Abstract
Purpose
Long noncoding RNAs (lncRNAs) are emerging as key regulators in cancer initiation and progression. LINC01137 is a recently identified lncRNA of which the functional role in the development of oral squamous cell carcinoma (OSCC) has not been determined yet.
Methods
We analyzed the expression of LINC01137 using a microarray-based OSCC gene expression dataset (GSE31056), and validated the results obtained using RT-qPCR in 26 pairs of primary OSCC tumor tissues and adjacent non-tumor tissues. The proliferative and invasive effects of LINC01137 on OSCC cells were determined using CCK-8, colony formation and transwell assays, respectively. Targeted binding between miR-22-3p and LINC01137 was verified using a dual luciferase reporter assay.
Results
We found that LINC01137 was significantly upregulated in primary OSCCs. LINC01137 knockdown inhibited OSCC cell proliferation, migration and invasion, whereas LINC01137 overexpression induced opposite effects. LINC01137 upregulation along with p53 inhibition enhanced the malignant transformation of oral cells. In addition, we found that miR-22-3p can directly target LINC01137 through interaction with a putative miR-22-3p-binding site present within the LINC01137 sequence. A significant negative correlation was observed between LINC01137 and miR-22-3p expression in primary OSCC specimens. Exogenous overexpression of miR-22-3p markedly reduced the endogenous expression level of LINC01137 in OSCC cells. Additional functional assays showed that miR-22-3p overexpression enhanced the inhibitory effect of siRNA-mediated LINC01137 silencing on OSCC cell proliferation, migration and invasion, whereas miR-22-3p inhibition had the opposite effect.
Conclusions
Our results indicate that LINC01137 functions as an oncogenic lncRNA in OSCC. miR-22-3p can directly target LINC01137 and negatively regulate its expression and function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Oral squamous cell carcinoma (OSCC) is one of the most invasive head and neck malignancies and is prone to lymph node and distant metastasis [1]. OSCC accounts for about 3 % of all malignant tumors worldwide, with 550,000 new cases every year [2]. The main risk factors for OSCC are smoking and alcohol use, as well as DNA oncogenic viruses and betel nut chewing [3]. Until now, the basic treatment for OSCC is surgical resection accompanied by radiotherapy and chemotherapy [4]. Although several advances have been made in surgical techniques, adjuvant therapies and general patient care, the prognosis of OSCC remains poor, with a 5 year overall survival rate of ~ 50 % [5, 6]. A deeper understanding of the molecular mechanisms underlying OSCC is considered important for a better early diagnosis, proper treatment and improved prognosis of patients suffering from this disease.
Whole genome and transcriptome sequencing efforts have shown that only ~ 2 % of the human genome encodes proteins, while > 90 % of it is transcribed as non-coding RNAs (ncRNAs) [7]. Among this vast number of ncRNAs, there are two types of regulatory RNAs, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), which play key roles in numerous physiological and pathological processes [8]. MiRNAs are highly conserved, single stranded, small ncRNAs that range in size from 19 to 25 nucleotides [9]. They regulate gene expression mainly through silencing of specific target messenger RNAs (mRNAs) at the post-transcriptional level [10, 11]. LncRNAs are single-stranded RNAs over 200 nucleotides in length without protein-coding ability [12, 13]. They act as potential regulators of various cell biological process, such as proliferation, programmed cell death, migration and differentiation [14]. Abnormal expression of lncRNAs has been related to diverse diseases, and specific lncRNAs have been found to play vital roles in the initiation and progression of various malignancies, including OSCC [15, 16]. It has for example been reported that lncRNA MORT is downregulated in OSCC, and that decreased MORT expression is correlated with a poor survival, whereas MORT overexpression leads to inhibition of OSCC cell proliferation via downregulation of ROCK1 [17]. LncRNA ANRIL has been found to be highly expressed in tumor tissues and sera of OSCC patients, and to promote the proliferation and suppress the apoptosis of OSCC cells by regulating the TGF-β/Smad signaling pathway [18].
Although research on miRNAs and lncRNAs mainly focuses on the regulation of protein coding genes, it has been shown that miRNAs and lncRNAs can also interact with each other to form extra post-transcriptional regulatory interaction networks. LINC01787 represents an example of this type of regulation. It binds pre-miR-125b, inhibits the binding between DICER and pre-miR-125b and, thereby, represses the generation of mature miR-125b [19]. Previously, we have shown that miR-96 can directly target lncRNA TP53TG1 and negatively regulate its expression, whereas TP53TG1 can act as a sponge for miR-96 and, thereby, repress the inhibitory effect of miR-96 on KRAS [20].
Long intergenic non-protein coding RNA 1137 (LINC01137, NCBI no: NR_038842.1) is a recently identified 1443-bp lncRNA of which the locus is located on chromosome 1p34.3. Following exposure to hydrogen peroxide, mercury II chloride and etoposide, the expression of LINC01137 has been found to be significantly upregulated in human HepG2 cells. These observations suggest that LINC01137 may serve as a useful indicator of chemical stress responses [21]. Smoking, alcohol consumption and betel nut chewing are all considered major risk factors for OSCC [22]. More than 60 toxic chemicals have been identified in tobacco and its use may cause epigenetic alterations in oral epithelial cells and induce defective DNA damage responses that appear to be strongly associated with tumor induction [23, 24]. Here, we hypothesize that the toxic metabolites and stimuli from tobacco, alcohol and betel nut may act as stress factors that affect LINC01137 expression, which in turn may play a role in the development and progression of OSCC.
In the present study, we investigated the putative role of LINC01137 in human OSCC as well as putative interactions between LINC01137 and miRNAs. We show that LINC01137 is significantly upregulated in primary OSCC tumor tissues and cell lines, and that it plays a role in promoting malignant processes in OSCC cells, including proliferation, migration and invasion. We also found that miR-22-3p can directly target LINC01137 through interaction with a putative miR-22-3p binding site within the LINC01137 sequence and, thereby, negatively regulate LINC01137 expression and function.
2 Materials and methods
2.1 Patient tissue samples and cell culture
Tissue samples were obtained from 26 cases diagnosed with OSCC at The Second Hospital of Hebei Medical University (Shijiazhuang, China), during 2015–2018. None of the patients received radiotherapy or chemotherapy before operation. Immediately after resection, the tissue samples were snap-frozen in liquid nitrogen and placed in a -80 °C freezer for long-term storage. All cases were confirmed as OSCC through pathological examination. This study was approved by the Research Ethics Committee of The Second Hospital of Hebei Medical University. Informed consents were signed by all patients before using the resected tissues for research purposes.
