Abstract
Solid-phase denitrification is one of the most promising methods for improving nitrate removal that relies on heterotrophic denitrifiers, and the availability of organic carbon significantly influences the denitrification efficiency. Synthetic carbon sources are often costly; on the other hand, natural carbon substrates are known for their abundant bioavailability. Selecting the right carbon source is crucial for achieving optimal denitrification efficiency. However, the underlying research on elucidating the efficiency of natural substrates for denitrification are limited. Therefore, the present study evaluated the denitrification potential of agricultural wastes, including rice straw (RS), wheat straw (WS), tender coconut shells (CS), and a combination of these thereof under batch and continuous mode. The study revealed that the continuous flow mode yielded the highest denitrification efficiency of 96.13%. This result outperformed the individual carbon sources’ efficiencies in batch mode, which were 72.6% (WS), 60.3% (CS), and 58.6% (RS). The superior efficiency of the continuous mode was attributed to factors such as increased biomass and stable biofilm formation. Physiochemical changes of lignocellulosic biomass in terms of distorted surface morphology following microbe-based cellulose hydrolysis were observed through scanning electron microscopy (SEM). Increase in elemental carbon and decrease in oxygen content, as evident from energy dispersive X-ray spectroscopy (EDS), further supported biofilm formation following denitrification. Changes in cellulose structure were also observed through changes in surface functional groups and crystallinity via fourier transform infrared spectroscopy (FTIR) and X-ray diffractogram (XRD) respectively. Initial visualization and microscopic identification of microbes from the denitrifying column showed the bacterial strains to form opaque, red, and yellow colonies with characteristic rod-shaped and gram-negative nature. Phylogenetic analysis through 16S rRNA sequencing identified the strains as Rhodococcus ruber and Cellulosimicrobium cellulans. Overall, the findings provide insights into the effective utilization of agricultural waste as a solid carbon source for enhancing denitrification efficiency in wastewater treatment processes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The rising population has led to a significant increase in water pollution, with nitrogen levels spiked in domestic wastewater. Agricultural runoffs, discharge of poorly treated domestic, and industrial wastewater, particularly effluents from mining activities, explosive factories, fertilizer industries, etc., are the main sources of nitrate pollution [1, 2]. In Europe, nitrate in groundwater comes from landfills, industry, and sewage, while in Asia, it is mainly due to septic tank seepage and agricultural runoff. The Mediterranean and certain parts of South Asia, driven by agriculture, face notable nitrate pollution [3]. The presence of nitrogen along with phosphorus in water bodies enriches them with anthropogenic nutrients and results in eutrophication [4]. While there are naturally occurring levels of nitrate in groundwater, quantities larger than 3 mg/L typically signify contamination [5]. The maximum contamination limit for nitrate has been set by the United States Environmental Protection Agency at 10 mg/L to prevent health hazards. But domestic discharge ranges from 30 to 70 mg/L NO3−-N, which is above the permissible limit [6]. Therefore, it is a pressing issue to limit the discharge of nitrates to the aquatic environment through the wastewater route. By eliminating nitrate from wastewater, the motto of “treat waste by waste” could be achieved [7].
Biological denitrification is the most economical and effective method for nitrate removal [8]. It is an anoxic process that requires organic carbon as an electron donor in which nitrates are the terminal electron acceptors used by denitrifying bacteria in place of oxygen [9]. Majority of wastewater lacks considerable amounts of organic carbon necessary for nitrate removal. Therefore, there is a need for an additional external carbon source to accomplish the required denitrification performance [10]. The nitrogen removal efficiency is directly influenced by the carbon source due to its chemical structure, dictating how electrons are supplied for nitrate reduction. The efficiency of heterotrophic denitrification process depends on the types and availability of organic carbons [11]. Although the denitrification efficiencies by using pure chemicals like glucose, methanol, or purified products of agricultural/industrial origin are significantly high, the high cost involved limits widespread application. Solid-phase denitrification, employing high-molecular-weight organic electron donors, has received extensive attention in the context of removing NO3− from groundwater and low carbon to nitrogen [C/N] ratio wastewater. Although high N2O emissions from inadequate C/N ratios and excessive sludge pose challenges for heterotrophic denitrification, that can be resolved by a combined approach of integrating both methods. This offers advantages including lower sulfate/total organic carbon (TOC) in effluent, stable pH, and reduced N2O emissions. Mixed electron donors exhibit better resilience to environmental changes [12].
