111B is highly expressed in ESCA tissues compared to normal tissues. We showed that FAM111B promotes the progression of ESCC cells by binding to GSDMA and that the trypsin protease domain is essential for the activity of FAM111B. Furthermore, we showed that the FAM111B/GSDMA axis regulates cisplatin sensitivity in ESCA.
Conclusions
Overall, we identified a novel FAM111B/GSDMA axis regulating ESCA tumorigenesis and chemosensitivity, at least in ESCC cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Esophageal cancer (ESCA) is a malignant tumor derived from the esophageal epithelium. Esophageal squamous cell carcinoma (ESCC) is the major histological subtype of ESCA and exhibits distinct characteristics [1]. Over the past two decades, although the development of new cancer treatment models such as targeted therapy and immunotherapy has benefited some ESCA patients, the overall prognosis of ESCA patients has not been significantly improved, and the 5-year survival rate is less than 20% [2]. Therefore, exploring new treatment strategies remains a hot topic in ESCA research. Chemotherapy drugs play an irreplaceable role in the first-line treatment and postoperative adjuvant therapy of ESCA patients. At present, the usual clinical treatments for ESCA patients with unresectable or relapsed tumors are cisplatin-based chemotherapy regimens [1, 3]. However, patients often develop drug resistance, leading to a poor prognosis [4]. Therefore, it is necessary to elucidate the molecular mechanisms of cisplatin resistance in ESCA to further improve the prognosis of patients.
Cisplatin resistance is regulated by complex mechanisms in malignant tumor biology, especially the activation of tumor-related signaling pathways [5]. However, the factors associated with cisplatin resistance in ESCA have not been fully elucidated. These factors include the involvement of multiple transporters, such as the solute carrier superfamily (SLC) [6], copper transporter 1/2 (CTR1/2) [7], and multidrug resistance protein subfamily (MPR) membrane transporters [8], in the influx/efflux of platinum agents that can induce resistance [9]. Similarly, platinum agents can lead to drug resistance through binding and inactivation of related proteins, such as glutathione (GSH), methionine, and metallothionein [10]. In addition, the nucleotide excision repair (NER) system in tumor cells can induce cisplatin resistance by removing damaged nucleotides and synthesizing DNA to reestablish genetic integrity [9, 11]. Other factors include apoptosis and autophagy [9]. Therefore, exploring the mechanisms of cisplatin resistance in ESCA will aid in the development of new therapeutic strategies.
The FAM111B gene is located on human chromosome 11q12.1 and is characterized as a protein with a trypsin-like cysteine/serine peptidase structural domain [12]. Initially, Mercier identified mutated FAM111B as the causative gene in a rare autosomal dominant disorder called Poikiloderma disease by whole-exome sequencing (WES) [13]. With the deepening of research, increasing evidence has shown that FAM111B can participate in occurrence and development as an oncogene or tumor suppressor gene and can thus affect prognosis [12, 14,15,16]. However, the pathological role and biological function of FAM111B in ESCA are not clear.
In this study, we found that FAM111B was upregulated in ESCA through bioinformatics analysis in the Gene Expression Profiling Interactive Analysis (GEPIA) web tool, ENCORI Pan-Cancer Analysis Platform and University of Alabama at Birmingham Cancer-The Cancer Genome Atlas (UALCAN-TCGA) database. Subsequent studies showed that knockdown of FAM111B in ESCA cells attenuated cell proliferation and migration. Then, RNA-seq was performed for FAM111B and showed that FAM111B silencing suppressed the proteasome signaling pathway, DNA replication and DNA damage repair pathways to block tumor growth. Furthermore, mass spectrometry indicated that FAM111B may bind to GSDMA, and RNA-seq was performed for GSDMA, which indicated that GSDMA silencing altered the chemical carcinogenesis DNA adduct pathway to sensitize ESCA cells to cisplatin. Therefore, we identified a FAM111B/GSDMA axis that modulates cisplatin sensitivity to regulate the malignant biological behavior of ESCA cells.
2 Method and Materials
2.1 Cell lines and cell culture
The ESCC cell lines KYSE410 (#SC0538, Shanghai, China) and TE-1 (#SC0150, Shanghai, China) were purchased from Yuchi biology and identified by short tandem repeat (STR) profiling. The KYSE410 and TE-1 cells were both cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (AC03L055, Shanghai Lifei Lab Biotech, China) and antibiotics. All cell lines were cultured at 37 °C in 5% carbon dioxide.
2.2 Antibodies and chemical agents
The primary antibodies were used in the study: FAM111B (#29407–1-AP, Proteintech, 1:1000 dilution), GSDMA (#ab230768, Abcam, 1:1000 dilution), SNAI1 (#13099–1-AP, Proteintech, 1:1000 dilution), cleaved caspase 3 (#9661, Cell Signaling Technology, 1:1000 dilution) and beta actin (#66009–1-Ig, Proteintech, 1:5000 dilution). The FAM111B and GSDMA shRNAs were purchased from GeneCopoeia (USA). The cisplatin (CDDP) was purchased from MedChemExpress (New Jersey, USA). The sequence of shRNAs as follows:
shFAM111B #1:
-
5'-CACCGCGACAGATTTCTCCTCAACTCGAGTTGAGGAGAAATCTGTCGC-3'; shFAM111B #2:
-
5'-CACCGGGTGAGACTATTGAAGGACTCGAGTCCTTCAATAGTCTCACCC-3'; shGSDMA #1:
-
5'-CACCGAAGGAGATGCAAGATCAACTCGAGTTGATCTTGCATCTCCTTC-3'; shGSDMA #2:
-
5'-CACCGCGCCATCCTCTATTTCGTCTCGAGACGAAATAGAGGATGGCGC-3'.