Human OSCC cell lines (HSC3 and HSC4) were obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB, Osaka, Japan). The human tongue squamous cell carcinoma cell line Tca8113 was purchased from the Tumor Cell Bank of the Chinese Academy of Medical Science (Beijing, China), whereas the human normal oral cell line HGF-1 (ATCC CRL-2014) was obtained from the American Type Culture Collection (Rockville, MD, USA). RPMI-1640 medium (Gibco, Grand Island, NY) supplemented with 10 % FBS (Gibco) was used for HSC3, HSC4 and Tca8113 cell culture, whereas Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10 % FBS was used for HGF-1 cell culture. Morphological examination, growth curve testing and mycoplasma detection were performed for all cell lines one month before the study, in accordance with the ATCC cell line verification test requirements. The four cell lines used in this study have also been authenticated by short tandem repeat profiling.
2.2 RNA isolation and RT-qPCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used for total RNA extraction from OSCC tissues and cells. A ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan) and a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) were employed for LINC01137 and miRNA detection, respectively. RT-qPCR was performed using SYBR Premix Ex Taq II (Qiagen, Hilden, Germany). The amplification conditions included initial pre-denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing at 60 °C for 1 min. Relative fold changes in gene expression were determined using the 2−ΔΔCT method. All primer sequences used are listed in Table 1.
2.3 siRNA and miRNA transfections
siRNAs specifically targeting LINC01137 or p53 [25] were purchased from Genepharma (Suzhou, Jiangsu, China). miRNAs that potentially bind to LINC01137 were predicted using miRcode software (http://www.mircode.org) [26]. Hsa-miRNA mimics and inhibitors were also purchased from Genepharma. Lipofectamine 2000 (Invitrogen) was used for siRNA or miRNA transfection. OSCC cells were transfected with 40 nM siRNA or 40 nM miRNA. The sequences of the siRNAs and miRNAs used in this study are listed in Table 2.
2.4 Plasmid construction and transfection
For overexpression of LINC01137 in HSC4 and HGF-1 cells, full-length LINC01137 was synthesized and subcloned into a pCDNA3.1 vector (Invitrogen), generating a pCDNA3.1-LINC01137 plasmid that was subsequently verified by DNA sequencing. Lipofectamine 2000 Reagent (Invitrogen) was used for plasmid transfection.
2.5 CCK-8 proliferation assay
Transfected OSCC cell suspensions were seeded into 96 well plates (2 × 103 cells/well) and cultured for 24, 48, 72, 96 and 120 hours, respectively, after which CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well. Next, the plates were incubated for 2 hours after which absorbance at 450 nm was measured.
2.6 Colony formation assay
Twenty-four hours after transfection, OSCC cells were seeded into six-well plates (103 cells/well) and cultured for 2 weeks. Subsequently, colonies were visualized by incubation with 1 % crystal violet and counted.
2.7 Migration and invasion assays
Transwell chambers (3422, Corning, Kennebunk, ME, USA) or Matrigel (BD Biosciences, San Jose, CA, USA) precoated Transwell chambers were employed for migration and invasion assays, respectively. Cells (2x105) in serum-free media were added to the upper chamber 24 h after transfection, and medium containing 10 % FBS was added to the lower chamber. After culture for 48 hours, the cells on the upper surface were scraped off and washed away, after which the chambers were immersed in 4 % paraformaldehyde for 30 minutes. Next, the cells that migrated/invaded to the lower surface were stained with 0.1 % crystal violet and counted under a microscope.
2.8 Western blot analysis
Cells were harvested, placed on ice and lysed in lysis buffer to collect total proteins. A BCA Protein Assay Kit (Solarbio, Beijing, China) was employed for measuring the protein content, after which 50 µg was loaded per lane on SDS-polyacrylamide gels, separated and transferred to PVDF membranes (Millipore, Billerica, MA, USA). For protein detection, the following antibodies were used: anti-MMP-9 (Affinity, Cincinnati, OH, USA), anti-Vimentin (Cell Signaling, Danvers, MA, USA), anti-Survivin (Cell Signaling), anti-CyclinD1 (Abcam, Cambridge, MA, USA), anti-Snail (Arigo Biolaboratories, Hsinchu, Taiwan), anti-β-actin (Santa Cruz Biotechnology, Dallas, TX, USA), anti-Bax (Abcam), anti-Bcl-2 (Abcam), anti-MMP-2 (Bioss, Woburn, MA, USA) and anti-Twsit (Affinity). Band densities were quantified using ImageJ software 1.46 (NIH, Bethesda, MD, USA) and normalized to β-actin.
2.9 Cytoplasmic/nuclear fraction isolation
NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA) were used for separation of nuclear and cytoplasmic fractions of OSCC cells following the manufacturer’s instructions. TRIzol reagent (Invitrogen) was used for RNA extraction from each subcellular fraction, after which RT-qPCR analyses were performed.
2.10 Luciferase reporter assay
Full-length LINC01137 was synthesized and inserted into a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) downstream of the luc2 reporter gene to generate a pmirGLO-LINC01137-WT vector. Site-directed mutagenesis of the putative miR-22-3p binding site within the LINC01137 sequence was performed to construct a pmirGLO-LINC01137-MUT vector. Both plasmids were verified by DNA sequencing. Wild-type or mutant pmirGLO-LINC01137 vectors were co-transfected with miRNA-22-3p mimic or control NC into OSCC cells using Lipofectamine 2000 (Invitrogen). Luciferase activity was detected using a dual-luciferase reporter assay system (Promega) 24 hours after transfection.
2.11 Statistics
SPSS 22.0 software (SPSS, Chicago, IL, USA) was employed for all statistical analyses in this study. Paired samples t-test was used to analyze expression differences between OSCC and matched normal tissues. Independent samples t-test was used to analyze changes in proliferation, migration and invasion after transfection. Spearman correlation test was used for expression correlation assessment. Each experiment was carried out at least three times independently. Data are shown as mean ± SD, and a p value < 0.05 was considered to be statistically significant.