Solid carbon sources used in the process of heterotrophic denitrification can be artificially synthesized biopolymers like poly (l-lactic acid) and polyvinyl alcohol [13] or in their natural forms like wood chips and agricultural waste. Primary sludge formed in any wastewater treatment plant can also be used as an external carbon source [14]. Agricultural waste can be a good carbon source on a large scale because of its enormous availability. It is a viable source due to its natural cellulose composition, also containing a substantial quantity of water-soluble compounds both on the surface and within the materials. Consequently, it releases carbon more quickly compared to artificial carbon sources [15, 16].
Sanchez et al. analyzed the efficiency of different lignocellulosic biomass during denitrification [17]. Similar to the above-mentioned study, Long et al. reported corncob hydrolysate could remove 97% of total nitrogen. The authors also identified potential microorganisms present in denitrifying organisms [18]. Feng et al. studied solid-phase denitrification in untreated and pre-treated rice hulls, rice straw, and corn cob samples and revealed the presence of denitrifiers and fermentative anaerobic bacteria [19]. Gao et al. and Feng et al. reported that the release of carbon substrate from lignocellulosic biomass declines with time-period [20, 21]. Zhang et al. and Sun et al. elucidated different pre-treatment techniques to make cellulose and hemicellulose, etc., more accessible to microbes, thereby increasing denitrification efficiency [22, 23].
Even though quite a number of studies have been done on denitrification process, the use of lignocellulosic biomass, as carbon substrates for denitrification under different modes of operation and its subsequent use as biofilm carrier, is limited. Thus, the present study aimed to evaluate the performance efficiency of three lignocellulosic biomass, i.e, rice straw (RS), wheat straw (WS), and coconut shell (CS) as well as their combination thereof in terms of nitrate removal from wastewater. Preliminary experiments were performed under batch mode followed by continuous mode to analyze their effect as biofilm carriers for heterotrophic denitrifying microorganisms. The study also identifies the potential microorganisms along with their biochemical characteristics to explore their underlying mechanism of action. Overall, the study portrays a cost-effective application of agricultural residues which are otherwise burnt, causing air-pollution and chronic diseases. The research would act as guideline for stakeholders to establish sustainable waste to resource recovery-based circular bio-economy.
2 Materials and methods
2.1 Organic carbon sources and nitrate solution
Agricultural wastes like rice straw (RS), wheat straw (WS), and tender coconut shells (CS) were collected from fields of villages like Rakhra and Dadhera near Patiala, which were then thoroughly washed, dried, and cut into smaller pieces prior to their use as the primary carbon sources for denitrifying bacteria. These specific carbon sources were chosen owing to their different degrees of biodegradation. A proper mix of easy and tough degrading substances was chosen for longevity in carbon supply for the denitrification process. Synthetic nitrate stock solution of 1000 ppm was prepared by dissolving 1.631 g of potassium nitrate (KNO3) to 1 L of water. Working volumes of 75 ppm nitrate was prepared by further diluting the stock solution. Since domestic wastewater contains 30 to 70 ppm nitrate, a slightly higher nitrate concentration was selected for denitrification experiments. The reason for selecting the specific nitrate concentration could furthermore be corroborated with the positive correlation between nitrate content and the abundance of denitrifiers, accompanied by a simultaneous decline in the population of nitrifying microorganisms as observed by Nair et al. [24].
2.2 Batch and column mode experimental design for denitrification
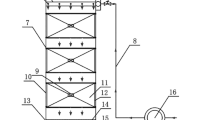
Five flasks were prepared with different quantities of RS, WS, and CS, and two flasks were taken with the mixture of these mentioned agricultural wastes as represented in Table 1 for batch-mode experiments. Twenty-five milliliters of collected sludge from a domestic sewage treatment plant was used as starter inoculum. Finally, 100 mL of working solution of 75 ppm nitrate solution was added in the whole mixture. The flasks were incubated at 120 rpm and at 37 °C. Similar experiments were replicated in closed wash bottles by adding 6 g of each agricultural wastes and 100 mL of 75 ppm nitrate solution under anoxic conditions. All the experiments were conducted in triplicates. In case of continuous mode of operation in column reactors, 27-g CS, 13-g WS, and 10-g RS were packed in a polyacrylic column to up to 10-cm height of total 50-cm length column having 10-cm diameter. Two hundred milliliters of sludge with mixed liquor suspended solids (MLSS) and mixed liquor volatile suspended solids (MLVSS) of 143.8 and 30.04 g/L respectively, were added to the column. It was left undisturbed to get acclimatized for 2–3 weeks for microbial growth and attachment to the solid carbon sources. The column was operated in up flow mode with a flow rate of 1 mL/min, resulting in a hydraulic retention time (HRT) of approximately 20 h. All the experimental set-ups are displayed in Fig. 1. Samples were withdrawn every 5 days and filtered prior to the analysis of residual nitrate concentrations. Nitrate removal efficiency (RE) was calculated using Eq. (1):
where, C0 and Ce are the influent and effluent nitrate concentrations (mg/L), respectively.