2.3 Co-immunoprecipitation and western blot analysis
For the co-immunoprecipitation (co-IP) experiment, 1 ml of RIPA buffer (#P0013, Beyotime, China) was added to ESCA cells in a 10 cm dish to lysis, fully blown and placed on ice for 30 min. Then, the lysed cells were transferred into a 1.5 ml Eppendorf tube and centrifuged at 12,000 × g for 15 min at 4 °C. After centrifugation, the cell supernatant was carefully drawn into a new 1.5 ml Eppendorf tube, incubated with protein A + G beads (#P2029, Beyotime, China) and a primary antibody or IgG (#A7007, Beyotime, China), and shaken at 4 °C overnight. The next day, the beads were centrifuged at 3000 rpm for 1 min, the supernatant was carefully discarded and repeatedly washed with RIPA buffer for 6 times. Then added 60 μl 1 × loading buffer. Finally, the beads containing 1 × loading buffer were boiled at 100 °C for 10 min.
For the western blotting experiment, the steps of lysing ESCA cells were consistent with the co-IP experiment. The differences were that the supernatant after centrifugation was carefully drawn into a new 1.5 ml Eppendorf tube, and then an appropriate amount of 5 × buffer was added. Next, the cell lysate containing 5 × buffer was boiled at 100 °C for 10 min. Finally, the boiled protein lysates were subjected to electrophoresis by using SDS-PAGE gels. The results were measured by Image J software.
2.4 Tissue microarray and immunohistochemistry (IHC)
Tissue microarray specimens (# D081ES01T, China) were purchased from Bioaitech, immunostained with FAM111B (#29,407–1-AP, Proteintech, 1:200 dilution) and GSDMA (#ab230768, Abcam, 1:1000 dilution). Transplanted tumors of nude mice immunostained with KI67 (#27,309–1-AP, Proteintech, 1:5000 dilution), FAM111B (#29,407–1-AP, Proteintech, 1:200 dilution), SNAI1 (#13,099–1-AP, Proteintech, 1:500 dilution) and cleaved caspase 3 (#9661, Cell Signaling Technology, 1:400 dilution). Dyeing intensity was graded/scored by blind method. 1 = showed weak staining (100 × magnification), but almost no staining (40 × magnification); 2 = showed medium staining (40 × magnification); 3 = showed strong staining (40 × magnification). The formula: staining intensity × percentage of positive cells to calculate the final staining index.
2.5 Quantitative real-time PCR (RT‒qPCR) analysis
The RT-qPCR method was reported previously [17]. Total cellular RNA was extracted using TRIzol reagent (#AG21102, Accurate Biotechnology, Hunan, China). Reverse transcription kits (#AG11728, Accurate Biotechnology, Hunan, China) and PCR kits (#AG11701, Accurate Biotechnology, Hunan, China) were used for RT-qPCR according to the instructions. All values were standardized by the corresponding β-actin values, and the 2-ΔΔCt method was used to quantify the fold change. The sequence of primers for RT-qPCR as follows:
-
FAM111B forward: 5′-GGAATGCCTCTCAAGTGCCT-3′;
-
FAM111B reverse: 5′-CTGGGAATTGTCTGGCGAGT-3′;
-
GSDMA forward: 5′-TACAGACCCAGTGAGCCCTA-3′;
-
GSDMA reverse: 5′-AGCTGTCTCACTCGAAAGGC-3′;
-
HUWE1 forward: 5′-CTAGCCGCATCTTCGGGG-3′;
-
HUWE1 reverse: 5′-GTGTAGTTGCACTGGGTGGA-3′;
-
KDM6A forward: 5′-GTGGAGCCTACCAATTCCCG-3′;
-
KDM6A reverse: 5′-CGTACCTGTGCAACTCCTGT-3′;
-
MUC16 forward: 5′-AGTCTCAGGGACTCTCTCGG-3′;
-
MUC16 reverse: 5′-TGGTTTGGGCAAAGACCACT-3′;
-
SNAIl forward: 5′-CCTGTCTGCGTGGGTTTTTG-3′;
-
SNAIl reverse: 5′-CTGAAATAGCTGCCTGGGCT-3′;
-
β-actin forward: 5′-AAAGACCTGTACGCCAACAC-3′;
-
β-actin reverse: 5′-GTCATACTCCTGCTTGCTGAT-3′;
Data presents as mean ± SD of three independent experiments.
2.6 Transwell assay
ESCA cells were divided into different treatment groups after intervention. Different groups of cells diluted by serum-free were added to the upper layer of Transwell chamber in 24-well plate (2 × 104 cells per well), and 500 μl of 15% FBS medium was added to the lower layer, placed in 37 °C, 5% CO2 incubator for 24 h. The next day, cells were washed twice with PBS and fixed with methanol for 30 min, then washed twice with PBS. Next, the cells were stained with 0.1% crystal violet for 30 min and washed with PBS three times. Finally, the results can be obtained by taking a photo count.
2.7 Cell counting Kit-8 (CCK-8) assay
The KYSE410 or TE-1 cells were seeded in 96-well plates (approximately 104 cells per well) and cultured at 37 °C in 5% CO2 for 24 h. Divided into different treatment groups, each group repeated at least 3 times. Each well was added with 10 μl CCK-8 reagent (#C0037, Beyotime, China) and incubated for 1 h. The absorbance values of different groups at 450 nm were measured by microplate reader, and the proliferation of different groups could be calculated.
2.8 Nude mouse xenograft assay
We selected half male and half female nude mice for the experiment (gender had no effect on the experimental results). BALB/C-nu/nu mice (6 weeks old, 22–24 g) were obtained from Hunan SJA Laboratory Animal Company, were raised in the animal center of Xiangya Hospital, humidity 60 ± 3%, temperature 22 ± 0.5 °C; automatic control of light and dark cycle. Food and water were freely available. The ESCA cells were subcutaneously injected on the left side of the back of the nude mice (about 1 × 107 cells/mouse). The length and width of the tumor were measured every 2 days, and the volume was calculated by formula (L × W2)/2. The drug was administered until the tumor reached about 100 mm3 (cisplatin, 1 mg/kg, twice a week). The mice were euthanized at appropriate time, and subcutaneous tumors were collected for further study. All animal experiments in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of Xiangya Hospital, Central South University.