3 Results
3.1 LINC01137 is upregulated in OSCC
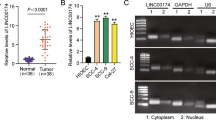
The ENCODE project was used to analyze LINC01137 activation and diffusion in different cancer cell lines. CHIP-seq data from the ENCODE database revealed 16 candidate cis-regulatory elements on human chromosome 1 (Fig. S1), which were close to or overlapped with the LINC01137 gene. The expression level of LINC01137 in most tumor cell lines tested was significantly higher than that in normal cells according to the RNA-seq data provided by the ENCODE database (Fig. S2). Next, we used the Genotype-Tissue Expression (GTEx) Project and TCGA RNA-seq public datasets to analyze LINC01137 expression in normal and tumor specimens. We found that among 25 common tumor types, the expression level of LINC01137 in most of them, including head and neck squamous cell carcinoma (HNSCC), was significantly higher than that in its corresponding normal tissues, whereas in only a few tumors LINC01137 expression was similar or lower than that in normal tissues (Fig. S3). Gene expression profiling analysis of GSE31056 from pubmed GEO DataSets showed that in 23 pairs of OSCC tumors and adjacent normal oral mucosa tissues, LINC01137 was one of the most differentially expressed lncRNAs, and that its expression in tumor tissues was significantly higher than that in adjacent normal oral mucosa tissues (Fig. 1a). To further verify this result, we examined the expression of LINC01137 in OSCC cell lines (HSC3, HSC4, Tca8113) using RT-qPCR and found that, compared with normal oral mucosa tissue, the OSCC cells showed markedly higher levels of LINC01137 expression (Fig. 1b). Next, we comparatively analyzed the expression of LINC01137 in 26 paired OSCC and adjacent normal oral mucosa tissues using RT-qPCR. We found that the expression of LINC01137 in the OSCC tissues was markedly increased compared to that in the normal oral mucosa tissues (Fig. 1c). Based on the median value, the 26 OSCC cases could be divided into two groups: those with a high LINC01137 expression level (n = 11) and those with a low LINC01137 expression level (n = 15). Subsequent statistical analysis revealed that the LINC01137 expression level significantly correlated with the occurrence of lymph node metastasis (Table S1, p < 0.05), but was not associated with other clinicopathological parameters, including gender, age, tumor size and tumor differentiation status (Table S1, p > 0.05).
LINC01137 is upregulated in OSCC tissues and cell lines. a LINC01137 expression analysis using NCBI/GEO/GSE31056 OSCC gene profiling datasets (n = 23). Since only one normal oral tissue could be found in patient No. 13, this information was not included in the analysis. Values are shown as median with 95 % CI. b RT-qPCR analysis of endogenous LINC01137 expression in OSCC cell lines and normal oral mucosa. The transcript levels were normalized to GAPDH expression. n = 3, *p < 0.05. c RT-qPCR analysis of LINC01137 expression in 26 pairs of OSCC tissues and corresponding normal oral mucosa. The transcript levels were normalized to GAPDH expression
The coding potential of LINC01137 was evaluated using CPAT (Coding-Potential Assessment Tool) software [27], and revealed that the possible open reading frame (ORF) of LINC01137 is very short and, thus, that LINC01137 does not appear to have coding capacity (Table 3). In addition, we used ExPaSy (https://web.expasy.org/translate/) to predict the protein coding ability of LINC01137. The predicted ORFs of LINC01137 are highlighted in red in Fig. S4. These ORFs are not only very short, but also discontinuous with several breakups, which further confirms that LINC01137 may not have a protein coding ability. In addition to an open reading frame, the ability of lncRNAs to be translated or not depends on the presence of ribosome binding sites. The predicted results of RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/index.html) indicate that there are no ribosome binding sites present within the LINC01137 sequence (Fig. S5). Together, these observations indicate that LINC01137 has no protein coding capacity.
3.2 LINC01137 knockdown inhibits OSCC cell proliferation
To evaluate the effect of LINC01137 on OSCC cell proliferation, we downregulated its expression by transfecting HSC3 and Tca8113, two cell lines in which LINC01137 expression is relatively high, with two different siRNAs directed against LINC01137 (siLINC01137-1 and siLINC01137-2). After 48 h, the knockdown efficiencies were detected by RT-qPCR. We found that LINC01137 expression was markedly decreased in the siLINC01137 groups compared to that in the control groups (Fig. 2a). Next, growth curves of OSCC cells transfected with siLINC01137s were plotted using a CCK-8 proliferation assay. The results showed that LINC01137 knockdown markedly inhibited the growth of both HSC3 and Tca8113 cells (Fig. 2b and Fig. S6). Subsequent colony formation assays confirmed that LINC01137 knockdown significantly inhibited the growth of HSC3 and Tca8113 cells (Fig. 2c). To investigate the mechanism underlying the effect of LINC01137 knockdown on OSCC cell proliferation, we next examined expression level changes of cell cycle and apoptosis-related proteins, including Cyclin D1, Survivin, Bcl-2 and Bax, using Western blotting. We found that the expression levels of Cyclin D1, Survivin and Bcl-2 in the LINC01137 knockdown groups were all significantly lower than those in the control group, while the expression level of Bax was significantly increased following LINC01137 downregulation (Fig. 2d), suggesting that LINC01137 knockdown decreases the proliferative ability of OSCC cells by inhibiting cell cycle progression and promoting apoptosis.