2.3 Isolation, identification, and characterization of denitrifying bacteria from column reactor
At the end of the continuous operation, partially degraded waste organic solid samples were collected from the column for analysis of inhabitant denitrifying microorganisms present in the biomass. After centrifuging a 10 mL sample at 10,000 rpm for 10 min at room temperature, appropriate dilution of the sample was spread plated using selective denitrifying media (HiMedia Nitrate Broth SKU- LQ083) and were incubated for 24–48 h for further growth. Colonies exhibiting most dominant presence were selected, and pure culture was established. Finally, pure culture of isolates was used for further biochemical characterization and molecular identification. Genomic DNA was extracted by using HiPurA Bacterial DNA purification spin column kit (MB505-250PR, HiMedia, India) and checked on 1% agarose gel electrophoresis. PCR amplification of bacterial specific 16s rRNA gene (1500 bp) was carried out by using primers F27 (5′AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTT ACGACTT 3′) [17]. The PCR reaction was performed in 25 μL volume containing 12.5 μL EmeraldAmp GT PCR Master Mix, 2× (Takara Bio USA), 1 μL DNA template (50–100 ng), 1.25 μL (10 μM) of each primer (forward and reverse), and 9 μL of free-nuclease water. PCR amplification was performed using Applied Biosystems Veriti Thermal Cycler as follows: denaturation at 94 °C for 5 min followed by 34 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1.30 min, and a final cycle at 72 °C for 7 min. PCR products were detected by staining with GelRed Nucleic Acid Gel Stain on 1% agarose electrophoresis gel in (1×) TBE buffer and visualized under UV transilluminator (Protein Simple Red Imager SA-1000). PCR product was purified using Exonuclease I and Shrimp Alkaline Phosphatase Purification Kit (New England Biolabs, Inc) and cycle sequenced using the BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, USA) with conditions as follows: denaturation at 96 °C for 1 min followed by 28 cycles of 96 °C for 1 min, 50 °C for 5 s, and 60 °C for 4 min. Cycle sequenced amplicons were purified using sodium acetate ethanol method (Thermo Fisher Scientific) and sequencing reactions were run on a 3500×L Genetic Analyzer (Applied Biosystems, USA). Sequencing files (.ab1) edited using BioEdit and further analyzed by Basic Local Alignment Search Tool (BLAST) with closest culture sequence retrieved from the National Center for Biotechnology Information (NCBI) database that finds regions of local similarity between sequences [25]. Finally, phylogenetic tree was constructed using neighbor-joining method in MEGA 2.0 software. The biochemical characterization of the pure isolates was done by KB002 HiAssortedTM Biochemical test kit (Himedia).
2.4 Analytical techniques
Nitrate measurement was done by sodium salicylate method in which 10 mL sample was taken and mixed with 1 mL of freshly prepared 1% sodium salicylate solution followed by evaporation at 100 °C. The residues were dissolved by adding 1 mL of concentrated H2SO4 followed by addition of 7 mL 10 M NaOH. The final volume was made up to 50 mL, and after 10 min of incubation time, optical density of the samples were measured at 410 nm using a UV-visible spectrophotometer [26, 27]. Surface elemental distribution/analysis of solid organic carbon sources before and after denitrification was monitored using field emission scanning electron microscope (FESEM), equipped with EDS at an accelerating voltage of 15 KV. The functional groups were analyzed via Fourier transform infrared spectroscopy (FTIR) spectra of potassium bromide (KBr) pellets containing organic carbon sample (2 mg sample in 200-mg KBr) in FTIR spectrophotometer by setting the scanning range of 500–4000 cm−1 with resolution of 4 cm−1 and a scanning rate of 64 scans per minute. The X-ray diffraction (XRD) analysis was carried out in the 2θ range of 15–70° in an X-ray diffractometer at room temperature, with Cu Kα radiation operated at 40-KV voltage, 40-mAmp current, scan speed of 3° min−1, and step size of 1 s.
