Abstract
Purpose
The analysis of breast cancer residual tumors after neoadjuvant chemotherapy (nCT) may be useful for identifying new biomarkers. MicroRNAs are known to be involved in oncogenic pathways and treatment resistance of breast cancer. Our aim was to determine the role of miR-18a, a member of the miR-17-92a cluster, in breast cancer behavior and outcome after nCT.
Methods
Pre- and post-nCT tumor miR-18a expression was retrospectively assessed by qRT-PCR in 121 patients treated with nCT and was correlated with survival outcomes and with clinical and pathological characteristics. Breast cancer-derived MCF-7 and MDA-MB-231 cell lines were transfected with miR-18a and anti-miR-18a to evaluate the biological effects of this molecule. In addition, whole-transcriptome expression analysis was performed.
Results
High miR-18a expression in post-nCT residual tumors was found to be associated with a significantly worse overall survival [hazard ratio (HR): 2.80, 95% confidence interval (CI): 1.01–7.76] and a strong trend towards a poorer disease-free survival (HR: 2.44, 95% CI: 0.99–5.02) compared to low miR-18a expressing post-nCT residual tumors. Clinical and experimental data were found to be in conformity with the proliferative effects of miR-18a, which showed a significant correlation with Ki67 and MYBL2 expression, both in pre- and post-nCT tumors and in public databases. In vitro analysis of the role of miR-18a in breast cancer-derived cell lines showed that a high expression of miR-18a was associated with a low expression of the estrogen receptor (ER), a decreased sensitivity to tamoxifen and an enrichment in luminal B and endocrine resistance gene expression signatures.
Conclusions
From our data we conclude that post-nCT miR-18a expression in breast cancer serves as a negative prognostic marker, especially in luminal tumors. Clinical, in vitro and in silico data support the role of miR-18a in breast cancer cell proliferation and endocrine resistance and suggest its potential utility as a biomarker for additional adjuvant treatment in patients without a pathologic complete response to neoadjuvant therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer (BC) is the most common type of malignant neoplasm among women worldwide [1]. Its high clinical heterogeneity is partially explained by its biological heterogeneity, even within well-characterized subtypes [2, 3]. Although clinical classification according to hormone receptor (HR) and HER2 status is the current basis for defining therapeutic strategies, the complexity of BC treatment in both early and advanced disease is further increased by the lack of reliable prognostic and predictive biomarkers. In fact, most new drugs introduced in recent years have been administered according to clinical criteria, and the identification of efficacy markers is a difficult challenge. Neoadjuvant therapy is considered an advantageous platform for clinical and translational research [4], and pathologic complete response (pCR) to neoadjuvant chemotherapy (nCT) is currently accepted as a surrogate endpoint for therapeutic benefit in HER2-positive and basal tumors [5, 6]. Research on new mechanisms of resistance and their related biomarkers has recently focused on the analysis of post-treatment residual tumors, which may provide guidance for biomarker-driven studies [7]. In luminal BC, additional prognostic classifications and accurate biomarkers, beyond the residual tumor burden [8], will be needed in the future to guide the inclusion of new therapies, such as immune therapy or targeted agents, as adjuvant treatment for patients with a high risk of recurrence after nCT [9].
MicroRNAs (miRNAs) are a class of small noncoding RNAs that can regulate coding genes through RNA transcript degradation and/or translational repression. Since the discovery of their important role in the pathogenesis of cancer [10,11,12], a global effort has been made to understand how dysregulation of miRNA expression levels can modulate the biological characteristics of tumor cells (such as invasiveness, aggressiveness, metastatic potential or resistance to antitumor agents) not only during the development of a neoplasm but also in response to treatment [13]. MiRNAs have been extensively studied in BC in recent years, both from a functional point of view [14, 15] and as biomarkers of therapeutic resistance [16], including the pre- and post-treatment neoadjuvant setting [17].
MicroRNA-18a (miR-18a; hsa-miR-18a-5p; MIMAT0000072) is a member of the miR-17-92 gene cluster [18,19,20,21,22,23]. This cluster is initially transcribed as a long noncoding RNA transcript (called pri-miRNA) that contains six miRNAs (miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a and miR-92a-1), is processed by the Drosha enzyme in the nucleus to form a large miRNA precursor (called pre-miRNA) and is subsequently exported to the cytosol and processed by an enzyme known as Dicer to yield mature products [24, 25]. MiR-17-92 and its paralogous gene clusters have been found to play roles in a wide variety of diseases, including cancer. The oncogenic potential of miR-17-92, sometimes referred to as an oncomiR, was first identified in mouse viral tumorigenesis screens [26,27,28] and explained its overexpression in diverse cancer types, including BC [29]. Constitutive activation of miR-17-92 in humans has been associated with non-Hodgkin’s lymphoma [30, 31], while deletion of the cluster is lethal in mice, producing lung and lymphoid cell developmental defects [32]. Specific overexpression of miR-18a leads to silencing of the tumor suppressor PTEN [33], and a high miR-18a level in normal breast tissue has been proposed to serve as a cancer risk biomarker [34]. The contribution of miR-18a to BC behavior seems to be complex: miR-18a is overexpressed in triple negative BC (TNBC) compared to the luminal A subtype [35, 36], and it represses estrogen receptor alpha expression (ER-α) in the MCF7 luminal cell line [37]. In TNBC, miR-18a may repress lung metastasis by downregulation of HIF1A [38], but it can also contribute to paclitaxel resistance through enhancement of autophagy via inhibition of the mTOR signaling pathway, as well as by silencing of Dicer expression [39, 40]. The association of high miR-18a expression with TNBC, proliferation markers and ER negativity has also been demonstrated in two series of untreated node-negative BCs [41, 42], indicating its potential role as a biomarker of basal-like tumors. Although its expression was not prognostic in that node-negative BC population [41], a bioinformatic approach suggested that miR-18a activity may serve as a prognostic marker in TNBC [43].
In this work, we analyzed associations between miR-18a expression in residual tumors and the clinical outcome of BC patients after treatment with nCT, and we experimentally explored its contribution to luminal BC behavior and response to treatment.
2 Materials and methods
2.1 Patients and clinical characteristics
A total of 121 consecutive patients with invasive BC (stages II–III) who received nCT (sequential anthracyclines and taxanes) were recruited in the Department of Hematology and Medical Oncology, University Hospital Morales Meseguer, Spain (Table 1). Clinical evaluation included physical examination, routine blood tests, chest X-ray, mammography, ultrasound breast exam, breast MRI and core biopsy of the primary tumor. Pre-nCT nodal status was determined by axillary and/or supraclavicular ultrasound-guided fine-needle aspiration. A sentinel lymph node biopsy (SLNB) was performed before chemotherapy in cases with a negative initial evaluation for nodal metastasis. In locally advanced tumors (defined as cT3N1, cN2–3 or cT4), whole-body bone scintigraphy and chest and abdomen CT were added to the staging workup. Dynamic breast MRI was performed after completion of chemotherapy and prior to surgery to determine the clinical response. Pathological complete response (pCR) was defined as ypT0/Tis N0. Endocrine resistance was defined according to established clinical criteria [44].
2.2 RNA extraction and RT-qPCR
Total RNA from formalin-fixed paraffin embedded (FFPE) biopsies was extracted using a RNeasy FFPE Kit (QIAGEN) following the supplier’s instructions. Total RNA from cells was extracted using RNAzol (MRC) reagent and a Direct-zol RNA MiniPrep kit (ZYMO Research). All miRNAs (without preamplification) and mRNAs (with preamplification) were retrotranscribed and amplified using TaqMan® Gene Expression Assays (Life Technologies) in a LightCycler® 480 Real-Time PCR System. Relative expression was calculated by the 2ΔCt method using U6 snRNA and ACTB as endogenous controls for miRNAs and mRNAs, respectively.