LINC01137 downregulation inhibits OSCC cell proliferation. a After transfecting HSC3 and Tca8113 cells with NC or LINC01137 siRNAs, RT-qPCR was performed to detect LINC01137 expression. The transcript levels were normalized to GAPDH expression. n = 3, **p < 0.01. b Growth of HSC3 and Tca8113 cells after LINC01137 knockdown determined by CCK-8 assays. n = 3, *p < 0.05,**p < 0.01. c Effect of LINC01137 downregulation on the colony forming abilities of HSC3 and Tca8113 cells. n = 3, **p < 0.01. d Expression of proliferation-related proteins (Cyclin D1, Survivin, Bcl-2 and Bax) detected by Western blotting after LINC01137 knockdown in HSC3 and Tca8113 cells. n = 3, *p < 0.05
3.3 LINC01137 knockdown inhibits OSCC cell migration and invasion
To quantify the effect of LINC01137 knockdown on the migratory ability of OSCC cells, transwell migration assays were performed. We randomly selected ten fields to count the number of HSC3 and Tca8113 cells that entered the lower chamber through the foramen. By doing so, we found that the average number of cells invading through the foramen of each visual field was significantly lower in the LINC01137 knockdown groups than that in the control groups (Fig. 3a), indicating that the migratory ability of the OSCC cells was impaired. To test the invasive ability of OSCC cells, transwell invasion assays were conducted, and the cells that crossed through the chamber coated with Matrigel were counted. We found that LINC01137 knockdown strongly suppressed the invasiveness of HSC3 and Tca8113 cells, in both cases being about half of that of the control cells (Fig. 3b). Collectively, these results suggest that LINC01137 can regulate the migration and invasion of OSCC cells. In order to assess the mechanism underlying the effect of LINC01137 on OSCC cell migration and invasion, we examined the expression of EMT markers by Western blotting after LINC01137 downregulation. We found that the expression of Vimentin, Twist, Snail, MMP-2 and MMP-9 was significantly decreased in the LINC01137 knockdown groups (Fig. 3c), suggesting that EMT may be involved in LINC01137-mediated regulation of OSCC cell migration and invasion.
LINC01137 downregulation inhibits OSCC cell migration and invasion. a Transwell chambers were used to detect the effect of LINC01137 knockdown on the migration of HSC3 and Tca8113 cells. n = 10 fields, **p < 0.01. b Transwell chambers precoated with Matrigel were used to detect the effect of LINC01137 knockdown on the invasion of HSC3 and Tca8113 cells. n = 10 fields, **p < 0.01. c Western blotting was performed to detect the expression of EMT related proteins (Vimentin, Snail, Twist, MMP-2 and MMP-9) after LINC01137 knockdown in HSC3 and Tca8113 cells. n = 3, *p < 0.05
3.4 LINC01137 overexpression promotes OSCC cell proliferation, migration and invasion
To further explore the biological effect of LINC01137 on OSCC cells, we exogenously overexpressed this lncRNA by transfection of a pcDNA3.1-LINC01137 plasmid into HSC4 cells, which show a relatively low endogenous level of LINC01137. Using RT-qPCR, we found that the expression of LINC01137 was drastically increased following pcDNA3.1-LINC01137 transfection compared to that in pcDNA3.1 empty vector transfected cells (Fig. 4a). Subsequent growth curves based on CCK-8 assays showed that LINC01137 upregulation markedly increased HSC4 cell proliferation (Fig. 4b). In addition, we found that the colony forming ability of HSC4 cells was markedly increased after LINC01137 overexpression, as indicated by larger and more colonies formed in the LINC01137 upregulation group (Fig. 4c). Additional transwell assays revealed that LINC01137 overexpression significantly enhanced the migrative and invasive capacities of HSC4 cells (Fig. 4d). These data indicate that LINC01137 may function as an oncogenic lncRNA enhancing the proliferation, migration and invasion of OSCC cells.
Overexpression of LINC01137 promotes proliferation, migration and invasion of OSCC cells. a After transfection of HSC4 cells with pcDNA3.1 empty vector or pcDNA3.1-LINC01137 vector, RT-qPCR was used to detect LINC01137 expression. The transcript levels were normalized to GAPDH expression. n = 3, **p < 0.01. b Growth of HSC4 cells after LINC01137 overexpression assessed by CCK-8 assays. n = 3, *p < 0.05,**p < 0.01. c Effect of LINC01137 overexpression on colony formation of HSC4 cells. n = 3, **p < 0.01. d Transwell chambers were used to detect the effect of LINC01137 overexpression on the migration of HSC4 cells. n = 10 fields, **p < 0.01. e Transwell chambers precoated with Matrigel were used to detect the effect of LINC01137 overexpression on the invasion of HSC4 cells. n = 10 fields, **p < 0.01
3.5 LINC01137 overexpression along with p53 inhibition promotes malignant transformation of normal oral cells
Most OSCC tumor tissues and cells harbor p53 mutations, which are likely to be involved in the development of OSCC [28, 29]. In order to test whether the “two hit hypothesis” applies to LINC01137 in the initiation of OSCC, we knocked down p53 and simultaneously overexpressed LINC01137 in normal oral HGF-1 cells. We found that the p53 mRNA level was markedly decreased after transfection with a siRNA targeting p53 (Fig. 5a). Subsequent CCK-8 assays showed that p53 downregulation or LINC01137 upregulation alone could promote HGF-1 cell growth, whereas a more profound increase in proliferation was seen after simultaneous p53 knockdown and LINC01137 overexpression (Fig. 5b). The colony forming ability of HGF-1 cells was also most profoundly increased in the p53 knockdown/LINC01137 overexpression group, compared to p53 knockdown or LINC01137 overexpression alone (Fig. 5c). Using transwell assays we found that both p53 inhibition and LINC01137 upregulation had the most significant effect on HGF-1 cell migration and invasion, whereas p53 knockdown or LINC01137 overexpression alone only moderately increased the migrative and invasive abilities of HGF-1 cells (Fig. 5d-e). Overall, these observations imply that LINC01137 upregulation along with p53 inhibition may enhance the malignant transformation of normal oral cells.