3 Results and discussion
3.1 Denitrification assessment under batch mode of operation
3.1.1 Denitrification studies in flasks
Nitrate concentrations in the flasks were monitored for 95 days, primarily at intervals of 5 days as shown in Fig. 2. On the days indicated by an asterisk (*), 75 ppm of nitrate solution was added because these flasks were operated in batch mode. All the waste solid carbon sources effectively reduced nitrate concentration in the first 25 days; RS had the highest efficiency of nitrate removal corresponding to a value of 94.9%. In contrast to RS, WS is a delayed carbon release waste organic source. Despite the presence of sufficient carbon sources, the smooth surface of WS hindered the attachment of hydrolysis and denitrifying bacteria, resulting in inadequate nitrate removal efficiency [28]. Due to this, flasks 1 and 3 maintained removal efficiencies that were significantly greater, at 75.2% and 68.6%, respectively. RS exhibits a greater total organic carbon (TOC) release than WS, signifying a more rapid carbon release. This leads to the gradual detachment of carbon-containing small molecules adhered to the material’s surface and the release of easily soluble small molecules. As the carbon-containing small molecules within the carbon source materials decompose further and leach into the water, complex decomposable compounds such as hemi lignin and lignin accumulate. Consequently, this decreases the decomposition rate over time [29]. For the first 15 days, flask with all three substrates removed nitrate more effectively than the flask with just two substrates, but after that, the trend reversed. Because WS and CS were present in higher concentration in flask 5 for the remaining days, and bacteria probably only relied on them after the carbon supply from RS was exhausted, flask 6 eliminated nitrate more effectively. When batch modes of identical solid carbon sources (SCS) were examined, one with the higher concentration was more effective. Because of this, flasks 3 and 4 (Fig. 2b) both attained their maximum efficiency of 91.8% (WS) and 89.9% (CS). In a comparable investigation conducted by Yuan et al. it was demonstrated that the utilization of Ca(OH)2-pretreated rice straw facilitated the denitrification process in the treatment of swine wastewater. This enhancement significantly improved the nitrogen removal efficiency of the reactors. The application of Ca(OH)2-pretreated rice straw resulted in a notable increase of 13.54% in the removal efficiency, reaching up to 96.39%. This was in stark contrast to the untreated rice straw, which exhibited a denitrification efficiency of 82.85% [30].
3.1.2 Denitrification studies in wash bottles
Under anoxic condition, WS demonstrated the best sustained nitrate removal capability, with a maximum efficiency of 72.6%. After day 13, the addition of nitrate solution causes the nitrate concentration to rise. The highest levels of efficiency that CS and RS could reach were 60.3% and 58.6%, respectively as demonstrated in Fig. 2d. Because the breakdown of the carbon source gives the nitrate solution a dark appearance, greater optical density (OD) values were observed. Since RS degrades more quickly than the other two agricultural wastes, a greater OD value was seen, which corresponded to a higher concentration of coloring substances produced during biodegradation of RS. Likewise, in a study conducted by Feng et al. alkali-treated rice straw systems were employed to enhance the performance of NO3-N removal. The research revealed a consistently stable operational phase with nitrate removal efficiencies exceeding 90%. However, it was observed that the use of alkali-treated rice straw had detrimental effects on the long-term operational performance for nitrate removal, leading to a decline to below 80% over time [19].
3.2 Denitrification assessment under column mode of operation
The continuous mode of experiment was continued over a period of 40–45 days. In the first 10 days, denitrifying efficiency of the column was very low, with the effluent’s nitrate concentration being 46.7 and 33 mg/L on day 7 and day 10 respectively. On day 20, a maximum denitrification efficiency of 96.13% was achieved as shown in Fig. 3. This demonstrates that continuous operation improves nitrate removal because of the stable biofilm establishment inside the column. A considerably higher nitrate concentration in effluent was seen whenever the process stopped. However, after the first 10 days, an efficiency of more than 90% was achieved in maintaining nitrate concentrations lower than 10 mg/L throughout the following 41 days.