2.9 RNA sequencing analysis
The KYSE410 cells were transfected with siNC or siFAM111B/siGSDMA, and the cells were collected after 48 h. The RNA sequencing (RNA-seq) was subsequently performed by Novogene (Beijing, China). In short, 1 μg RNA per sample was used as the starting material for RNA-seq. The NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) was used to generate sequencing libraries according to the manufacturer's instructions, and an index code was added to each sample to assign sequence attributes. TruSeq PE Cluster Kit v3-cBot-HS (Illumina) was used to cluster samples on the cBot Cluster Generation System according to the manufacturer's instructions. After clustering, the library was sequenced on the Illumina Novaseq platform, resulting in a paired-end reads of 150 bp. The number of reads mapped to each gene was counted using Feature Counts v1.5.0-p3. The DESeq2R package (1.16.1) was used for differential expression analysis, differentially expressed genes (DEGs) were statistically enriched in the KEGG pathways through the clusterProfiler R package.
2.10 Statistical analysis
The experimental data were expressed as the mean ± SD. The sample size (n) for each statistical analysis was provided in the legend. p values were calculated using GraphPad Prism 5 software. The difference was considered statistically significant when the p < 0.05.
3 Results
3.1 FAM111B is abnormally overexpressed in ESCA
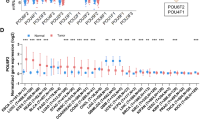
The cancer-related role of FAM111B in ESCA is unclear. First, we performed bioinformatics analysis in the GEPIA web tool, ENCORI Pan-Cancer Analysis Platform and UALCAN-TCGA database, and found that FAM111B expression was upregulated in ESCA tissues compared to nontumor esophageal tissues (Fig. 1A-C). Then, we analyzed the protein levels of FAM111B in samples derived from patients with ESCC. We also found that FAM111B was upregulated in ESCC tissues compared with adjacent tissues (p < 0.05) (Fig. 1D, E). Moreover, we performed IHC staining to measure the protein level of FAM111B in tumor tissues and nontumor tissues in a tissue microarray including samples from a cohort of patients with ESCC (N = 15). Representative images are shown in Fig. 1F. Similar to the above findings, FAM111B had a higher expression level in ESCC tissues than in nontumor esophageal tissues (Fig. 1G). We also noticed that the protein and mRNA expression levels of FAM111B in ESCA cell lines were greater than those in HET-1A cells, which are derived from normal esophageal tissue (Fig. 1H, I). Finally, analyses of the Gene Expression Omnibus (GEO) dataset GSE53622 showed that ESCA patients (patients were divided into high and low FAM111B expression groups based on the optimal cut-off value) with high expression of FAM111B have shorter overall survival (OS) time (p = 0.036, Fig. 1J). In addition, to further explore the correlation between high expression of FAM111B in ESCA patients and clinical prognosis, we performed univariate and multivariate Cox regression analyses using the Gene Expression Omnibus (GEO) dataset GSE53622. However, the results of the univariate Cox analysis showed that only ESCA stage (hazard ratio: 2.123, 95% CI: 1.152 − 3.912, p = 0.016) was a poor prognostic indicator for OS (Fig. S1A). Multivariate Cox analysis showed that the expression of FAM111B (hazard ratio: 2.12, 95% CI: 1.06 − 4. 2, p = 0.033) and ESCA stage (hazard ratio: 3.67, 95% CI: 1.13 − 11.9, p = 0.03) were poor prognostic indicators for OS (Fig. S1B). Moreover, we collected relevant clinicopathological information from 76 ESCC patients in this study (Table 1). Together, our results suggest that FAM111B is abnormally overexpressed in ESCA.
FAM111B is highly expressed in ESCA. A-C, Differential expression analysis of FAM111B between tumor and normal tissues by GEPIA web tool, ENCORI Pan-Cancer Analysis Platform and UALCAN-TCGA database. *, p < 0.05. D and E, The protein expression levels of FAM111B in the adjacent non-tumor esophageal tissues (n = 20) and ESCA tissues (n = 20) were analyzed by the western blot. The protein levels of FAM111B were quantified by the image J software. F and G, The IHC staining was performed in the tissue microarray of ESCA and normal esophageal tissues by using the FAM111B antibodies. H and I, The esophageal normal or cancer cell lines were harvested for Western blot and RT-qPCR analysis. Data presents as mean ± SD with three replicates. ***, p < 0.001. J, The prognosis of FAM111B in ESCA cancer was determined by the GEO dataset GSE53622. p value as indicated
3.2 FAM111B promotes the proliferation and invasion of ESCA cells
Since the clinicopathological features related to FAM111B expression indicated that FAM111B is an oncogenic protein in ESCA, we investigated the biological effect of FAM111B in ESCA cells. Two different shRNAs were applied to knockdown FAM111B in KYSE410 and TE-1 cells (Fig. 2A, B). The Cell Counting Kit-8 (CCK-8) assay, Transwell assay and angiogenesis assay indicated that FAM111B silencing blocked ESCC cell growth, invasion and angiogenesis respectively (Fig. 2C-F). In contrast, overexpression of FAM111B enhanced the growth, invasion and angiogenesis of ESCC cells (Fig. 2G-L). Moreover, the nude mouse xenograft experiments showed that knockdown of FAM111B inhibited the growth of tumors derived from KYSE410 and TE-1 cells, and immunohistochemical (IHC) staining of nude mouse xenografts showed reduced expression levels of FAM111B and a tumor cell proliferation marker (Ki-67) in nude mouse xenografts of the shFAM111B group (Fig. 3A-J). To further validate this conclusion, we injected KYSE410 cells transfected with shFAM111B or empty vector into the inner left thigh of each mouse to generate a lymph node metastasis model. The left popliteal lymph nodes were isolated 21 days after injection. The lymph nodes of mice in the knockdown FAM111B group were significantly smaller than those in the control group (Fig. S1C, D). Then, we analyzed the RNA levels of EMT-associated proteins based on RNA-seq data of cells with different level of FAM111B, and we found that only the RNA level of SNAI1 (also known as snail) was decreased after knockdown of FAM111B (Fig. S1E), which was further verified by RT-qPCR, Western blotting and immunohistochemistry (Fig. S1F-H). Therefore, our data indicate that knockdown of FAM111B inhibits the progression of ESCA in cells and in mice.