2.3 Cell cultures
MCF7 (ATCC: HTB-22), MDA-MB-231 (ATCC HTB-26) and EA.hy926 (ATCC: CRL-2922) cell lines were obtained from Servicio de Apoyo a la Investigación (SAI, University of Murcia, Spain) and maintained and sub-cultured in DMEM-GlutaMAX with 1 g/l glucose and 10% fetal bovine serum (FBS, GIBCO). Mycoplasma security controls were carried out using a Venor®GeM Mycoplasma PCR Detection kit (Minerva Biolabs). Cell line authentication testing was performed in our laboratory through analysis of the ATCC recommended STR loci (TH01, TPOX, vWA, CSF1PO, D16S539, D7S820, D13S317 and D5S818 plus Amelogenin).
2.4 Transient transfection assay
Transient transfection was carried out using siPORT NeoFX Transfection Agent (Invitrogen) and mirVana™ miRNA Mimics (Life Technologies) or miRCURY LNA™ microRNA Inhibitors (EXIQON) following the manufacturer’s protocol (including a negative control in each transfection).
2.5 Proliferation assays
Proliferation was assayed by cell counting using a TC10 Automated Cell Counter (Bio-Rad) and an XTT Cell Proliferation kit II (Roche). Evaluation was performed after transient transfection with miR-18a mimic or silencer (50–200 nM) plus controls in 96-well plates (eight wells per condition) from 0 to 72 h in a Biotek Synergy HT reader.
2.6 Immunohistochemistry (IHC) from biopsies and cell blocks
Evaluation of ERα expression in tumor biopsies from clinical samples was carried out by pathologists at University Hospital Morales Meseguer using clinically standardized methods. To evaluate ERα expression in MCF7 cells by IHC, we fixed centrifuged cell pellets in formalin solution for at least 24 h after which we paraffin embedded the samples. IHC was performed in an AutostainerLink 48 using a FLEX Monoclonal Rabbit Anti-Human Estrogen Receptor α antibody (IS151).
2.7 Apoptosis assay
Apoptosis was evaluated using an Apo-ONE® Homogeneous Caspase-3/7 Assay following the manufacturer’s instructions in 100 nM miR-18a/control-transfected MCF7 cells after 48 h of incubation. Fluorescence was measured in 96-well black plates in a Biotek Synergy HT reader.
2.8 Electrophoresis and Western blotting
After cell lysis using RIPA buffer with phosphatase and protease inhibitors (Thermo-Fisher), protein concentrations were determined by bicinchoninic acid assays. SDS-PAGE was carried out with 25 μg of whole protein lysates per well in a Bio-Rad electrophoresis system. Protein transfer was performed using Amersham TE 77 PWR for one hour. Western blotting was performed using 5% milk in PBST as a blocking agent and anti-ERα (ab32063) and anti-β-actin (A5441–2ML) primary antibodies. PVDF membranes were evaluated using Amersham™ ECL™ Prime in ImageQuant LAS4000 (GE Healthcare).
2.9 Angiogenesis assays
A total of 15,000 EA.hy926 endothelial cells and 100 μl conditioned medium from growing MCF7 cells transfected with miR-18a mimic/silencer and controls were added (in octuplicate per condition) to 96-well plates containing BD Matrigel™ Basement Membrane Matrix. After 24 h of incubation, the length of the formed vessels was measured by image analysis using the Angiogenesis Analyzer plugin for ImageJ (http://image.bio.methods.free.fr/ImageJ/).
2.10 Whole-transcriptome expression profiling and gene-set enrichment analysis (GSEA)
After 48 h of transient transfection, triplicates of MCF7 cells with miR-18a overexpression or silencing (100 nM) plus controls were lysed using RNAzol, and the total RNA extracted was assessed for integrity and quality before microarray analysis. Whole transcriptomic expression analysis was carried out on an Affymetrix platform (SAI-UM) using Human Gene 2.1 ST Array Strips. Microarray data analysis was performed using Partek and GSEA software [45, 46]. The molecular signatures in the GSEA were used to test for endocrine therapy resistance [47] and BC luminal B subtype [48]. The microarray data sets used in this work are deposited in the Gene Expression Omnibus (GSE102121).
2.11 Effects of hormone therapy on miR-18a-transfected cells
After 24 h of transient transfection (50 nM scrambled or miR-18a mimic) in charcoal serum-supplemented medium, cells were seeded at a density of 25,000 cells per well (24-well plates). After attachment, the cells were stimulated with 10 nM estradiol and treated in parallel with 1 μM 4-OH-tamoxifen, 1 μM everolimus, 1 μM palbociclib, and their combinations. After treatment, the cells were detached and counted at 24, 48 and 72 h.
2.12 Analysis of validation series
A series of frozen tumors (n = 100) from the University Hospital Santa Lucía, Cartagena, Spain, was used as internal validation for biological correlations (Cartagena series) (Electronic supplementary material, Table S1). Public biological and clinical data from TCGA [49], Oxford (GSE22220) [16] and Oslo (GSE19536) [14] series were used for external validation.
2.13 Functional enrichment analysis
For functional enrichment analysis, GeneMANIA software was employed [50]. Using TCGA BC data [49], we constructed two sets of different genes based on the top genes directly correlated with miR-18a expression and the top inversely related genes. Genes with a high significant Pearson correlation coefficient (positive or negative) were chosen, and the limit of genes introduced in the application was based on the capacity limit of the application.
2.14 Statistical methods
Quartiles of expression were selected as cut-off points. The normal distribution of each variable was analyzed using one-sample Kolmogorov-Smirnov tests with Lilliefors significance correction. The association of miR-18a levels with clinical, pathological or biological data was evaluated using non-parametric tests (Mann-Whitney U test, Wilcoxon signed rank test, Kruskal-Wallis test and Spearman’s rank correlation coefficient). For multiple comparisons, a post hoc correction method (Dunn’s test or FDR for gene expression data) was applied. Kaplan-Meier curves, log-rank test, Wilcoxon test (for early differences between survival curves) and Cox proportional hazard regression multivariate models were used for disease-free survival (DFS), distant relapse-free survival (DRFS) and breast cancer-specific overall survival (OS) analyses. MiR-18a median pre-nCT and post-nCT expression was arbitrarily chosen as a predefined cut-off point for survival comparisons. Statistical analysis and graphics were performed using SPSS 21 and GraphPad Prism 4.0. All experiments were replicated at least 3 times.
3 Results
3.1 miR-18a expression associates with poor clinicopathological BC features
The expression of miR-18a was examined in pre- and post-treatment paraffin-embedded core biopsies from a series of 121 BC patients treated with nCT (Table 1). A pCR rate of 17.6% was obtained for the whole group, and the pCR for primary breast tumors was 21.0%. At the time of the last follow-up, after a median follow-up of 116 months for the whole group (n = 119), 35 recurrences and 27 deaths occurred. The median OS, DFS and DRFS were not reached. The two-year OS, DFS and DRFS were 95%, 88.1% and 88.1%, respectively. The five-year OS, DFS and DRFS were 87.4%, 75.1% and 77.1%, respectively.