3.3 Characterization of waste organic solids before and after denitrification
3.3.1 Surface morphology analysis by scanning electron microscopy (SEM)
The development of biofilm is significantly impacted by waste organic solid’s surface properties. Analyzing the variations between a carbon source’s surface before and after denitrification reveals information about the solid carbon source’s degradation, porosity, microbial proliferation, and evenness [10]. Untreated RS’s smooth surface signifies lignin coverage, while a sample from a column’s rough surface denotes the breakdown of lignin walls, which enhances the porosity of RS fibers (Fig. 4a). Broken cellulose and pores in cell walls were visible in treated WS’s images (Fig. 4b). In such damaged WS cells, further magnification revealed the presence of microorganisms. The presence of roughness on the surface of the RS leads to an increase in its outer surface area, hence facilitating the production of bacterial biofilms. When comparing fresh straw with biofilm-detached straw, it was observed that the latter had a rough and uneven surface, characterized by the presence of dents and hollows resulting from the deterioration caused by microorganisms. The findings of this study indicate that both materials exhibit favorable characteristics as biofilm carriers for the development and utilization of microorganisms [31].
3.3.2 Energy dispersive X-ray spectroscopy (EDS) characterization of surface elemental content
As evident from Table 2, the results revealed that the carbon content increased in both RS and WS samples after denitrification as compared to the samples before denitrification. The carbon content of rice straw-raw (RSR) and wheat straw-raw (WSR) was measured to be 24.63% and 43.19%, respectively whereas the corresponding carbon content of rice straw-column (RSC) and wheat straw-column (WSC) was observed to be 45.69% and 49.8%, respectively. The observed trend is obvious since the formation of biofilm on the surface of waste organic sources contributed to the overall carbon content of the substrates [32]. This increased carbon content of the RSC and WSC is further supporting the biofilm formation as evidenced from the SEM images. Unlike the carbon content, both RSC and WSC displayed a reduced mass percentage of potassium (K), suggesting that the potassium within the biomass utilized would likely be released into the water during the immersion process [33].
3.3.3 Functional group analysis through Fourier transform infrared (FTIR) spectroscopy
The diverse molecular bending and stretching of chemical groups in the lignocellulosic biomass before and after denitrification are represented by different FTIR peaks. In WS, the peaks between 2900 and 2850 cm−1 represent aliphatic groups; the difference in intensities of these peaks indicates either pure or amorphous cellulose [34]. Peak at 1640 cm−1 corresponds to different concentrations of carboxylates. Peaks around 1500 cm−1 indicate lignin content. O–H stretching of hydrogen bonds is represented by a broad band of tiny peaks between 3200 and 3400 cm−1 [35, 36]. C–N bond stretching has been attributed to the peak near 2920 cm−1 [37]. These bands signify modifications to the cellulose structure. Peak 1445 cm−1 is associated with CH2 bending. In RS, lignin is seen towards the peaks of 1300 cm−1. Sharp peaks around 1000 cm−1 as shown in Fig. 5 imply glycosidic interactions [38]. Peak at 1630 cm−1 indicates formation of carboxylic acids and esters, and a significant rise in that peak is observed on treated RS. These results show the utilization of the lignin and hemicellulose from the RS and WS fibers. Microbial biomass deposition has the potential to elevate specific FTIR peaks in lignocellulosic biomass. As microorganisms utilize the lignocellulosic biomass as substrates and generate their own biomass during solid-phase denitrification, the resulting microbial biomass introduces unique composition and functional groups, leading to additional peaks in the FTIR spectrum. Consequently, when analyzing FTIR spectra of lignocellulosic biomass post solid-state fermentation, it is essential to consider both the original biomass composition and the supplementary peaks arising from microbial biomass. This comprehensive approach allows for a better understanding of the overall changes occurring during the solid phase denitrification process.
3.3.4 Surface crystallinity through XRD (X-ray diffraction) analysis
The change in crystallinity or amorphous nature of the waste organic solids before and after denitrification was monitored via X-ray diffraction. The diffractograms of RSR and RSC exhibited two major reflections at 2ϴ around 22 °C and 35 °C as depicted in Fig. 6 [39]. Similarly, the X-ray diffractograms of WSR and WSC also exhibited two major reflections at 2ϴ around 22 and 35 °C. The peak at 22 °C indicated the higher exposure of cellulose in both the samples whereas the broad hump appeared around 35 °C might be due to amorphous cellulose [40]. The variations in the crystal structures of the waste organic solids might be attributed to the microbial enzymatic mediated biodegradation of the lignocellulosic biomass [41]. When microbial biomass is deposited on lignocellulosic biomass, microbial communities can produce various enzymes, such as cellulases and hemicellulases, that can degrade the lignocellulosic components. The enzymatic degradation can lead to the removal of amorphous regions, breaking down hemicellulose and lignin, and exposing more crystalline cellulose. As a result, the crystallinity index of the lignocellulosic biomass may increase [42]. This finding further supports the utilization of the waste organic solids as carbon sources for heterotrophic denitrification.