Knockdown of FAM111B attenuates the progression of ESCA in vitro. A-F, KYSE410 and TE-1 cells were infected with shControl, shFAM111B #1, or shFAM111B #2 for 72 h. Cells were collected for Western blot analysis (A), RT-qPCR analysis (B), CCK-8 assay (C), Transwell assay (D) and angiogenesis (E, F). Data presents as mean ± SD with three replicates. **, p < 0.01; ***, p < 0.001. G-L, KYSE410 and TE-1 cells were transfected with HA-FAM111B for 24 h. Cells were collected for Western blot and RT-qPCR analysis (G), CCK-8 assay (H), Transwell assay (I, J) and angiogenesis (K, L). Data presents as mean ± SD with three replicates. **, p < 0.01; ***, p < 0.001
Knockdown of FAM111B attenuates the progression of ESCA in vivo. A-E, KYSE410 cells were transfected with indicated shRNAs for 72 h. After puromycin selection, cells were subcutaneously injected into the nude mice. The tumor image was shown in panel A, the tumor mass was shown in panel B, the tumor growth curve was shown in panel C. The excised tumors were subjected to IHC staining of FAM111B (D) or Ki-67 (E). Data presents as mean ± SD with six replicates. ***, p < 0.001. F-J, TE-1 cells were transfected with indicated shRNAs for 72 h. After puromycin selection, cells were subcutaneously injected into the nude mice. The tumor image was shown in panel F, the tumor mass was shown in panel G, the tumor growth curve was shown in panel H. The excised tumors were subjected to IHC staining of FAM111B (I) or Ki-67 (J). Data presents as mean ± SD with six replicates. ***, p < 0.001
3.3 FAM111B activates the proteasome signaling pathway, DNA replication and DNA damage repair pathways in ESCA
To further explore the mechanism of the tumor-promoting effect of FAM111B in ESCA, RNA-seq was performed after knockdown of FAM111B in KYSE410 cells (Fig. 4A). KEGG pathway enrichment analysis showed that FAM111B silencing suppressed numerous signaling pathways, such as the MAPK signaling pathway, proteoglycans in cancer, PI3K-Akt signaling pathway, and ubiquitin-mediated proteolysis pathway in ESCA cells (Fig. 4B). Given that FAM111B is reported to degrade p16 to promote the progression of lung adenocarcinoma [18], our finding was consistent with previous reports. On the other hand, gene set enrichment analysis (GSEA) also showed that knockdown of FAM111B was involved in the reduction of the proteasome signaling pathway, nucleotide excision repair signaling pathway, mismatch repair signaling pathway, and homologous recombination signaling pathway (Fig. 4C). Moreover, we performed GSEA of the TCGA-ESCA database to study the pathways in which FAM111B was involved, patients were divided into high and low FAM111B expression groups based on the median value (Fig. 4D). Similarly, we demonstrated that the DNA replication signaling pathway, homologous recombination pathway, Fanconi anemia pathway, nucleotide excision pathway, mismatch pathway, and non-homologous end-joining pathway had the most prominent positive association with FAM111B in ESCA (Fig. 4E). Consistently, GSEA of the TCGA-ESCA database showed that differentially expressed genes related to FAM111B were involved in the activation of the ubiquitin-mediated proteolysis pathway, cell cycle, mismatch repair, and homologous recombination signaling pathway in ESCA (Fig. 4F). It has been well documented that the DNA replication signaling pathway and cell cycle pathway represent tumor growth ability. Meanwhile, the homologous recombination pathway, Fanconi anemia pathway, nucleotide excision pathway, mismatch pathway, and non-homologous end-joining pathway are all involved in DNA damage repair. Thus, our results showed that FAM111B plays an important role in modulating the proteasome signaling pathway, DNA replication and DNA damage repair pathways in ESCA.
FAM111B activates the proteasome signaling pathway, DNA replication and DNA damage repair pathways in ESCA. A, KYSE410 cells were infected with shControl or shFAM111B for 72 h. Cells were subjected to RNA-seq analysis. B, KEGG pathway enrichment analysis indicated that the pathways were changed after knockdown of FAM111B in KYSE410 cells. C, GSEA analysis showed proteasome signaling pathway, nucleotide excision repair pathway, mismatch repair pathway, and homologous recombination pathway were activated by knocking down FAM111B in KYSE410 cells. D, Heatmap to show the differentially expressed genes between the FAM111B-high and FAM111B-low expression groups. E, Pathway enrichment analysis of the TCGA-ESCA database. F, GSEA enrichment analysis of the TCGA database
3.4 FAM111B interacts with GSDMA and degrades GSDMA expression in ESCA
Since downregulation of FAM111B is important for the progression of ESCA, the regulatory mechanism of FAM111B in ESCA is worthy of further study. We performed mass spectrometry to identify the potential binding partners of FAM111B, revealing that FAM111B may bind to GSDMA (Fig. 5A). This finding was confirmed by immunoprecipitation in both KYSE410 and TE-1 cells (Fig. 5B). We further evaluated the relationship between FAM111B and GSDMA in ESCA cells after knocking down FAM111B. We found that FAM111B silencing increased the protein level of GSDMA and had no effect on the mRNA level of GSDMA in ESCA cells (Fig. 5C, D). In contrast, overexpression of FAM111B reduced the level of GSDMA protein in ESCA cells and had no effect on the mRNA level (Fig. 5E, F). Moreover, we designed a plasmid that removed the peptidase domain of the FAM111B gene (HA-FAM111B ∆TPD) and found that HA-FAM111B ∆TPD had no effect on the protein level of GSDMA in either KYSE410 or TE-1 cells (Fig. 5G-I). We also measured the protein levels of FAM111B and GSDMA in an ESCA tissue microarray by IHC (Fig. 5J, K). Our results indicated that FAM111B expression was negatively correlated with GSDMA expression. Together, these findings support that FAM111B is a binding partner of GSDMA and degrades GSDMA in ESCA cells. More importantly, the trypsin protease domain is essential for the activity of FAM111B in ESCA cells.