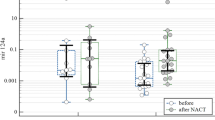
MiRNA and RNA quantification was possible in 95 pre- and 100 post-treatment BC biopsies. In both settings, miR-18a expression was found to be associated with aggressive clinical features: grade 3 (p = 0.016 and p = 0.008; Fig. 1a) and the triple negative phenotype (p = 0.001 and p = 0.034; Fig. 1b). A significantly lower expression of miR-18a was also observed in HR + HER2- tumors, both before and after treatment, compared to other tumor subtypes (Electronic supplementary material, Fig. S1). An association with extensive lymph node metastases was also observed in post-chemotherapy biopsies (p = 0.004), with a nonsignificant trend in pretreatment samples (p = 0.054; Fig. 1c). No association was found between baseline miR-18a expression and tumor size. Treatment with nCT consistently increased the overall miR-18a expression (p = 0.001) (Fig. 1d), and no association was observed between pre-nCT miR-18a expression and pCR [odds ratio (OR) = 2.185; 95% confidence interval (CI) = 0.783–6.097; p = 0.136; Fig. 1e], suggesting that miR-18a expression is not involved in resistance to chemotherapy. When miR-18a expression was analyzed according to immunohistochemical surrogate subtypes, pre-nCT expression was found to be significantly increased in triple negative tumors compared to that in HR + HER2- tumors (p = 0.0004) (Fig. 1f), an association that was maintained in post-chemotherapy samples (Electronic supplementary material, Fig. S2). These findings were replicated in an external validation cohort (Electronic supplementary material, Table S1) and in public databases (Electronic supplementary material, Fig. S3). Additional in vitro analysis showed that miR-18a expression was significantly higher in the triple negative MDA-MB-231 cell line than in the MCF7 luminal cell line (Electronic supplementary material, Fig. S4).
Association of clinical and pathological features with miR-18a expression before and after treatment with neoadjuvant chemotherapy. (a) MiR-18a expression was significantly increased in patients with poorly differentiated tumors (pre-CT Grades 1–2, n = 36; pre-CT Grade 3, n = 53; post-CT Grades 1–2, n = 39; post-CT Grade 3, n = 50), (b) the triple negative phenotype (pre-CT no TNBC, n = 72; pre-CT TNBC, n = 23; post-CT no TNBC, n = 74; post-CT TNBC, n = 22) and (c) extensive (cN2–3) lymph node involvement (pre-CT cN0–1, n = 62; pre-CT cN2–3, n = 33; post-CT cN0–1, n = 64; post-CT cN2–3, n = 36). (d) Chemotherapy-induced increase in miR-18a expression (n = 70). (e) MiR-18a expression was not associated with pathological complete response (OR = 2.185; 95% CI = 0.783–6.097; p = 0.136). (f) MiR-18a expression among different breast cancer subtypes before chemotherapy (HR + HER2-, n = 45; HR + HER2+, n = 14; HR-HER2+, n = 13; TNBC n = 23)
3.2 High post-chemotherapy miR-18a expression affects BC survival
Next, we analyzed whether post-nCT miR-18a expression in breast residual tumors was associated with patient prognosis. Median miR-18a post-nCT expression was arbitrarily predefined as the cut-off point to distinguish two groups of patients: the miR-18a high and miR-18a low groups. We found that a high miR-18a expression in the residual tumors of patients without pCR predicted a significantly worse OS (p = 0.03), DFS (p = 0.026) and DRFS (p = 0.047) in the univariate analysis (log-rank test; Fig. 2a). The prognostic impact of miR-18a expression in the residual tumors after chemotherapy was confirmed in multivariate Cox proportional hazard regression models that also included other clinical and pathological variables that were significant or nearly significant in previous univariate analyses: tumor subtype (TNBC vs. other), post-nCT node involvement (ypN+ vs. ypN0) and grade 3 (vs. grades 1–2) for DFS (Table 2). For DRFS and OS, only miR-18a expression, post-nCT node involvement and grade 3 were included in the multivariate model. By doing so, high miR-18a expression in the residual tumor was found to serve as a significant marker of worse OS (HR 2.80, 95% CI: 1.01–7.76; p = 0.048), whereas a nonsignificant trend toward worse DFS and DRFS was observed (Table 2). Survival analysis according to post-nCT subtypes showed that the main impact of high miR-18a expression on prognosis occurred in HR+ tumors (n = 58) (Fig. 2b), although statistical significance was not reached, and the effect was more evident in the early follow-up (log-rank test: 0.07, 0.05 and 0.11 for OS, DFS and DRFS; Wilcoxon test: 0.03, 0.04 and 0.08 for OS, DFS and DRFS). An exploratory comparison of the type of recurrence according to clinical definitions of endocrine resistance showed that 9/10 recurrences in the group with high miR-18a expression fulfilled the criteria for primary or secondary endocrine resistance compared to only 2/6 recurrences in the group with low miR-18a expression (Fisher’s exact test; p = 0.04). No prognostic differences were found for post-nCT miR-18a expression in HR- tumors (n = 17) (log-rank test: p = 0.61, 0.53 and 0.53 for OS, DFS and DRFS). In contrast to the results obtained in the post-chemotherapy setting, pretreatment expression of miR-18a was not found to be associated with different prognoses: no significant survival differences were found between patients with low and high pretreatment miR-18a levels (log-rank; OS: p = 0.18, DFS: p = 0.48, DRFS: p = 0.26) (Fig. 2c).
Survival analysis according to low or high tumor miR-18a expression. (a) Kaplan-Meier survival curves for DFS, DRFS and OS according to post-chemotherapy (post-nCT) expression of miR-18a (high vs. low using the post-nCT median value as the cut-off point) in patients with residual breast tumors (n = 80). (b) Kaplan-Meier survival curves for DFS, DRFS and OS according to post-chemotherapy (post-nCT) expression of miR-18a (high vs. low) in patients with residual breast tumors and hormone receptor expression (HR+) (n = 58). (c) Kaplan-Meier survival curves for DFS, DRFS and OS according to pretreatment (pre-nCT) expression of miR-18a (high vs. low using the pre-nCT median value as the cut-off point) in the whole group (n = 94). p values correspond to the log-rank test
3.3 MiR-18a expression is associated with proliferation in BC
To explore potential mechanisms to explain the association of miR-18a expression with a poor prognosis, we used those cases with miR-18a information (n = 685) from the publicly available TCGA BC database [47]. Using a cut-off point of ± 0.4 for the Pearson test, we built two gene sets based on the top miR-18a correlated genes (positive and negative), after which we performed a functional enrichment analysis using GeneMANIA software [48]. Genes associated with miR-18a expression were mainly found to be related to mitotic features and cell cycle checkpoints. A nonsignificant trend towards an inverse association between miR-18a expression and estrogen response was also observed (Table 3). The gene sets used as inputs are deposited as Electronic supplementary material (Spreadsheet S1).
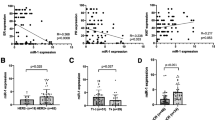
Next, we validated the association of miR-18a expression with proliferation in our cohort of patients. The mRNA expression of MKI67 and MYBL2, two well-known genes involved in proliferation, was found to significantly correlate with miR-18a expression, both in pre- and post-chemotherapy biopsies (Table 4). We subsequently validated the association with MKI67 using an external series of untreated frozen BC samples (n = 100) from another center (Hospital Universitario Santa Lucía, Cartagena, Spain). In this cohort, we found that miR-18a expression was significantly increased in patients with high IHC Ki67 scores (using both 14% and 20% as cut-off points) (Electronic supplementary material, Fig. S5). Additional validation for both MKI67 and MYBL2 was obtained in three public external series (Table 4). An exploratory analysis of the correlation between ESR1 and miR-18a expression in the same databases showed an inverse correlation between both markers (Table 4).