3.3.5 Isolation and characterization of denitrifying microorganisms
Two dominant colonies were isolated during the sampling from column bioreactor. Both isolates can be seen visibly based on their colony pigmentation as shown in Fig. 7. One isolate was found to be of yellow color whereas the other was of red color. Testing for the generation of hydrogen sulfide, methyl red, citric acid consumption, and carbohydrate fermentation were performed. The yellow color isolates exhibited nitrate reduction and carbohydrate utilization ability (Table 3). Likewise, the red color isolate exhibited amino acid utilization, nitrate reduction, and carbohydrate utilization ability as shown in Table 3. The nitrate reduction ability of both the strain further supports the experimental evidence of denitrification of this study [43]. Both strains were subjected to DNA extraction and amplification in this study using the universal primers 27F and 1492R. The resulting PCR products were used to construct phylogenetic trees, presented in Fig. 8. By performing BLASTN homology searches on the NCBI database, it was found that strain PRM-2 exhibited a close relationship with Rhodococcus species, while strain PRM-1 was closely related to Cellulosimicrobium species. Further analysis using BLASTN revealed that strain PRM-2 had a 99.93% homology with Rhodococcus ruber (Accession no. NR_026185.1). A distinct linkage between strain PRM-2 and Rhodococcus electrodiphilus (Accession no. NR_179575.1) with 99.86% homology was also observed. On the other hand, strain PRM-1 exhibited 99.59% homology with Cellulosimicrobium cellulans (Accession no. NR_119095) according to BLASTN search results. The phylogenetic tree analysis indicated that strain PRM-1 showed a close evolutionary relationship with Cellulosimicrobium aquatile, with 99.45% homology. Phylogenetic tree has been commonly used in detecting homology between different evolutionary-related microbial species [44]. Rout et al. showed a close linkage of the identified Bacillus cereus GS-5 strain for heterotrophic nitrification and aerobic denitrification (HNAD) with Bacillus cereus PNA-2 using phylogenetic analysis [45]. 16s rRNA sequencing and maximum likelihood analysis resulted in the identification of Chryseobacterium haifense from abbatoir wastewater with capacity of simultaneous heterotrophic nitrification and aerobic denitrification [46].
4 Conclusions
Agricultural residues like rice straw, wheat straw, and coconut shell have potential to act as promising carbon substrates and biofilm carriers during solid phase denitrification in batch and continuous mode. Under aerobic/anoxic conditions in flasks under batch mode, the nitrate removal efficiency varied from 89.9% (CS) to 94.9% (WS). The efficiency was found to decline further in wash bottles under anoxic mode with sludge being replaced with effluent from shake flasks, owing to decline in organic load or inoculum amount. Maximum nitrate removal efficiency of 96.13% was achieved under continuous mode of operation in denitrifying column. Increased efficiency could be attributed to the presence of more biomass, stable biofilm formation, and the presence of both difficult and easy to degrade carbon sources, which ensure the immobilized microbes the access to both fast and slow released carbon. The existence of mixed waste organic solids, a favorable C/N ratio, and the development of a stable biofilm of denitrifying bacteria mandated nitrate removal in both early and later days, contributed to the experiment’s longevity in the column. The biodegradation of lignocellulosic biomass resulting in biofilm formation was further confirmed through changes in surface morphology and microbial deposition as observed in SEM. Increased carbon and decreased elemental content of carbon and oxygen in EDS also corroborated the development of biofilm during denitrification. Changes in surface functional groups and crystallinity of cellulose analyzed via FTIR and XRD also revealed hydrolysis of biomass, thereby promoting microbial growth favoring denitrification. Microbial analysis of the column after the completion of experimental period revealed the involvement of limited consortium of active denitrifiers (red and yellow isolates). Biochemical analysis revealed the nitrate reduction potential of both red and yellow isolates, thereby confirming the experimental evidences of denitrification of this study. Microbial sequencing and phylogenetic analysis showed the presence of Rhodococcus ruber and Cellulosimicrobium cellulans. The study establishes the use of waste agricultural residues to act as a low-cost solid state carbon substrate for promoting denitrification, thereby facilitating easier scale-up of efficient wastewater treatment strategies.