FAM111B interacts with GSDMA and decreases GSDMA expression in ESCA. A, The mass spectrometry results showed that FAM111B and GSDMA were bound to each other. B, KYSE410 and TE-1 cells were harvested and immunoprecipitated with IgG and FAM111B or GSDMA antibodies. C and D, KYSE410 and TE-1 cells were infected with indicated shRNAs for 72 h. Cells were harvested for Western blot (C) and RT-qPCR analysis (D). Data presents as mean ± SD with three replicates. Ns, not significant. E and F, KYSE410 and TE-1 cells were transfected with HA-FAM111B for 24 h. Cells were collected for Western blot analysis (E) and RT-qPCR analysis (F). Data presents as mean ± SD with three replicates. Ns, not significant. G, The structure difference of FAM111B and FAM111B ∆TPD. H and I, KYSE410 and TE-1 cells were transfected with indicated HA-FAM111B/HA-FAM111B ∆TPD plasmids for 48 h. Cells were collected for Western blot analysis. Ns, not significant. ***, p < 0.001. J and K, The IHC staining was performed in the tissue microarray of ESCA by using the FAM111B and GSDMA antibodies
3.5 The downregulation of GSDMA promotes the proliferation and invasion of ESCA cells
Then, we studied the biological role of GSDMA in ESCA cells. First, RNA-seq was performed after knockdown of GSDMA in KYSE410 cells (Fig. 6A). KEGG pathway enrichment analysis of the RNA-seq database showed that knockdown of GSDMA activated numerous cellular processes and signaling pathways (Fig. 6B). Among them, the chemical carcinogenesis DNA adduct pathway and drug metabolism-other enzymes pathway were activated after GSDMA silencing (Fig. 6B), which was opposite to FAM111B (Fig. 4E). The expression level of GSDMA was decreased after transfection with GSDMA-specific shRNAs (Fig. 6C, D). We showed that knockdown of GSDMA promoted cancer cell invasion and proliferation by Transwell assay and CCK-8 assay in both KYSE410 and TE-1 cells (Fig. 6E, F). We further found that co-transfection of cells with shGSDMA and shFAM111B attenuated the induction of cancer cell proliferation and invasion mediated by knockdown of FAM111B alone (Fig. 6G-I). Moreover, the nude mouse xenograft experiments showed that co-transfection of shGSDMA and shFAM111B in KYSE410 cells attenuated the proliferation of tumors mediated by knockdown of FAM111B alone (Fig. S2A, B). On the other hand, transfection of KYSE410 and TE-1 cells with HA-FAM111B plasmids did not further enhance the cell proliferation and invasion abilities compared to knockdown of GSDMA alone (Fig. 6J, K). Together, our data suggest that knockdown of GSDMA promotes the proliferation and invasion of ESCA cells and that GSMDA contributes to mediating the cancer-related function of FAM111B in ESCA.
The downregulation of GSDMA promotes the proliferation and invasion of ESCA cells. A, KYSE410 cells were infected with shControl or shGSDMA for 72 h. Cells were subjected to RNA-seq analysis. B, KEGG pathway enrichment analysis indicated that the pathways were changed after knockdown of GSDMA in KYSE410 cells. C-F, KYSE410 and TE-1 cells were infected with shControl, shGSDMA #1, or shGSDMA #2 for 72 h. Cells were collected for Western blot analysis (C), RT-qPCR analysis (D), Transwell assay (E) and CCK-8 assay (F). Data presents as mean ± SD with three replicates. **, p < 0.01; ***, p < 0.001. G-M, KYSE410 and TE-1 cells were transfected with indicated constructs for 72 h or 24 h. Cells were collected for Western blot analysis (G, J), Transwell assay (H, L) and CCK-8 assay (I, M). Data presents as mean ± SD with three replicates. Ns, not significant. ***, p < 0.001
3.6 The FAM111B/GSDMA axis regulates the sensitivity of ESCA cells to cisplatin
Next, we searched for genes commonly regulated by both FAM111B and GSDMA through RNA-seq analysis, and the results are shown in Fig. 7A, B, C. Seventeen genes (ERLIN1, DUT, ACKR3, GBP1, ZFHX3, LUZP1, CBL, KDM6A, AGO2, SCAF4, MUC16, ACTG1, HUWE1, B3GALT5, SPEN, CCN2, NAV2) were regulated by both FAM111B and GSDMA in KYSE410 cells. The above results indicated that drug metabolism-associated pathways are commonly modulated by FAM111B and GSDMA (Fig. 4E and 6B). Given that cisplatin is the major agent for the chemotherapy of ESCA, dysregulation of DNA damage repair associated pathways is involved in regulating the ESCA cells to cisplatin. Our results also demonstrated that FAM111B was closely correlated with the activation of DNA damage repair pathways (Fig. 4B, 4C, 4E, 4F). Notably, previous studies have shown that three genes (HUWE1, KDM6A and MUC16) commonly regulated by FAM111B and GSDMA are associated with cisplatin resistance [19,20,21]. Thus, we investigated whether the FAM111B/GSDMA axis regulates the sensitivity of ESCA cells to cisplatin. First, we demonstrated that HUWE1, KDM6A and MUC16 were regulated by FAM111B and GSDMA in both TE-1 and KYSE410 cells (Fig. 7D). Then, we showed that knockdown of FAM111B enhanced the sensitivity of TE-1 and KYSE410 cells to cisplatin, but inhibition of GSDMA increased the IC50 values of cisplatin in TE-1 and KYSE410 cells (Fig. 7E). We also indicated that depletion of GSDMA attenuated the change in cisplatin IC50 values induced by knockdown of FAM111B in both KYSE410 and TE-1 cells (Fig. 7E). Then, we showed that knockdown of FAM111B increased the cleaved caspase 3 levels compared to those in the control group in KYSE410 and TE-1 cells after cisplatin treatment. Knockdown of GSDMA reduced the cleaved caspase 3 levels compared to those in the control group in KYSE410 and TE-1 cells after cisplatin treatment (Fig. S2C, D). Moreover, co-knockdown of FAM111B and GSDMA attenuated the changes in cleaved caspase 3 compared to knockdown of FAM111B alone in KYSE410 and TE-1 cells after cisplatin treatment (Fig. S2C, D). Subsequent in vivo cell proliferation studies showed that knockdown of GSDMA blocked FAM111B-mediated regulation of the sensitivity of ESCA cells to cisplatin (Fig. 7F-H). In addition, IHC staining assay by detecting cleaved caspase 3 in nude mice transplanted tumors also showed that GSDMA diminished the effect of FAM111B on regulating the apoptosis induced by cisplatin treatment (Fig. S2E, F). In conclusion, our results suggest that FAM111B regulates the sensitivity of ESCA cells to cisplatin via GSDMA.