3.4 miR-18a induces in vitro changes in proliferation, gene expression and other cellular processes
To better understand the processes by which miR-18a induces proliferation in luminal BC, we employed expression microarray analyses after transient transfection of MCF7 cells with 100 nM miR-18a mimic, anti-miR-18a and two controls. Efficient miRNA transfection was confirmed by RT-qPCR (Electronic supplementary material, Fig. S6). Hierarchical cluster analysis showing the best up- or downregulated markers after miR-18a overexpression is depicted in Fig. 3a, and the protein-coding genes with the strongest alterations are shown in Fig. 3b. Analysis performed at the exonic level revealed that miR-18a may be involved in biogenesis and cellular component organization in MCF7 cells (Fig. 4).
Transcriptome analysis of MCF7 cells after overexpression and silencing of miR-18a. (a) Microarray matrix comparing each condition (two controls vs. overexpression vs. silencing; n = 3/group) and showing the list of top candidate genes. (b) Top down- and up-regulated protein-coding genes in our experimental model
Pathway analysis also showed that miR-18a may be involved in proliferative pathways, such as the cell cycle (p < 0.001), estrogen signaling (p = 0.001), DNA replication (p = 0.002), PI3K-Akt signaling (p = 0.02) or p53 signaling (p = 0.04) pathways (Table 5). These results were substantiated by GO enrichment analysis, showing that miR-18a acts in biological processes such as the mitotic cell cycle (p < 0.001), organelle fission (p < 0.001) and nuclear division (p < 0.001), thereby suggesting a complex role and multiple associations with proliferative networks, together with other altered pathways (Table 5). Finally, we conducted a search for expression changes in other candidate genes that have previously been reported to act in pathways mediating the effects of miR-18a on cancer cells, without finding new associations (Electronic supplementary material, Table S2).
To further understand the relevance of miR-18a in luminal BC, we evaluated the direct effects of experimental overexpression of miR-18a in MCF7 cells. As predicted by in silico analysis and by our data on MKI67 and MYBL2 expression, overexpression of miR-18a increased the proliferation of MCF7 cells in an XTT proliferation assay after 24 h (Fig. 5a). In addition, we explored alternative mechanisms of miR-18a-induced biological aggressiveness in luminal BCs. First, no differences were observed in vimentin or E-cadherin expression after transfection with miR-18a or anti-miR18a, suggesting that epithelial-mesenchymal transition (EMT) is not induced by miR-18a overexpression or silencing (Electronic supplementary material, Fig. S7). Conditioned medium from MCF7 cells overexpressing miR-18a did not enhance angiogenesis, but decreased microvessel density (MVD) in an in vitro angiogenesis assay using EA.hy926 as an endothelial cell line model (p = 0.001). With anti-miR-18a, the effect in MCF7 cells was the opposite, with a slight increase in the MVD of EA.hy926 (p = 0.027) (Fig. 5b), which suggests a mild antiangiogenic effect of miR-18a. Western blot analyses of angiogenic markers did not show any miR-18a-related changes in angiogenic markers, such as VEGFA, THBS1, PDGFBB or HIF1A (Electronic supplementary material, Fig. S8). We also did not find any differences in the apoptotic index after transfection of MCF7 cells with either miR-18a or anti-miR-18a (Fig. 5c). Finally, no miR-18a-related differences were observed in the low migration capacity of MCF7 cells (p = 0.200) (Fig. 5d) or in its invasive ability (p = 0.506) (Fig. 5e). The same in vitro analyses were performed on MDA-MB-231, a basal-like cell line with a high expression of miR-18a, with similar results, although the increase in proliferation was slight and occurred later (Electronic supplementary material, Fig. S8). Together, these results are consistent with induction of proliferation by miR-18a expression without a substantial modification of apoptosis, migration and/or invasion.
Analysis of the effects of miR-18a on MCF7 in vitro. (a) Overexpression of miR-18a showed a slight but significant increase in the proliferation index of MCF7 cells. (b) MiR-18a showed a slight but significant antiangiogenic effect in the EA.hy926 endothelial cell model. (c) MiR-18a showed no effect on the apoptotic index of MCF7 cells. (d) MiR-18a did not affect the migration of MCF7 cells. (e) MiR-18a did not affect the invasive features of MCF7 cells
3.5 MiR-18a represses ERα by blocking protein translation and is associated with low ERα expression in both untreated and nCT-treated BC samples
As previously reported, ERα is a target of miR-18a. To elucidate its effect in BC, we first performed transient transfection assays with miR-18a mimic and control mimic in MCF7cells. By doing so, we found that overexpression of miR-18a did not affect the estrogen receptor (ESR1) mRNA levels (Fig. 6a). Second, we confirmed by Western blot analysis that miR-18a inhibited protein expression of both the 66 kDa and the 36 kDa Erα isoforms (Fig. 6b). This result was confirmed by IHC quantification of the same transfected cells (Fig. 6c). We also confirmed an inverse correlation between miR-18a and both ERα mRNA and protein expression levels in pre‐chemotherapy and post-chemotherapy BC biopsies in our clinical series (Electronic supplementary material, Fig. S9).
MiR-18a affects ERα expression only at the protein level in MCF7 cells. (a). Overexpression or silencing of miR-18a in MCF7 cells did not affect ESR1 mRNA levels. (b) MiR-18a impaired both ER66 and ER36 isoform levels of ERα. (c) This result was confirmed using IHC in formalin-fixed, paraffin-embedded and previously transfected MCF7 cells. The proportion of ERα nuclei was significantly reduced after transfection with miR-18a using a clinically validated antibody
3.6 Luminal BC cells transfected with miR-18a show in vitro resistance to tamoxifen and are enriched in luminal B and endocrine resistance signatures
To determine the potential therapeutic relevance of miR-18a expression in luminal BC, we tested the effect of tamoxifen, as a sole agent or combined with everolimus or palbociclib, on MCF7 cells supplemented with estradiol (10 nM) after transfection with miR-18a. Compared to control cells (scrambled), we found that miR-18a-transfected cells did not slow their growth after exposure to tamoxifen (p = ns), while this resistance was reversed by combination of tamoxifen with everolimus or palbociclib (p = 0.044 and p = 0.024, respectively), which showed a response pattern similar to that found in control cells (Fig. 7). Additionally, GSEA of MCF7 cells transfected with miR-18a showed enrichment in endocrine resistance (FDR < 0.001) (Fig. 8a) and luminal B (FDR = 0.021) signatures (Fig. 8b).