References
How SW, Ting CX, Yap JY, Kwang CY, Tan CK, Yoochatchaval W et al (2021) Effect of carbon-to-nitrogen ratio on high-rate nitrate removal in an upflow sludge blanket reactor for polluted raw water pre-treatment application. Sustain Environ Res 31(1):1–10. https://doi.org/10.1186/s42834-021-00086-8
Zhang X, Zhang Y, Shi P, Bi Z, Shan Z, Ren L (2021) The deep challenge of nitrate pollution in river water of China. Sci Total Environ 770:144674. https://doi.org/10.1016/j.scitotenv.2020.144674
Abascal E, Gómez-Coma L, Ortiz I, Ortiz A (2022) Global diagnosis of nitrate pollution in groundwater and review of removal technologies. Sci Total Environ 810:152233. https://doi.org/10.1016/j.scitotenv.2021.152233
Shen LQ, Amatulli G, Sethi T, Raymond P, Domisch S (2020) Estimating nitrogen and phosphorus concentrations in streams and rivers, within a machine learning framework. Sci Data 7(1):161. https://doi.org/10.1038/s41597-020-0478-7
Madison RJ, Brunett JO (1985) Overview of the occurrence of nitrate in groundwater of the United States. US Geol Survey Water Suppl Paper 2275:93–105
Subhashree Devasena S, Padmavathy P, Rani V, Ganesan P, Jaculine Pereira J (2023) Thermally modified fish scale as potential adsorbents for the remediation of nitrite, nitrate and ammonium ions in wastewater. Biomass Convers Biorefin 1-17. https://doi.org/10.1007/s13399-023-04298-3
Clarridge JE III (2004) Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 17(4):840–862. https://doi.org/10.1128/cmr.17.4.840-862.2004
Shen Z, Zhou Y, Wang J (2013) Comparison of denitrification performance and microbial diversity using starch/polylactic acid blends and ethanol as electron donor for nitrate removal. Bioresour Technol 131:33–39. https://doi.org/10.1016/j.biortech.2012.12.169
Wang J, Chu L (2016) Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol Adv 34(6):1103–1112. https://doi.org/10.1016/j.biotechadv.2016.07.001
Kim E, Shin SG, Jannat MAH, Tongco JV, Hwang S (2017) Use of food waste-recycling wastewater as an alternative carbon source for denitrification process: a full-scale study. Bioresour Technol 245:1016–1021. https://doi.org/10.1016/j.biortech.2017.08.168
Rout PR, Bhunia P, Dash RR (2017) Assessing possible applications of waste organic solid substances as carbon sources and biofilm substrates for elimination of nitrate toxicity from wastewater. J Hazard Toxic Radioactive Waste 21(3):04016027. https://doi.org/10.1061/(asce)hz.2153-5515.0000350
Pang Y, Wang J (2021) Various electron donors for biological nitrate removal: a review. Sci Total Environ 794:148699. https://doi.org/10.1016/j.scitotenv.2021.148699
Xu Z, Dai X, Chai X (2018) Effect of influent pH on biological denitrification using biodegradable PHBV/PLA blends as electron donor. Biochem Eng J 131:24–30. https://doi.org/10.1016/j.bej.2017.12.008
Cao S, Du R, Zhou Y (2020) Development of a denitrification system using primary sludge as solid carbon source–potential to couple with anammox process. Sci Total Environ 737:140315. https://doi.org/10.1016/j.scitotenv.2020.140315
Feng Y, Wang L, Yin Z, Cui Z, Qu K, Wang D et al (2023) Comparative investigation on heterotrophic denitrification driven by different biodegradable polymers for nitrate removal in mariculture wastewater: organic carbon release, denitrification performance, and microbial community. Front Microbiol 14:1141362. https://doi.org/10.3389/fmicb.2023.1141362
Peng Z, Jiang K, Lou T, Niu N, Wang J (2021) Enhanced denitrification of secondary effluent using composite solid carbon source based on agricultural wastes and synthetic polymers. Water Sci Technol 83(4):886–893. https://doi.org/10.2166/wst.2021.023