LINC01137 overexpression along with p53 inhibition can promote malignant transformation of normal oral cells. a After transfection of HGF-1 cells with sip53, RT-qPCR was used to detect p53 mRNA expression. The transcript levels were normalized to GAPDH expression. n = 3, **p < 0.01. b HGF-1 cells were transfected with sip53 or pcDNA3.1-LINC01137 alone or in combination, after which their growth was assessed using CCK-8 assays. n = 3, *p < 0.05,**p < 0.01. c Colony formation assays were employed to detect the growth ability of HGF-1 cells after transfection with sip53 or pcDNA3.1-LINC01137 alone or in combination. n = 3, **p < 0.01. d Transwell chambers were used to detect the migrative ability of HGF-1 cells after transfection with sip53 or pcDNA3.1-LINC01137 alone or in combination. n = 10 fields, **p < 0.01. e Transwell chambers precoated with Matrigel were used to detect the invasive ability of HGF-1 cells after transfection with sip53 or pcDNA3.1-LINC01137 alone or in combination. n = 10 fields, **p < 0.01
3.6 miR-22-3p negatively regulates LINC01137 expression
Recently, it has been found that lncRNAs may exert their functions by binding specific miRNAs. To explore whether LINC01137 acts similarly, miRcode software (http://www.mircode.org) [22] was used to search for putative LINC01137-miRNA interactions. Several miRNAs were predicted to have binding sites for LINC01137 (Fig. S7). We subsequently transfected HSC3 and Tca8113 cells with the respective miRNA mimics and found that miR-22-3p exhibited the greatest effect on the expression of LINC01137 (Fig. S8 and Fig. 6b). This miRNA was chosen for further analysis. The “seed sequences” between miR-22-3p and LINC01137 showed perfect base pairing and were found to be conserved across species (Fig. 6a). Using RT-qPCR, we found that transfection of a miR-22-3p mimic significantly reduced the expression of endogenous LINC01137 in HSC3 and Tca8113 cells, suggesting that miR-22-3p may negatively regulate LINC01137 expression (Fig. 6b). To further explore the relationship between miR-22-3p and LINC01137, the expression level of miR-22-3p in clinical specimens was examined. RT-qPCR data showed that, contrary to the high expression of LINC01137 in OSCC, the expression of miR-22-3p in the same tumor samples was low, thus indicating a marked negative correlation between LINC01137 and miR-22-3p expression (Fig. 6c, r = -0.7420, p < 0.001). Among the 26 OSCC cases tested, 12 exhibited high miR-22-3p levels and 14 exhibited low miR-22-3p levels, relative to the median value. We found that the low miR-22-3p levels were significantly associated with lymph node metastasis (Table S1, p < 0.05). In addition, we detected LINC01137 and miR-22-3p expression in the nuclear and cytoplasmic fractions of HSC3 and Tca8113 cells by RT-qPCR. The results showed that the expression of LINC01137 and miR-22-3p in the cytoplasm of HSC3 and Tca8113 cells was significantly higher than that in the nucleus (Fig. 6d), indicating that both of them were mainly located in the cytoplasm, which is a prerequisite for their interaction.
Validation of LINC01137 as a direct target of miR-22-3p in OSCC. a Predicted binding sites of miR-22-3p within the LINC01137 sequence are shown. The area within the frame represents conserved complementary nucleotides of the miR-22-3p seed sequence in various mammals. b RT-qPCR was performed to determine the expression of LINC01137 after miR-22-3p overexpression in HSC3 and Tca8113 cells. n = 3, **p < 0.01. c RT-qPCR was performed to determine the expression of miR-22-3p in OSCC tissues and corresponding normal oral mucosa, after which Spearman correlation analysis was performed to explore the correlation between miR-22-3p and LINC01137 expression (r = -0.7420, p < 0.001). d Percentages of LINC01137, miR-22-3p, GAPDH and MALAT1 levels in cytoplasmic and nuclear fractions of HSC3 and Tca8113 cells detected by RT-qPCR. GAPDH serves as a cytoplasmic localization marker, and MALAT1 as a nuclear localization marker. e Schematic representation of wild-type (pmirGLO-LINC01137-WT) and mutant (pmirGLO-LINC01137-MUT) dual luciferase reporter constructs. f Luciferase activity in HSC3 and Tca8113 cells co-transfected with miR-22-3p mimic and pmirGLO-LINC01137-WT or pmirGLO-LINC01137-MUT vectors detected by dual-luciferase reporter assay. n = 3, *p < 0.05
3.7 miR-22-3p directly targets LINC01137
To verify whether LINC01137 is a bona fide target of miR-22-3p, full-length wild-type and mutant LINC01137 sequences were separately inserted into a pmirGLO vector, generating pmirGLO-LINC01137-WT and pmirGLO-LINC01137-MUT vectors (Fig. S9 and Fig. 6e). Next, we transfected each of these with a miR-22-3p mimic or a negative control (NC) into OSCC cells. Using a dual luciferase reporter assay we found that miR-22-3p upregulation markedly reduced the luciferase activity, and that directed mutagenesis of the miR-22-3p-binding region in the LINC01137 sequence abolished the repressive effect of miR-22-3p on the luciferase activity (Fig. 6f), which suggests that miR-22-3p can directly target LINC01137.
3.8 miR-22-3p functionally regulates LINC01137
Since we found that LINC01137 silencing resulted in proliferation, migration and invasion inhibition of OSCC cells, the tumor suppressive effect of LINC01137 silencing might be regulated by miR-22-3p, as miR-22-3p can directly target LINC01137. To test this hypothesis, the proliferative, migrative and invasive abilities of OSCC cells were evaluated after co-transfection of siLINC01137 and miR-22-3p mimic or inhibitor. Using a colony forming assay we found that, compared with the siLINC01137 transfected group, OSCC cells had a lower cell proliferation ability when co-transfected with siLINC01137 and miR-22-3p mimic, and a higher cell proliferation ability when co-transfected with siLINC01137 and miR-22-3p inhibitor (Fig. 7a). Concordantly, we found that the migrative and invasive abilities of the OSCC cells were decreased following co-transfection with siLINC01137 and miR-22-3p mimic, and increased following co-transfection with siLINC01137 and miR-22-3p inhibitor, when compared with the LINC01137 only downregulated group (Fig. 7b-c). Overall, these data indicate that overexpression of miR-22-3p can enhance the inhibitory effect of siLINC01137 on OSCC cell proliferation, migration and invasion, while knockdown of miR-22-3p can at least partially rescue the effect of siLINC01137 on the proliferation, migration and invasion of OSCC cells.
miR-22-3p inhibits LINC01137 activity. a HSC3 and Tca8113 cells were co-transfected with siLINC01137 and miR-22-3p mimic or miR-22-3p inhibitor, after which cell viability was detected using colony formation assays. n = 3, *p < 0.05. b HSC3 and Tca8113 cells were co-transfected with siLINC01137 and miR-22-3p mimic or miR-22-3p inhibitor, after which cell migration was detected using transwell assays. n = 10 fields, *p < 0.05. c HSC3 and Tca8113 cells were co-transfected with siLINC01137 and miR-22-3p mimic or miR-22-3p inhibitor, after which their invasive ability was determined using transwell chambers precoated with Matrigel. n = 10 fields, *p < 0.05
4 Discussion
It has been well documented that lncRNAs play critical roles in the development of various tumors, and increasing numbers of tumor suppressive or oncogenic lncRNAs are emerging as novel diagnostic, prognostic and/or therapeutic markers [30,31,32,33]. Also, aberrant expression patterns and functional effects of lncRNAs in OSCC have begun to be revealed. lncRNA BLACAT1 has, for example, been found to be highly expressed in OSCC and its knockdown to inhibit the viability, migration and invasion of OSCC cells [34]. LncRNA CEBPA-AS1, which is localized in peri-nuclear and cytoplasm domains, has been found to function as a potential oncogenic lncRNA in OSCC and to be related with a poor differentiation and high clinical stages [35]. Silencing of lncRNA CRNDE has been found to inhibit EMT via inactivation of the Wnt/β‑catenin signaling pathway, thereby decreasing the migration and invasion of human OSCC cells [36]. In spite of these results, the functional role of lncRNAs in OSCC initiation and progression is far from well-characterized.