Sensitivity of MCF7 cells transfected with miR-18a to hormone therapy agents. MCF7 cells transfected with scrambled (a) or miR-18a (b) were stimulated with estradiol (E2, 10 nM) and treated with tamoxifen (TAM), tamoxifen plus everolimus (EVE) or tamoxifen plus palbociclib (PAL). Cell growth was evaluated for 72 h. * Indicates adjusted p value < 0.05
4 Discussion
In this work, we found that high miR-18a expression may be a marker of a poor OS and DFS after nCT, especially in luminal tumors. In silico and in vitro data suggest that this impaired prognosis may be related to miR-18a involvement in increased proliferation and loss of estrogen dependence in BC, thereby suggesting the potential utility of miR-18a as a prognostic and predictive marker. In our clinical series, we found that miR-18a expression in post-chemotherapy residual tumors, a likely representation of the resistant fraction of the primary tumor [7], correlates with the expression of proliferation markers. This finding does not seem to depend only on its association with grade 3 or the triple negative subtype, both of which are well-known tumor phenotypes characterized by a high proliferation rate. In addition to these associations, exogenous overexpression of miR-18a in the luminal MCF7 cell model, as well as in the basal MDA-MB-231 cell model, significantly increased the proliferation rate. Our approach does not allow for the identification of a single target of miR-18a that is responsible for the activation of proliferation processes, but since a single miRNA may regulate the expression of up to 200 genes [51], we aimed to assess global transcriptomic associations. Using a pathway-based approach [52] to evaluate changes in global mRNA expression, we revealed the activation of some key pathways for proliferation due to miR-18a overexpression in the MCF7 luminal model. These results were consistent with a previous bioinformatic approach using TCGA data in which, by selecting the top correlated genes with miR-18a (directly or indirectly) and using pathway and GO enrichment analysis, we found a direct correlation with proliferation. We performed an additional search for expression changes in other genes, such as PTEN, KLF4 and ATM, previously reported to be related to biological effects of miR-18a on cancer, without finding further associations. The role of the miR-17-92 cluster and miR-18a in proliferation has been extensively studied in vitro and in vivo in some cancer types as well as in normal tissues [53,54,55,56], and previous work also demonstrated an association of its expression with a high proliferation rate in untreated node-negative BC [41]. Our data, in addition to establishing further biological correlations, extend this association to node-positive disease and post-treatment biopsies.
Importantly, in the luminal model, we found that miR-18a induced an increase in proliferation without substantially modifying other relevant characteristics of tumor behavior, such as invasiveness or migration. Only angiogenesis was partially impaired by miR-18a expression, a finding that is in agreement with other reports [57], although we did not find any changes in the expression of THBS1 or other angiogenic markers. These data suggest that the angiogenic role of miR-18a differs from that found for other members of the miR-17-92 cluster, such as miR-20a, which are associated with pro-angiogenic effects [58]. A second relevant finding is that, although miR-18a expression was associated with increased expression of proliferation markers such as MKI67, a high miR-18a baseline expression was not associated with a different rate of pCR. This finding is somewhat paradoxical since proliferation-related gene expression signatures have been reported to be the main components of predictive signatures of chemotherapy responses [59, 60]. Taken together, these results suggest a role of miR-18a in tumor behavior that goes beyond a pure increase in proliferation and that is not clearly associated with sensitivity to chemotherapy.
The miR-18a-related increase in BC proliferation also seems to be independent of ER-associated stimuli, occurs both in luminal BC and in TNBC, and has been confirmed in clinical series without any selection for BC subtypes. However, the role of miR-18a in ER expression and signaling seems to be complex. First, our data together with previous work shows miR-18a-mediated silencing of ER expression through a direct protein translation blockage. In fact, miR-18a expression has a strong inverse correlation with ESR1 mRNA expression in our series as well as in the TCGA BC database, while showing a strong direct correlation with proliferative markers such as MKI67 or MYBL2. Determination of whether the acquisition of luminal B genomic signatures is related only to ESR1 loss or to a wider reprogramming of estrogen signaling programs requires other experimental approaches. Second, it has been reported that pri-mir-17–92 expression is induced by ER after 17-β-estradiol stimuli in ER+ cells, but that these changes do not correlate with increased levels of miR-18a or other miRNAs in the cluster, which suggests inhibition during miRNA biogenesis and different processing to miR-18a according to the BC subtype [61, 62]. MiR-17-92 expression is induced primarily by MYC in some cancer types [63, 64], as well as by other proteins, such as VEGF, through ERK/ELK1 activation in endothelial cells [65]. These mechanisms may explain upregulation of miR-18a in TNBC [66, 67]. Independent of the precise mechanisms underlying these changes, the prognostic impact on HR-positive tumors (all of them with adjuvant endocrine treatment) and the significant association of high miR-18a expression with clinical endocrine resistance suggest a potential interaction of miR-18a with hormonal therapy. Additionally, our experimental data are consistent with the hypothesis that MCF7 cells overexpressing miR-18a are at least partially resistant to treatment with tamoxifen. These results seem to contradict other work showing participation of miR-18a in UCA1-dependent tamoxifen resistance [68], although our transcriptomic data on MCF7 further support the induction of endocrine resistance signatures by miR-18a. However, the combination of tamoxifen with everolimus, especially with palbociclib, showed a response similar to that found in control MCF7 cells, which may suggest that patients with luminal tumors overexpressing miR-18a could derive maximum benefit from schemes combining hormone therapy and CDK4/6 inhibitors.
MiR-17-92 and specifically miR-18a expression is considered as a marker of TNBC [41]. Here, although the expression of miR-18a is low in luminal BC, we found that a relatively high miR-18a expression was associated with a poor prognosis, especially in the luminal subtype. Unfortunately, the development of new drugs for metastatic luminal BC has not been paralleled with the introduction of validated biomarkers. In the neoadjuvant setting, the burden of residual disease is the only reliable prognostic marker [8], and new biomarkers to guide therapeutic strategies are needed, especially when new molecularly targeted agents such as CDK4/6 inhibitors finally reach the adjuvant setting. Thus, the characterization of miR-18a as a prognostic biomarker related to high proliferation and endocrine resistance, if further validated, may be instrumental for the identification of high-risk luminal BC patients with residual disease after nCT. In addition to patient selection for new adjuvant strategies, miR-18a may also be an interesting target for silencing in aggressive luminal tumors. Furthermore, a prognostic value of miR-18a plasma levels, already proposed as a diagnostic tool for BC [69], may be explored in the luminal BC population.
Our work has several limitations. First, the apparent prognostic relevance of miR-18a in luminal tumors may be related to the small sample size. Thus, our findings support the prognostic impact of miR-18a in HR-positive tumors, but are probably not sufficient to exclude a similar effect in HR-negative BC. Second, although our experimental results seem to be in concordance with our clinical and pathological findings, further insight may be obtained with a more wide-ranging analysis of activated or silenced miR-18a targets, with a more comprehensive analysis of miR-18a relevance for endocrine resistance or with a more precise classification of tumors according to molecular subtypes.
In conclusion, miR-18a may serve as a new prognostic factor in luminal BC with residual tumors after nCT. Its relationship with high proliferation, ER downregulation and transcriptomic changes associated with luminal B signatures, together with preliminary experimental data showing resistance to tamoxifen without impaired sensitivity to mTOR or CDK4/6 inhibitors, indicates that it is also a potential biomarker for new targeted agents, both in adjuvant and neoadjuvant settings. Further insights into the biological mechanisms underlying these findings and additional validation in larger clinical series are needed to definitely establish the relevance and usefulness of miR-18a in the management of BC.