The FAM111B/GSDMA axis regulates the sensitivity of cisplatin in ESCA. A-C, The genes intersection of the RNA-seq of FAM111B and GSDMA. D, KYSE410 and TE-1 cells were transfected with indicated constructs for 72 h or 24 h. Cells were collected for RT-qPCR analysis. Data presents as mean ± SD with three replicates. Ns, not significant; **, p < 0.01; ***, p < 0.001. E, TE-1 and KYSE410 cells were infected with indicated shRNAs for 72 h. Then, these cells were treated with cisplatin and harvested for CCK-8 assay. F and G, TE-1 cells were transfected with indicated shRNAs for 72 h. Cells were collected and subcutaneously injected into the nude mice. These mice were treated with or without cisplatin. The tumor growth curve was shown in panel G and H. Data presents as mean ± SD with six replicates. ***, p < 0.001
4 Discussion
Cisplatin is the main agent in ESCA chemotherapy. Therefore, exploring the underlying mechanism that regulates the sensitivity of ESCA cells to cisplatin is important for prolonging the survival time of patients with ESCA. Previous studies have shown that HUWE1 can enhance the toxicity of platinum drugs to cells through the DNA damage repair pathway [19, 22]. Our results also proved that FAM111B is closely related to the activation of DNA damage repair pathways in the RNA-seq analysis of ESCA cells. He et al. found that KDM6A acts as a demethylating enzyme to degrade H3K27me3, which in turn leads to resistance to cisplatin in osteosarcoma [20]. In addition, studies have shown that patients with elevated expression of MUC16 in lung cancer, gastric cancer, ovarian cancer and prostate cancer exhibit resistance to cisplatin [21, 23,24,25]. Interestingly, our study showed that knockdown of FAM111B reduced the mRNA levels of HUWE1, KDM6A and MUC16 in ESCA cells. However, knocking down GSDMA led to the opposite results. Therefore, we further explored the relationship between the FAM111B/GSDMA axis and cisplatin in KYSE410 and TE-1 cells. We found that knockdown of FAM111B improved sensitivity to cisplatin, while inhibition of GSDMA increased the IC50 values of cisplatin in ESCA cells, and depletion of GSDMA attenuated the change induced by knockdown of FAM111B. Subsequent nude mouse xenograft experiments also demonstrated that depletion of GSDMA attenuated the FAM111B-mediated regulation of cisplatin sensitivity in ESCA cells. Our results revealed that the FAM111B/GSDMA axis was critical for the sensitivity of ESCA cells to cisplatin. However, the cancer-related function and associated mechanism of the FAM111B/GSDMA axis should be studied in the future.
Pyroptosis, a novel inflammatory form of programmed cell death, is mediated by pore-forming proteins, such as gasdermin family proteins (GSDMs). Recent studies suggest that the role of pyroptosis in tumor progression is controversial. On the one hand, inducing pyroptosis can kill tumor cells, but the resulting inflammatory environment can promote tumor proliferation and metastasis [26]. However, the association between pyroptosis and ESCA is poorly understood. GSDMs are pore-forming proteins, mainly gasdermins A-E and pejvakin [27]. GSDMD was originally discovered as a downstream effector of the inflammasome by Shi et al. Upon caspase cleavage, GSDMD releases the gasdermin-N and gasdermin-C structural domains. The gasdermin-N structural domain binds to membrane phospholipids, which in turn disrupts membrane permeability and leads to cell swelling [28]. In addition to GSDMD, other members of the GSDM family can also penetrate cell membranes and activate pyroptosis [29]. Gasdermin A participates in the apoptosis of tumor cells by regulating mitochondrial homeostasis, which in turn regulates tumor development [30]. SpeB, a streptococcal protease, can induce cell pyroptosis by lysing human GSDMA and murine GSDMA [31]. Furthermore, pyroptosis-induced inflammation can act synergistically with checkpoint blockade to induce antitumor immunity [32, 33]. Here, we showed that GSDMA suppressed the progression of ESCA, which might be mediated by pyroptosis. However, we did not further explore the relationship between GSDMA and pyroptosis in this study. In future exploration, we will focus on the specific role and related molecular mechanisms of GSDMA and pyroptosis in the progression of ESCA.
FAM111B is a protein containing the structural domain of a trypsin-like cysteine/serine peptidase [12]. With the deepening of research, increasing evidence shows that aberrant expression of FAM111B is closely related to tumorigenesis and tumor progression [14, 16]. Kawasaki et al. found that FAM111B enhances the proliferation and migration of lung adenocarcinoma cells by degrading p16 [18], our findings indicate that FAM111B can regulate the progression of ESCA by degrading GSDMA and the trypsin protease domain is essential for the activity of FAM111B, which is consistent with previous report. In this study, we found that FAM111B is abnormally highly expressed in ESCA. Subsequently, in vitro and in vivo experiments showed that knockdown of FAM111B inhibited the growth and lymph node metastasis of ESCC cells. We need more experiments to show whether knockdown of FAM111B can inhibit the liver or lung metastasis of ESCC cells. Moreover, we performed univariate and multivariate Cox analyses of FAM111B expression in ESCA with the TCGA-ESCA dataset. Although the result of univariate COX analysis of the GEO dataset GSE53622 did not support FAM111B expression as an independent risk predictor in ESCA, we still believe that abnormal FAM111B expression has some value in the evaluating clinical prognosis and deciding on a treatment strategy for ESCA patients, and we hope to support our finding through more investigations in the future.