Through the analysis of microarray-based gene expression profiling data (GSE31056), we found that LINC01137 was one of the most highly expressed lncRNAs in OSCC. Subsequent RT-qPCR analyses confirmed that the expression of LINC01137 was indeed markedly increased in primary OSCC tissues and cell lines. Additional CCK-8 and colony formation assays showed that LINC01137 knockdown significantly reduced the proliferative ability of OSCC cells, while transwell assays revealed that LINC01137 downregulation strongly suppressed the migrative and invasive abilities of OSCC cells. Conversely, we found that LINC01137 overexpression enhanced the proliferation, migration and invasion of OSCC cells. In addition, we found that LINC01137 upregulation along with p53 inhibition appeared to enhance the malignant transformation of normal oral cells. Together, these findings indicate that LINC01137 plays an oncogenic role in OSCC, and that the high expression of LINC01137 in OSCC may contribute to tumor progression. We also found that after LINC01137 downregulation the expression of Cyclin D1, Survivin, Bcl-2 and that of several EMT-related proteins including Vimentin, Twist, Snail, MMP-2 and MMP-9, was significantly decreased, while the expression of Bax was markedly increased, suggesting that LINC01137 knockdown may reduce the proliferative, migrative and invasive abilities of OSCC cells by inhibiting cell cycle progression, promoting apoptosis and inhibiting EMT. As yet, however, the specific molecular events downstream of LINC01137 remain to be explored in further detail.
It has been reported that besides protein-coding genes, lncRNAs may serve as targets of miRNAs [8]. LncRNAs may function as competing endogenous RNAs (ceRNAs) through competitively binding miRNAs and, thereby, reducing the inhibitory effects of miRNAs on their targets [37,38,39]. Conversely, miRNAs may regulate lncRNAs via binding to target sequences, which is similar to miRNA-mediated targeted mRNA silencing. For instance, miR-196a can directly bind to the seventh exon of GAS5 and, thereby, inhibit the expression of GAS5 [40]. Also, miR-125b has been found to regulate HOTTIP expression at the post-transcriptional level, i.e., the endogenous HOTTIP level and the HOTTIP-coupled luciferase activity could both be inhibited by miR-125b overexpression [41]. Another miRNA, miR-141 can directly bind to HOTAIR and suppress its expression and function in an Ago2-dependent manner [42].
In order to explore whether LINC01137 can interact with miRNAs, a search for miRNAs that may possibly target LINC01137 was performed and, by doing so, 6 miRNAs were predicted to exhibit complementary base pairing with LINC01137. After evaluation of LINC01137 expression changes upon overexpression of these miRNAs, miR-22-3p was selected for further analysis. It showed the greatest effect on LINC01137 expression and the predicted miR-22-3p binding sites within the LINC01137 sequence were found to be highly conserved across species. It has recently been reported that miR-22-3p is downregulated in many tumors, including hepatocellular carcinoma, retinoblastoma, gastrointestinal stromal tumor and lung adenocarcinoma, and may function as a tumor suppressor in these tumors [43,44,45,46]. Here, we found that the expression of miR-22-3p was decreased in OSCC tissues, and to be significantly negatively correlated with LINC01137 expression. Exogenous expression of miR-22-3p resulted in a significant downregulation of LINC01137 in OSCC cells, suggesting that miR-22-3p may negatively regulate LINC01137 expression. In addition, we found that both LINC01137 and miR-22-3p were mainly located in the cytoplasm of OSCC cells, and that miR-22-3p can directly target LINC01137 in a sequence-specific manner. Additional, functional assays revealed that exogenous miRNA-22-3p expression enhanced the repressive effect of siLINC01137 on OSCC cell proliferation, migration and invasion, whereas miR-22-3p knockdown partially rescued the repressive effect of siLINC01137 on OSCC cells. The results of our present study are consistent with recent reports on the direct regulation of lncRNAs by miRNAs.
In conclusion, we found that lncRNA LINC01137 is upregulated in OSCC and may play a role as an oncogene through the promotion of OSCC cell proliferation, migration and invasion. miR-22-3p directly targets LINC01137 and suppresses LINC01137 expression and function. The high expression of LINC01137 in OSCC may at least partly contribute to the observed downregulation of miR-22-3p. The direct regulation of lncRNAs by other regulatory RNAs such as miRNAs greatly increases the complexity of gene expression regulation, and may provide new insights into the mechanisms underlying OSCC initiation and progression.