References
L.A. Torre, F. Bray, R.L. Siegel, J. Ferlay, J. Lortet-Tieulent, A. Jemal, Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108 (2015)
C.M. Perou, T. Sørlie, M.B. Eisen, M. van de Rijn, S.S. Jeffrey, C.A. Rees, J.R. Pollack, D.T. Ross, H. Johnsen, L.A. Akslen, Ø. Fluge, A. Pergamenschikov, C. Williams, S.X. Zhu, P.E. Lønning, A.-L. Børresen-Dale, P.O. Brown, D. Botstein, Molecular portraits of human breast tumours. Nature 406, 747–752 (2000)
T. Sorlie, C.M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M.B. Eisen, M. van de Rijn, S.S. Jeffrey, T. Thorsen, H. Quist, J.C. Matese, P.O. Brown, D. Botstein, P.E. Lonning, A.-L. Borresen-Dale, Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98, 10869–10874 (2001)
M. Kaufmann, G. von Minckwitz, E.P. Mamounas, D. Cameron, L.A. Carey, M. Cristofanilli, C. Denkert, W. Eiermann, M. Gnant, J.R. Harris, T. Karn, C. Liedtke, D. Mauri, R. Rouzier, E. Ruckhaeberle, V. Semiglazov, W.F. Symmans, A. Tutt, L. Pusztai, Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol 19, 1508–1516 (2012)
P. Cortazar, L. Zhang, M. Untch, K. Mehta, J.P. Costantino, N. Wolmark, H. Bonnefoi, D. Cameron, L. Gianni, P. Valagussa, S.M. Swain, T. Prowell, S. Loibl, D.L. Wickerham, J. Bogaerts, J. Baselga, C. Perou, G. Blumenthal, J. Blohmer, E.P. Mamounas, J. Bergh, V. Semiglazov, R. Justice, H. Eidtmann, S. Paik, M. Piccart, R. Sridhara, P.A. Fasching, L. Slaets, S. Tang, B. Gerber, C.E. Geyer, R. Pazdur, N. Ditsch, P. Rastogi, W. Eiermann, G. von Minckwitz, Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 384, 164–172 (2014)
S. Di Cosimo, G. Arpino, D. Generali, Neoadjuvant treatment of HER2 and hormone-receptor positive breast cancer - moving beyond pathological complete response. Breast 23, 188–192 (2014)
J.M. Balko, J.M. Giltnane, K. Wang, L.J. Schwarz, C.D. Young, R.S. Cook, P. Owens, M.E. Sanders, M.G. Kuba, V. Sánchez, R. Kurupi, P.D. Moore, J.A. Pinto, F.D. Doimi, H. Gómez, D. Horiuchi, A. Goga, B.D. Lehmann, J.A. Bauer, J.A. Pietenpol, J.S. Ross, G.A. Palmer, R. Yelensky, M. Cronin, V.A. Miller, P.J. Stephens, C.L. Arteaga, Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 4, 232–245 (2014)
M. Choi, Y.H. Park, J.S. Ahn, Y.-H. Im, S.J. Nam, S.Y. Cho, E.Y. Cho, Assessment of pathologic response and long-term outcome in locally advanced breast cancers after neoadjuvant chemotherapy: Comparison of pathologic classification systems. Breast Cancer Res Treat 160, 475–489 (2016)
P. Samadi, S. Saki, M. Pourjafar, M. Saidijam, Emerging ways to treat breast cancer: Will promises be met? Cell Oncol 41, 605–621 (2018)
M. Negrini, M.S. Nicoloso, G.A. Calin, MicroRNAs and cancer — New paradigms in molecular oncology. Curr Opin Cell Biol 21, 470–479 (2009)
G.A. Calin, C.M. Croce, MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866 (2006)
R.I. Gregory, R. Shiekhattar, R.I. Gregory, R. Shiekhattar, MicroRNA biogenesis and cancer microRNA biogenesis and cancer. Cancer Res 65, 3509–3512 (2005)
G.S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C. Polytarchou, K.B. Marcu, A.G. Papavassiliou, R. Sandaltzopoulos, E. Kolettas, A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol 40, 303–339 (2017)
E. Enerly, I. Steinfeld, K. Kleivi, S.K. Leivonen, M.R. Aure, H.G. Russnes, J.A. Rønneberg, H. Johnsen, R. Navon, E. Rødland, R. Mäkelä, B. Naume, M. Perälä, O. Kallioniemi, V.N. Kristensen, Z. Yakhini, A.L. Børresen-Dale, miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One 6, e16915–e16915 (2011)
A.J. Lowery, N. Miller, A. Devaney, R.E. McNeill, P.A. Davoren, C. Lemetre, V. Benes, S. Schmidt, J. Blake, G. Ball, M.J. Kerin, MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 11, R27–R27 (2009)
F.M. Buffa, C. Camps, L. Winchester, F.M. Buffa, C. Camps, L. Winchester, C.E. Snell, H.E. Gee, microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast Cancer Therapeutic Targets Identified by Integrated mRNA and microRNA Expression Profiling in Breast Cancer. Cancer Res 71, 5635–5645 (2011)
M. Raychaudhuri, H. Bronger, T. Buchner, M. Kiechle, W. Weichert, S. Avril, MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 162, 511–521 (2017)
Z. Mourelatos, J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, G. Dreyfuss, miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16, 720–728 (2002)
S. Griffiths-Jones, The microRNA registry. Nucleic Acids Res 32, 109D–111D (2004)
S. Griffiths-Jones, R.J. Grocock, S. van Dongen, A. Bateman, A.J. Enright, miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34, D140–D144 (2006)
A. Kozomara, S. Griffiths-Jones, miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39, D152–D157 (2011)
A. Kozomara, S. Griffiths-Jones, MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–D73 (2014)
A. Tanzer, P.F. Stadler, Molecular evolution of a microRNA cluster. J Mol Biol 339, 327–335 (2004)
V. Ambros, microRNAsTiny regulators with great potential. Cell 107, 823–826 (2001)
M. Lagos-Quintana, R. Rauhut, W. Lendeckel, T. Tuschl, Identification of novel genes coding for RNAs of small expressed RNAs. Science 294, 853–858 (2001)
S. Landais, S. Landry, P. Legault, E. Rassart, Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res 67, 5699–5707 (2007)
C.L. Wang, B.B. Wang, G. Bartha, L. Li, N. Channa, M. Klinger, N. Killeen, M. Wabl, Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci U S A 103, 18680–18684 (2006)
H.C. Hwang, C.P. Martins, Y. Bronkhorst, E. Randel, A. Berns, M. Fero, B.E. Clurman, Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc Natl Acad Sci U S A 99, 11293–11298 (2002)
M. JT, miRiad roles for the miR-17[sim]92 cluster in development and disease. Cell 133, 217–222 (2008)
A. Rinaldi, G. Poretti, I. Kwee, E. Zucca, C. Catapano, M.G. Tibiletti, F. Bertoni, Concomitant MYC and microRNA cluster miR-17-92 (C13orf25) amplification in human mantle cell lymphoma [2]. Leuk Lymphoma 48, 410–412 (2007)
A. Ota, H. Tagawa, S. Karnan, A. Ota, H. Tagawa, S. Karnan, S. Tsuzuki, A. Karpas, S. Kira, Y. Yoshida, M. Seto, Identification and characterization of a novel gene , C13orf25 , as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64, 3087–3095 (2004)
A. Ventura, A.G. Young, M.M. Winslow, L. Lintault, A. Meissner, S.J. Erkeland, J. Newman, R.T. Bronson, D. Crowley, J.R. Stone, R. Jaenisch, P.A. Sharp, Targeted deletion reveals essential and overlapping functions of the miR-17w92 family of miRNA clusters. Cell 132, 875–886 (2008)
J.K. Mouw, Y. Yui, L. Damiano, R.O. Bainer, J.N. Lakins, I. Acerbi, G. Ou, A.C. Wijekoon, K.R. Levental, P.M. Gilbert, E.S. Hwang, Y.Y. Chen, V.M. Weaver, Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med 20, 360–367 (2014)