Studies on FAM111B gene mutations have mainly focused on hereditary-related diseases and have shown that autosomal dominant mutations in the FAM111B gene can lead to hereditary fibrosing poikiloderma with tendon contractures, myopathy, and pulmonary fibrosis (POIKTMP) [34, 35]. The POIKTMP mutation is in the hinge or enzyme domain within the FAM111B cluster [36]. Arowolo et al. showed that mutations in the protease domain of FAM111B have a more severe disease phenotype than those in the hinge [37]. However, Sandra et al. showed that FAM111B mutation is associated with increased susceptibility to pancreatic cancer [15], suggesting that the FAM111B mutation may promote cancer formation. Recent studies have found that disease-related mutations in FAM111B cause excessive activation [38], indicating that tumorigenesis may be due to dysfunctional protease activity. Our findings suggest that FAM111B is aberrantly overexpressed in ESCA and that FAM111B can activate the proteasome signaling pathway, DNA replication and DNA damage repair pathways to participate in the development of ESCA as an oncogene. However, more experiments are needed to prove whether there is a highly mutated FAM111B in ESCA. In future studies, we will focus on this issue and explore the correlation between highly mutated FAM111B and ESCA progression and cisplatin resistance. Overall, the shortcoming of this study is that due to the limitation of clinical samples, we only used ESCC cell lines (KYSE410 and TE-1) and ESCC tissues to prove our hypothesis. In the future, we will explore the association of FAM111B with esophageal adenocarcinoma, as well as other types of esophageal tumors, and continue to investigate the relationship between the FAM111B/GSDMA axis and ESCA progression. In addition, we will pay more attention to the relationship between the FAM111B/GSDMA axis and the proteasome signaling pathway, DNA replication and DNA damage repair pathways and pyroptosis in ESCA development and treatment.
Collectively, we demonstrated that FAM111B is overexpressed in ESCA. Then, we found that knockdown of FAM111B inhibits the progression of ESCC cells. Moreover, analysis of FAM111B RNA-seq data indicated that FAM111B can activate the proteasome signaling pathway, DNA replication and DNA damage repair pathways to participate in the development of ESCA. Furthermore, FAM111B interacts with GSDMA and regulates its stability. More importantly, we revealed that the trypsin protease domain is essential for the activity of FAM111B. Subsequently, our data suggest that knockdown of GSDMA promotes the proliferation and invasion of ESCA cells. Moreover, analysis of GSDMA RNA-seq data indicated that GSDMA silencing altered the chemical carcinogenesis DNA adduct pathway. Finally, we showed that FAM111B regulates the sensitivity of ESCA cells to cisplatin via GSDMA. In summary, we identified a novel FAM111B/GSDMA axis modulating the tumorigenesis and chemosensitivity of ESCA (Fig. 8), at least in ESCC cells.
Data availability
The databases used and/or analyzed during the current study are available from the corresponding authors (Xiaoxiong Xiao, E-mail: xiaoxx1988@csu.edu.cn) on reasonable request.
References
H.C. Puhr, G.W. Prager, A. Ilhan-Mutlu, How we treat esophageal squamous cell carcinoma. ESMO Open 8(1), 100789 (2023)
M. Sheikh et al., Current status and future prospects for esophageal cancer. Cancers (Basel) 15(3), 765 (2023)
J. An et al., Natural products for esophageal cancer therapy: from traditional medicine to modern drug discovery. Int J Mol Sci 23(21), 13558 (2022)
Y. Baba et al., Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci 111(9), 3132–3141 (2020)
Y. Jia et al., Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer 20(1), 162 (2021)
E. Perland, R. Fredriksson, Classification systems of secondary active transporters. Trends Pharmacol Sci 38(3), 305–315 (2017)
A.K. Holzer, S.B. Howell, The internalization and degradation of human copper transporter 1 following cisplatin exposure. Cancer Res 66(22), 10944–10952 (2006)
A. Yaneff et al., MRP4/ABCC4 As a new therapeutic target: meta-analysis to determine cAMP binding sites as a tool for drug design. Curr Med Chem 26(7), 1270–1307 (2019)
J. Zhou et al., The drug-resistance mechanisms of five platinum-based antitumor agents. Front Pharmacol 11, 343 (2020)
S. Dilruba, G.V. Kalayda, Platinum-based drugs: past, present and future. Cancer Chemother Pharmacol 77(6), 1103–1124 (2016)
W.P. Roos, B. Kaina, DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 332(2), 237–248 (2013)
X. Zhu et al., DNMT3B-mediated FAM111B methylation promotes papillary thyroid tumor glycolysis, growth and metastasis. Int J Biol Sci 18(11), 4372–4387 (2022)
S. Mercier et al., Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, myopathy, and pulmonary fibrosis. Am J Hum Genet 93(6), 1100–1107 (2013)
H. Sun et al., FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco Targets Ther 12, 2829–2842 (2019)
S. Mercier et al., FAM111B mutation is associated with pancreatic cancer predisposition. Pancreas 48(5), e41–e42 (2019)
H. Wu, C. Liang, Pan-cancer analysis of the tumorigenic effect and prognostic diagnostic value of fam111b in human carcinomas. Int J Gen Med 16, 1845–1865 (2023)
X. Xiao et al., Silencing of UAP1L1 inhibits proliferation and induces apoptosis in esophageal squamous cell carcinoma. Mol Carcinog 60(3), 179–187 (2021)
K. Kawasaki et al., FAM111B enhances proliferation of KRAS-driven lung adenocarcinoma by degrading p16. Cancer Sci 111(7), 2635–2646 (2020)
S. Wenmaekers et al., A Potential Role for HUWE1 in Modulating Cisplatin Sensitivity. Cells 10(5), 1262 (2021)
C. He et al., Elevated H3K27me3 levels sensitize osteosarcoma to cisplatin. Clin Epigenetics 11(1), 8 (2019)