References
P.R. Brocklehurst, S.R. Baker, P.M. Speight, Oral cancer screening: what have we learnt and what is there still to achieve? Future Oncol. 6, 299–304 (2010)
D.M. Parkin, F. Bray, J. Ferlay, P. Pisani, Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 (2005)
Y. Ghantous, I. Abu Elnaaj, Global incidence and risk factors of oral cancer. Harefuah 156, 645–649 (2017)
S.A. Gharat, M. Momin, Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit. Rev. Ther. Drug Carrier Syst. 33, 363–400 (2016)
S.D. da Silva, A. Ferlito, R.P. Takes, R.H. Brakenhoff, M.D. Valentin, J.A. Woolgar, C.R. Bradford, J.P. Rodrigo, A. Rinaldo, M.P. Hier, L.P. Kowalski, Advances and applications of oral cancer basic research. Oral Oncol. 47, 783–791 (2011)
J.Y. Wu, C. Yi, H.R. Chung, D.J. Wang, W.C. Chang, S.Y. Lee, C.T. Lin, Y.C. Yang, W.C. Yang, Potential biomarkers in saliva for oral squamous cell carcinoma. Oral Oncol. 46, 226–231 (2010)
M.K. Iyer, Y.S. Niknafs, R. Malik, U. Singhal, A. Sahu, Y. Hosono, T.R. Barrette, J.R. Prensner, J.R. Evans, S. Zhao, A. Poliakov, X. Cao, S.M. Dhanasekaran, Y.M. Wu, D.R. Robinson, D.G. Beer, F.Y. Feng, H.K. Iyer, A.M. Chinnaiyan, The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015)
V. Taucher, H. Mangge, J. Haybaeck, Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell. Oncol. 39, 295–318 (2016)
R. Rupaimoole, F.J. Slack, MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug. Discov. 16, 203–222 (2017)
T. Mittag, N.L. Fawzi, Protein quality and miRNA slicing get into phase. Nat. Cell Biol. 20, 635–637 (2018)
W. Xiao, C. Wang, K. Chen, T. Wang, J. Xing, X. Zhang, X. Wang, MiR-765 functions as a tumour suppressor and eliminates lipids in clear cell renal cell carcinoma by downregulating PLP2. EBioMedicine 51, 102622 (2020)
J.J. Quinn, H.Y. Chang, Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016)
J. Wang, J. Sun, F. Yang, The role of long non-coding RNA H19 in breast cancer. Oncol. Lett. 19, 7–16 (2020)
T.R. Mercer, M.E. Dinger, J.S. Mattick, Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159 (2009)
P.J. Batista, H.Y. Chang, Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013)
L. Zhang, X. Meng, X.W. Zhu, D.C. Yang, R. Chen, Y. Jiang, T. Xu, Long non-coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol. Cancer 18, 102 (2019)
Z. Jin, S. Jiang, S. Jian, Z. Shang, Long noncoding RNA MORT overexpression inhibits cancer cell proliferation in oral squamous cell carcinoma by downregulating ROCK1. J. Cell. Biochem. (2019). https://doi.org/10.1002/jcb.28449
L. Liu, S.B. Ning, S. Fu, Y. Mao, M. Xiao, B. Guo, Effects of lncRNA ANRIL on proliferation and apoptosis of oral squamous cell carcinoma cells by regulating TGF-beta/Smad pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 6194–6201 (2019)
Y. Li, Y. Song, Z. Wang, Z. Zhang, M. Lu, Y. Wang, Long non-coding RNA LINC01787 drives breast cancer progression via disrupting miR-125b generation. Front. Oncol. 9, 1140 (2019)
Y. Zhang, H. Yang, Y. Du, P. Liu, J. Zhang, Y. Li, H. Shen, L. Xing, X. Xue, J. Chen, X. Zhang, Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer Sci. 110, 2760–2772 (2019)
H. Tani, A. Numajiri, M. Aoki, T. Umemura, T. Nakazato, Short-lived long noncoding RNAs as surrogate indicators for chemical stress in HepG2 cells and their degradation by nuclear RNases. Sci. Rep. 9, 20299 (2019)
A. Capote-Moreno, P. Brabyn, M.F. Muñoz-Guerra, J. Sastre-Pérez, V. Escorial-Hernandez, F.J. Rodríguez-Campo, T. García, Naval-Gías, Oral squamous cell carcinoma: epidemiological study and risk factor assessment based on a 39-year series. Int. J. Oral Maxillofac. Surg. 49, 1525–1534 (2020)
X. Jiang, J. Wu, J. Wang, R. Huang, Tobacco and oral squamous cell carcinoma: A review of carcinogenic pathways. Tob. Induc. Dis. 17, 29 (2019)
G. Frohwitter, O.L. Zimmermann, K. Kreutzer, C. Doll, C.M. Rendenbach, H. Dommisch, K.D. Wolff, M.R. Kesting, M. Heiland, S. Koerdt, Oxidative and nitrosative stress in oral squamous cell carcinoma. Cells Tissues Organs 209, 120–127 (2020)
M. Gasparyan, M.C. Lo, H. Jiang, C.C. Lin, D. Sun, Combined P53/PTEN deficiency activates expression of mesenchyme homeobox 1 (MEOX1) required for growth of triple-negative breast cancer. J. Biol. Chem. 295, 12188–12202 (2020)
A. Jeggari, D.S. Marks, E. Larsson, miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063 (2012)
L. Wang, H.J. Park, S. Dasari, S. Wang, J.P. Kocher, W. Li, CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 (2013)
E. Sakai, N. Tsuchida, Most human squamous cell carcinomas in the oral cavity contain mutated p53 tumor-suppressor genes. Oncogene 7, 927–933 (1992)
E. Sakai, K. Rikimaru, M. Ueda, Y. Matsumoto, N. Ishii, S. Enomoto, H. Yamamoto, N. Tsuchida, The p53 tumor-suppressor gene and ras oncogene mutations in oral squamous-cell carcinoma. Int. J. Cancer 52, 867–872 (1992)
B. Zhong, Q. Wang, J. He, Y. Xiong, J. Cao, LncRNA LOC285194 modulates gastric carcinoma progression through activating Wnt/β-catenin signaling pathway. Cancer Med. 9, 2181–2189 (2020)
M.C. Jiang, J.J. Ni, W.Y. Cui, B.Y. Wang, W. Zhuo, Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 9, 1354–1366 (2019)
W. Zhao, Y. Liu, C. Zhang, C. Duan, Multiple roles of exosomal long noncoding RNAs in cancers. Biomed. Res. Int. 2019, 1460572 (2019)
C. Zhang, Y. Qu, H. Xiao, W. Xiao, J. Liu, Y. Gao, M. Li, J. Liu, LncRNA SNHG3 promotes clear cell renal cell carcinoma proliferation and migration by upregulating TOP2A. Exp. Cell Res. 384, 111595 (2019)
D. Dai, X.D. Feng, W.Q. Zhu, Y.N. Bao, LncRNA BLACAT1 regulates the viability, migration and invasion of oral squamous cell carcinoma cells by targeting miR-142-5p. Eur. Rev. Med. Pharmacol. Sci. 23, 10313–10323 (2019)