A. Shidfar, F.F. Costa, D. Scholtens, J.M. Bischof, M.E. Sullivan, D.Z. Ivancic, E.F. Vanin, M.B. Soares, J. Wang, S.A. Khan, Expression of miR-18a and miR-210 in normal breast tissue as candidate biomarkers of breast cancer risk. Cancer Prev Res 10, 89–97 (2017)
C.M.C. Calvano Filho, D.C. Calvano-Mendes, K.C. Carvalho, G.A. Maciel, M.D. Ricci, A.P. Torres, J.R. Filassi, E.C. Baracat, Triple-negative and luminal A breast tumors: Differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumor Biol 35, 7733–7741 (2014)
N. Yoshimoto, T. Toyama, S. Takahashi, H. Sugiura, Y. Endo, M. Iwasa, Y. Fujii, H. Yamashita, Distinct expressions of microRNAs that directly target estrogen receptor α in human breast cancer. Breast Cancer Res Treat 130, 331–339 (2011)
S.-K. Leivonen, R. Mäkelä, P. Östling, P. Kohonen, S. Haapa-Paananen, K. Kleivi, E. Enerly, A. Aakula, K. Hellström, N. Sahlberg, V.N. Kristensen, A.-L. Børresen-Dale, P. Saviranta, M. Perälä, O. Kallioniemi, Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 28, 3926–3936 (2009)
R. Krutilina, W. Sun, A. Sethuraman, M. Brown, T.N. Seagroves, L.M. Pfeffer, T. Ignatova, M. Fan, MicroRNA-18a inhibits hypoxia-inducible factor 1α activity and lung metastasis in basal breast cancers. Breast Cancer Res 16, R78–R78 (2014)
Y.-X. Fan, Y.-Z. Dai, X.-L. Wang, Y.-Q. Ren, J.-J. Han, H. Zhang, MiR-18a upregulation enhances autophagy in triple negative cancer cells via inhibiting mTOR signaling pathway. Eur Rev Med Pharmacol Sci 20, 2194–2200 (2016)
L.-Y. Sha, Y. Zhang, W. Wang, X. Sui, S.-K. Liu, T. Wang, H. Zhang, MiR-18a upregulation decreases dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur Rev Med Pharmacol Sci 20, 2201–2208 (2016)
K. Jonsdottir, S.R. Janssen, F.C. Da Rosa, E. Gudlaugsson, I. Skaland, J.P.A. Baak, E.A.M. Janssen, Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS One 7, e48692–e48692 (2012)
E.A.M. Janssen, A. Slewa, E. Gudlaugsson, K. Jonsdottir, I. Skaland, H. Søiland, J.P.A. Baak, Biologic profiling of lymph node negative breast cancers by means of microRNA expression. Mod Pathol 23, 1567–1576 (2010)
E. Lee, K. Ito, Y. Zhao, E.E. Schadt, H.Y. Irie, J. Zhu, Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics 32, 96–105 (2016)
F. Cardoso, A. Costa, E. Senkus, M. Aapro, F. André, C.H. Barrios, J. Bergh, G. Bhattacharyya, L. Biganzoli, M.J. Cardoso, L. Carey, D. Corneliussen-James, G. Curigliano, V. Dieras, N. El Saghir, A. Eniu, L. Fallowfield, D. Fenech, P. Francis, K. Gelmon, A. Gennari, N. Harbeck, C. Hudis, B. Kaufman, I. Krop, M. Mayer, H. Meijer, S. Mertz, S. Ohno, O. Pagani, E. Papadopoulos, F. Peccatori, F. Penault-Llorca, M.J. Piccart, J.Y. Pierga, H. Rugo, L. Shockney, G. Sledge, S. Swain, C. Thomssen, A. Tutt, D. Vorobiof, B. Xu, L. Norton, E. Winer, 3rd ESO-ESMO international consensus guidelines for advanced breast Cancer (ABC 3). Breast 31, 244–259 (2017)
A. Subramanian, P. Tamayo, V.K. Mootha, S. Mukherjee, B.L. Ebert, M.A. Gillette, A. Paulovich, S.L. Pomeroy, T.R. Golub, E.S. Lander, J.P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005)
V.K. Mootha, C.M. Lindgren, K.-F. Eriksson, A. Subramanian, S. Sihag, J. Lehar, P. Puigserver, E. Carlsson, M. Ridderstråle, E. Laurila, N. Houstis, M.J. Daly, N. Patterson, J.P. Mesirov, T.R. Golub, P. Tamayo, B. Spiegelman, E.S. Lander, J.N. Hirschhorn, D. Altshuler, L.C. Groop, PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273 (2003)
C.J. Creighton, S. Massarweh, S. Huang, A. Tsimelzon, S.G. Hilsenbeck, C.K. Osborne, J. Shou, L. Malorni, R. Schiff, Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res 68, 7493–7501 (2008)
M. Smid, Y. Wang, Y. Zhang, A.M. Sieuwerts, J. Yu, J.G.M. Klijn, J.A. Foekens, J.W.M. Martens, Subtypes of breast cancer show preferential site of relapse. Cancer Res 68, 3108–3114 (2008)
Cancer Genome Atlas N, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012)
D. Warde-Farley, S.L. Donaldson, O. Comes, K. Zuberi, R. Badrawi, P. Chao, M. Franz, C. Grouios, F. Kazi, C.T. Lopes, A. Maitland, S. Mostafavi, J. Montojo, Q. Shao, G. Wright, G.D. Bader, Q. Morris, The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, W214–W220 (2010)
W. Wu, M. Sun, G.M. Zou, J. Chen, MicroRNA and cancer: Current status and prospective. Int J Cancer 120, 953–960 (2007)
J. Hayes, P. Peruzzi, S. Lawler, MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med 20, 460–469 (2014)
Y. Chen, L. Tian, S. Wan, Y. Xie, X. Chen, X. Ji, Q. Zhao, C. Wang, K. Zhang, J.M. Hock, H. Tian, X. Yu, MicroRNA-17-92 cluster regulates pancreatic beta-cell proliferation and adaptation. Mol Cell Endocrinol 437, 213–223 (2016)
W. Zhang, C. Lei, J. Fan, J. Wang, miR-18a promotes cell proliferation of esophageal squamous cell carcinoma cells by increasing cylin D1 via regulating PTEN-PI3K-AKT-mTOR signaling axis. Biochem Biophys Res Commun 477, 144–149 (2016)
M.D. Bo, R. Bomben, L. Hernández, V. Gattei, The MYC/miR-17-92 axis in lymphoproliferative disorders: A common pathway with therapeutic potential. Oncotarget 6, 19381–19392 (2015)
Y. Li, P.S. Choi, S.C. Casey, D.L. Dill, D.W. Felsher, MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell 26, 262–272 (2014)
R. Ferreira, T. Santos, A. Amar, S.M. Tahara, T.C. Chen, S.L. Giannotta, F.M. Hofman, MicroRNA-18a improves human cerebral arteriovenous malformation endothelial cell function. Stroke 45, 293–297 (2014)
G. Luengo-Gil, E. Gonzalez-Billalabeitia, S.A. Perez-Henarejos, E. Navarro Manzano, A. Chaves-Benito, E. Garcia-Martinez, E. Garcia-Garre, V. Vicente, F. Ayala de la Peña, Angiogenic role of miR-20a in breast cancer. PLoS One 13, e0194638 (2018)
D.G. Stover, J.L. Coloff, W.T. Barry, J.S. Brugge, E.P. Winer, L.M. Selfors, The role of proliferation in determining response to neoadjuvant chemotherapy in breast cancer: A gene expression-based meta-analysis. Clin Cancer Res 22, 6039–6050 (2016)
P. Wirapati, C. Sotiriou, S. Kunkel, P. Farmer, S. Pradervand, B. Haibe-Kains, C. Desmedt, M. Ignatiadis, T. Sengstag, F. Schütz, D.R. Goldstein, M. Piccart, M. Delorenzi, Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10, R65 (2008)
L. Castellano, G. Giamas, J. Jacob, R.C. Coombes, W. Lucchesi, P. Thiruchelvam, G. Barton, L.R. Jiao, R. Wait, J. Waxman, G.J. Hannon, J. Stebbing, The estrogen receptor- -induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A 106, 15732–15737 (2009)
S. Guil, J.F. Cáceres, The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14, 591–596 (2007)
T.V. Bui, J.T. Mendell, Myc: Maestro of MicroRNAs. Genes and Cancer 1, 568–575 (2010)
K.A. O’Donnell, E.A. Wentzel, K.I. Zeller, C.V. Dang, J.T. Mendell, c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005)