I. Lakshmanan et al., MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing p53. Clin Cancer Res 23(14), 3906–3917 (2017)
X. Gong et al., The structure and regulation of the E3 ubiquitin ligase HUWE1 and its biological functions in cancer. Invest New Drugs 38(2), 515–524 (2020)
C. Guo et al., Integrated analysis of multi-omics alteration, immune profile, and pharmacological landscape of pyroptosis-derived lncRNA pairs in gastric cancer. Front Cell Dev Biol 10, 816153 (2022)
M. Boivin et al., CA125 (MUC16) tumor antigen selectively modulates the sensitivity of ovarian cancer cells to genotoxic drug-induced apoptosis. Gynecol Oncol 115(3), 407–413 (2009)
M. Liu et al., Predictive biomarkers of dicycloplatin resistance or susceptibility in prostate cancer. Front Genet 12, 669605 (2021)
X. Lu, T. Guo, X. Zhang, Pyroptosis in Cancer: Friend or Foe? Cancers (Basel) 13(14), 3620 (2021)
X. Ouyang et al., Pyroptosis, inflammasome, and gasdermins in tumor immunity. Innate Immun 29(1–2), 3–13 (2023)
J. Shi et al., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526(7575), 660–665 (2015)
K.W. Chen et al., Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. Embo J 38(10), e101638 (2019)
Q. Wang et al., A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 579(7799), 421–426 (2020)
W. Deng et al., Streptococcal pyrogenic exotoxin B cleaves GSDMA and triggers pyroptosis. Nature 602(7897), 496–502 (2022)
W. Zou, J.D. Wolchok, L. Chen, PD-L1 (B7–H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 8(328), 328rv4 (2016)
A. Ribas, J.D. Wolchok, Cancer immunotherapy using checkpoint blockade. Science 359(6382), 1350–1355 (2018)
M. Macchiaiolo et al., Expanding phenotype of FAM111B-related disease focusing on liver involvement: Literature review, report of a case with end-stage liver disease and proposal for a new acronym. Am J Med Genet A 188(10), 2920–2931 (2022)
A. Seo et al., FAM111B mutation is associated with inherited exocrine pancreatic dysfunction. Pancreas 45(6), 858–862 (2016)
A.L. Welter, Y.J. Machida, Functions and evolution of FAM111 serine proteases. Front Mol Biosci 9, 1081166 (2022)
A. Arowolo, C. Rhoda, N. Khumalo, Mutations within the putative protease domain of the human FAM111B gene may predict disease severity and poor prognosis: A review of POIKTMP cases. Exp Dermatol 31(5), 648–654 (2022)
S. Hoffmann et al., FAM111 protease activity undermines cellular fitness and is amplified by gain-of-function mutations in human disease. EMBO Rep 21(10), e50662 (2020)
Acknowledgements
Not applicable.
Funding
This work were supported by grants from the National Natural Science Youth Foundation of China (Grant No. 82203838 (Xiaoxiong Xiao)); Natural Science Foundation General Program of Hunan Province (Grant No. 2022JJ40830 (Xiaoxiong Xiao)); Natural Science Foundation General Program of Changsha City (Grant No. kq2014290 (Xiaoxiong Xiao)); National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases (Lung Cancer, grant number: z027002 (Xiaoxiong Xiao)); National Natural Science Foundation of China (Grant No. 81772800, 82072945 (Pian Liu)).
Author information
Authors and Affiliations
Contributions
Haiqin Wang. supervised the experiments, and was responsible for bioinformatics and statistical analysis. Haohui Wang. carried out the in vitro and in vivo experiments, and wrote the original draft. Jiajing Chen. collaborated to collect specimens and record clinical information. Pian Liu. conducted the major revision work, and provided critical reviews. Xiaoxiong Xiao. generated the study design, provided the clinical specimens and revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted in accordance with the principles of the Declaration of Helsinki principles. It was approved by the Animal Use and Care Committees at Xiangya hospital, Central South University.
Consent for publication
All subjects have written informed consent.
Conflict of interest
The authors have declared that no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Haiqin Wang and Haohui Wang contributed equally.
Supplementary Information
Below is the link to the electronic supplementary material.
13402_2023_871_MOESM1_ESM.tif
Supplementary file1 Supplementary figure 1 A and B, The univariate and multivariate Cox regression analyses through the GEO dataset GSE53622. p values as indicated. C and D, Lymph node metastasis of KYSE410 cells with FAM111B knockdown. E, The expression of SNAI1 in RNA-seq of FAM111B. Fand G, KYSE410 and TE-1 cells were infected with shControl, shFAM111B #1, or shFAM111B #2 for 72 h. Cells were collected for RT-qPCR analysis (F), and Western blot analysis (G). H, The excised subcutaneous tumors of nude mice were subjected to IHC staining of SNAI1. Data presents as mean ± SD with six replicates. ***, p < 0.001 (TIF 35268 KB)
13402_2023_871_MOESM2_ESM.tif
Supplementary file2 Supplementary figure 2 A and B, KYSE410 cells were transfected with indicated shRNAs for 72 h. Cells were collected and subcutaneously injected into the nude mice. The tumor growth curve was shown in panel B. Data presents as mean ± SD with six replicates. ***, p < 0.001. C and D, KYSE410 and TE-1 cells were transfected with indicated shRNAs for 72 h and treated with cisplatin, cells were collected for Western blot analysis to measure the expression of cleaved caspase 3. ***, p < 0.001. E and F, The excised subcutaneous tumors of nude mice were subjected to IHC staining of cleaved caspase 3. Data presents as mean ± SD with six replicates. ***, p < 0.001 (TIF 47227 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, H., Wang, H., Chen, J. et al. Overexpressed FAM111B degrades GSDMA to promote esophageal cancer tumorigenesis and cisplatin resistance. Cell Oncol. 47, 343–359 (2024). https://doi.org/10.1007/s13402-023-00871-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00871-0