Y. Guo, Y. Ma, X. Hu, R. Song, L. Zhu, M. Zhong, Long non-coding RNA CEBPA-AS1 correlates with poor prognosis and promotes tumorigenesis via CEBPA/Bcl2 in oral squamous cell carcinoma. Cancer Biol. Ther. 19, 205–213 (2018)
J. Dai, J.W. Mu, H. Mu, Long non-coding RNA CRNDE regulates cell proliferation, migration, invasion, epithelial-mesenchymal transition and apoptosis in oral squamous cell carcinoma. Oncol. Lett. 17, 3330–3340 (2019)
L. Salmena, L. Poliseno, Y. Tay, L. Kats, P.P. Pandolfi, A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011)
J. Zhang, N. Li, J. Fu, W. Zhou, Long noncoding RNA HOTAIR promotes medulloblastoma growth, migration and invasion by sponging miR-1/miR-206 and targeting YY1. Biomed. Pharmacother. 124, 109887 (2020)
Y. Han, N. Wu, F. Xia, S. Liu, D. Jia, Long noncoding RNA GAS5 regulates myocardial ischemiareperfusion injury through the PI3K/AKT apoptosis pathway by sponging miR5325p. Int. J. Mol. Med. 45, 858–872 (2020)
K. Wang, J. Li, G. Xiong, G. He, X. Guan, K. Yang, Y. Bai, Negative regulation of lncRNA GAS5 by miR-196a inhibits esophageal squamous cell carcinoma growth. Biochem. Biophys. Res. Commun. 495, 1151–1157 (2018)
F.H. Tsang, S.L. Au, L. Wei, D.N. Fan, J.M. Lee, C.C. Wong, I.O. Ng, C.M. Wong, Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 35, 1597–1606 (2015)
T. Chiyomaru, S. Fukuhara, S. Saini, S. Majid, G. Deng, V. Shahryari, I. Chang, Y. Tanaka, H. Enokida, M. Nakagawa, R. Dahiya, S. Yamamura, Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J. Biol. Chem. 289, 12550–12565 (2014)
J. Chen, F.X. Wu, H.L. Luo, J.J. Liu, T. Luo, T. Bai, L.Q. Li, X.H. Fan, Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am. J. Transl. Res. 8, 4932–4941 (2016)
Y. Liu, H. Li, Y. Liu, Z. Zhu, MiR-22-3p targeting alpha-enolase 1 regulates the proliferation of retinoblastoma cells. Biomed. Pharmacother. 105, 805–812 (2018)
Y. Xu, M. Cheng, L. Mi, Y. Qiu, W. Hao, L. Li, Mir-22-3p enhances the chemosensitivity of gastrointestinal stromal tumor cell lines to cisplatin through PTEN/PI3K/Akt pathway. Iran. J. Allergy Asthma Immunol. 17, 318–325 (2018)
H.X. Dong, R. Wang, X.Y. Jin, J. Zeng, J. Pan, LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J. Cell. Physiol. 233, 4126–4136 (2018)
Funding
This study was supported by the National Nature Science Foundation of China (81702299) and the Natural Science Foundation of Hebei province, China (H2017206022).
Author information
Authors and Affiliations
Contributions
XZ contributed to study design and study supervision; YD and HY contributed to study design, acquisition of data and drafting of the manuscript; YL, WG and YZ contributed to experiments and data analysis; LX, HS, YL and WW contributed to technical and material support. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
All experiments were approved by the Research Ethics Committee of the second hospital of Hebei Medical University.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Table S1
(DOCX 19 kb)
Fig. S1
Sixteen candidate cis-Regulatory Elements were screened out on human chromosome 1 by CHIP-seq data from ENCODE database. (PNG 771 kb)
Fig. S2
The relative expression of LINC01137 in tumor cell lines and normal cells was analyzed by RNA-seq data from ENCODE database. (PNG 46 kb)
Fig. S3
The distribution of LINC01137 in normal and tumor specimens was analyzed though the Genotype-Tissue Expression (GTEx) Project and TCGA RNA-seq public datasets. The area above the abscissa represents the expression of LINC01137 in normal tissues, and the area below the abscissa represents the expression of LINC01137 in tumor tissues. TPM, Transcripts Per Million. A invasive breast carcinoma; B prostate adenocarcinoma; C ovarian serous cystadenocarcinoma; D pheochromocytoma and paraganglioma; E pleomorphic glioblastoma; F head and neck squamous cell carcinoma; G endometrial carcinoma; H cervical sarcoma; I cervical cancer; J diffuse large B-cell lymphoma; K acute myeloid leukemia; L thyroid carcinoma; M cutaneous melanoma; N testicular germ cell tumor; O Colon adenomyoma; P sarcoma; Q hepatocellular carcinoma; R lung squamous cell carcinoma; S adrenocortical carcinoma; T papillary renal cell carcinoma; U clear cell renal cell carcinoma; V gastric adenomyoma; W pancreatic cancer; X brain low-grade glioma; Y esophageal cancer. (PNG 764 kb)
Fig. S4
The open reading frames of LINC01137 predicted by ExPaSy. (PNG 7686 kb)
Fig. S5
The ribosome binding sites and open reading frames of LINC01137 predicted by RegRNA2.0. (PNG 1034 kb)
Fig. S6
RT-qPCR was used to detect LINC01137 expression in HSC-3 (A) and Tca-8113 (B) cells at 1, 2, 3, 4, 5, 6 days after transfected with NC or with LINC01137 siRNAs. Transcript levels were normalized to GAPDH expression. n = 3, **p < 0.01. (PNG 86 kb)
Fig. S7
Several miRNAs were predicted to be bound to LINC01137 (Ensembl: ENSG00000233621) by miRcode software. (PNG 2862 kb)
Fig. S8
RT-qPCR was used to detect the expression of LINC01137 after overexpression of miR-146a-5p (a), miR-148a-3p (b), miR-3619-5p (c), miR-23b-3p (d) and miR-10a-5p (e) in OSCC cells HSC3 and Tca8113. **P < 0.01. (PNG 314 kb)
Fig. S9
Schematic diagram of pmirGLO-LINC01137 dual luciferase reporter constructs. (PNG 1636 kb)
Rights and permissions
About this article
Cite this article
Du, Y., Yang, H., Li, Y. et al. Long non‐coding RNA LINC01137 contributes to oral squamous cell carcinoma development and is negatively regulated by miR-22-3p. Cell Oncol. 44, 595–609 (2021). https://doi.org/10.1007/s13402-021-00586-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-021-00586-0