A. Chamorro-Jorganes, M.Y. Lee, E. Araldi, S. Landskroner-Eiger, M. Fernández-Fuertes, M. Sahraei, M. Quiles Del Rey, C. Van Solingen, J. Yu, C. Fernández-Hernando, W.C. Sessa, Y. Suárez, VEGF-induced expression of miR-17-92 cluster in endothelial cells is mediated by ERK/ELK1 activation and regulates angiogenesis. Circ Res 118, 38–47 (2016)
J. Xu, Y. Chen, O.I. Olopade, MYC and breast Cancer. Genes Cancer 1, 629–640 (2010)
G. Luengo-Gil, E. González-Billalabeitia, A. Chaves-Benito, E. García Martínez, E. García Garre, V. Vicente, F. Ayala de la Peña, Effects of conventional neoadjuvant chemotherapy for breast cancer on tumor angiogenesis. Breast Cancer Res Treat 151, 577–587 (2015)
X. Li, Y. Wu, A. Liu, X. Tang, Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumour Biol 37, 14733–14743 (2016)
A.R. Kodahl, M.B. Lyng, H. Binder, S. Cold, K. Gravgaard, A.S. Knoop, H.J. Ditzel, Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: A case control study. Mol Oncol 8, 874–883 (2014)
Acknowledgments
This study was funded by Instituto de Salud Carlos III (Projects PI12/02877 and PI15/1499), including FEDER funding by the European Union, and by Fundación Salud 2000 (Ayudas Merck Serono de Investigación 2012). ENM was funded by the Ministerio de Educación, Cultura y Deporte (Spain) (FPU16/06537). We acknowledge technical assistance by Dr. María José López Andreo (SAI-UM), Dr. Fara Sáez-Belmonte (SAI-UM), Charo Martínez-Marín (BIOBANC-MUR-IMIB), Lorena Velázquez (CRH), Nuria García Barberá (CRH-IMIB), Dr. Irene Martínez-Martínez (CRH-IMIB) and Natalia Bohdan (CRH-IMIB). We also acknowledge data managing support by Jose Antonio López Oliva and Natalia Andúgar (Hospital G. Universitario Morales Meseguer, Murcia, Spain).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure of potential conflict of interest
All authors declare that they have no conflict of interest.
Informed consent
All procedures performed in this study involving patients were in accordance with the ethical standards of the institutional research committee and with the Declaration of Helsinki. Written informed consent was obtained from all patients included in the study, and the study was approved by the hospital Ethics and Clinical Research Committee (CEIC Hospital G. Universitario Morales Meseguer; reference number: ESTU-23/12).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Comparative analysis of miR-18a expression (before and after chemotherapy) depending on hormone receptors (HR) and HER2 status. (a) MiR-18a expression was significantly decreased in HR + HER2- tumors before and after treatment (pre-CT n: yes = 45, no = 50, p = 0.001; post-CT n: yes = 50, no = 46; p = 0.013). (b) There were no differences in miR-18a expression in HR + HER2+ tumors (pre-CT n: yes = 14, no = 57, p = 0.170; post-CT n: yes = 13, no = 56; p = 0.771). (c) There were no differences in miR-18a expression in HR-HER2+ tumors (pre-CT n: yes = 13, no = 82, p = 0.119; post-CT n: yes = 11, no = 85; p = 0.726). (JPG 1110 kb)
Fig. S2
MiR-18a expression among different breast cancer subtypes after chemotherapy. After treatment, significant differences were observed only between HR + HER2- and TNBC tumors (Kruskal-Wallis Test, p = 0.0443; HR + HER2- n = 50, HR + HER2+ n = 13, HR-HER2+ n = 11 and TNBC n = 22). (JPG 396 kb)
Fig. S3
Validation analysis of miR-18a expression by breast cancer subtype. (a) In our validation series (Cartagena) (Kruskal-Wallis Test, p = 0.019; luminal A, n = 18; luminal B, n = 42; HR + HER2+, n = 15; HR-HER2+, n = 8; TNBC, n = 14). (b) In Oslo external series (GSE19536) (Kruskal-Wallis Test, p = 0.003; luminal A, n = 41; luminal B, n = 12; HER2+, n = 17; basal, n = 15). (c) In TCGA external series (Kruskal-Wallis Test, p < 0.0001; luminal A, n = 342; luminal B, n = 152; HER2+, n = 55; basal, n = 109). (JPG 1386 kb)
Fig. S4
Baseline expression of miR-18a in different cell lines. MiR-18a expression in the luminal model, MCF7, was significantly lower than that in the triple negative model, MDA-MB-231 (p = 0.049); expression in the luminal model, MCF7, was significantly lower than that in the HER2+ model, SK-BR-3 (p = 0.049); the HER2+ model, SK-BR-3 did not significantly differ from the triple negative model, MDA-MB-231 (p = 0.513) (n = 3 per group, three independent experiments). (JPG 591 kb)
Fig. S5
Expression of miR-18a depending on the level of ki67 (IHC). (a) Using a cutoff point of 14%. (b) Using a cutoff point of 20%. In both cases, the expression of miR-18a was associated with increased expression of ki67. N values for each group are shown in the figure. (JPG 441 kb)
Fig. S6
Efficient miR-18a transfection by RT-qPCR. Prior to array experiments, efficient transfection of MCF7 cells under all conditions was assessed by measuring the expression levels of miR-18a. (JPG 199 kb)
Fig. S7
Absence of changes in EMT markers after transfection of MCF7 cells with miR-18a and anti-miR18a. (a) Vimentin mRNA expression (n = 3 per group, three independent experiments). (b) Cadherin-1 mRNA expression (n = 3 per group, three independent experiments). (JPG 535 kb)
Fig. S8
Lack of modification of angiogenic marker expression (western blot) after transfection of MCF7 with miR-18a. (a) MiR-18a does not affect the protein expression of thrombospondin-1. (b) MiR-18a does not affect the protein expression of platelet-derived growth factor subunit B. (c) MiR-18a does not affect the protein expression of hypoxia-inducible factor 1-alpha. (d) MiR-18a does not affect the protein expression of vascular endothelial growth factor A. (JPG 2241 kb)
Fig. S9
Analysis of the effects of miR-18a on MDA-MB-231 in vitro. (a) Overexpression of miR-18a showed a slight but significant increase in the proliferation index of MDA. (b) MiR-18a conditioned medium from MDA showed no angiogenic effects on the EA.hy926 endothelial cell model. (c) Impaired migration was observed after transfection with both miR-18a and anti-miR-18a. (d) MiR-18a did not affect the invasive features of MDA (JPG 16905 kb)
Fig. S10
Association between ESR1/ERα and miR-18a expression. Expression of the estrogen receptor (mRNA: a, c; protein by immunohistochemistry: b, d) was inversely correlated with miR-18a in both prechemotherapy (a, b) and postchemotherapy (c, d) biopsies. (JPG 483 kb)
ESM1
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Luengo-Gil, G., García-Martínez, E., Chaves-Benito, A. et al. Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cell Oncol. 42, 627–644 (2019). https://doi.org/10.1007/s13402-019-00450-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-019-00450-2