Sánchez MP, Sulbarán-Rangel BC, Tejeda A, Zurita F (2020) Evaluation of three lignocellulosic wastes as a source of biodegradable carbon for denitrification in treatment wetlands. Int J Environ Sci Technol 17:4679–4692. https://doi.org/10.1007/s13762-020-02815-9
Long Y, Ma Y, Wan J, Wang Y, Tang M, Zheng Q, Ma Y (2023) Hydrolysate from the enzymatic treatment of corn cob as a carbon source for heterotrophic denitrification process. J Water Process Eng 51:103473. https://doi.org/10.1016/j.jwpe.2022.103473
Feng L, Chen K, Han D, Zhao J, Lu Y, Yang G et al (2017) Comparison of nitrogen removal and microbial properties in solid-phase denitrification systems for water purification with various pretreated lignocellulosic carriers. Bioresour Technol 224:236–245. https://doi.org/10.1016/j.biortech.2016.11.002
Gao L, Zhang L, Hou J, Wei Q, Fu F, Shao H (2013) Decomposition of macroalgal blooms influences phosphorus release from the sediments and implications for coastal restoration in Swan Lake, Shandong, China. Ecol Eng 60:19–28. https://doi.org/10.1016/j.ecoleng.2013.07.055
Feng L, Yang G, Yang Q, Zhu L, Xu X, Gao F (2015) Enhanced simultaneous nitrification and denitrification via addition of biodegradable carrier Phragmites communis in biofilm pretreatment reactor treating polluted source water. Ecol Eng 84:346–353. https://doi.org/10.1016/j.ecoleng.2015.09.041
Zhang Y, Chen X, Gu Y, Zhou X (2015) A physicochemical method for increasing methane production from rice straw: extrusion combined with alkali pretreatment. Appl Energy 160:39–48. https://doi.org/10.1016/j.apenergy.2015.09.011
Sun S, Sun S, Cao X, Sun R (2016) The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour Technol 199:49–58. https://doi.org/10.1016/j.biortech.2015.08.061
Nair RR, Dhamole PB, Lele SS, D’Souza SF (2007) Biological denitrification of high strength nitrate waste using preadapted denitrifying sludge. Chemosphere 67(8):1612–1617. https://doi.org/10.1016/j.chemosphere.2006.11.043
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/s0022-2836(05)80360-2
Amirsadat K, Sharififard H, Lashanizadegan A (2022) Adsorption of nitrate from municipal wastewater by synthesized chitosan/iron/activated carbon of orange peel composite. Biomass Convers Biorefin 1-17. https://doi.org/10.1007/s13399-022-03198-2
You H, Lin H, Li Y, Yang Y, Ma Y, Shang Z, Niu X (2022) Iron-aluminum and aluminum-single impregnated biochar composite for nitrate adsorption in rare earth wastewater: behavior and mechanism. Biomass Convers Biorefin:1–12. https://doi.org/10.1007/s13399-021-02148-8
Ling Y, Yan G, Wang H, Dong W, Wang H, Chang Y et al (2021) Release mechanism, secondary pollutants and denitrification performance comparison of six kinds of agricultural wastes as solid carbon sources for nitrate removal. Int J Environ Res Public Health 18(3):1232. https://doi.org/10.3390/ijerph18031232
Guan X, Ji G, Xu S, Yun Y, Liu H (2019) Selection of agricultural straws as sustained-release carbon source for denitrification in a drawer-type biological filter. Water Air Soil Pollut 230:1–11. https://doi.org/10.1016/j.jclepro.2022.130776
Yuan L, Tan L, Shen Z, Zhou Y, He X, Chen X (2022) Enhanced denitrification of dispersed swine wastewater using Ca(OH)2-pretreated rice straw as a solid carbon source. Chemosphere 305:135316. https://doi.org/10.1016/j.chemosphere.2022.135316
Xia L, Li X, Fan W, Wang J (2022) Denitrification performance and microbial community of bioreactor packed with PHBV/PLA/rice hulls composite. Sci Total Environ 803:150033. https://doi.org/10.1016/j.scitotenv.2021.150033
Zhang L, Guo H, Zhao D, Qiu S, Li M, Liang J et al (2022) Preparation and mechanism of modified quaternary amine straw for efficient nitrate removal from aqueous solution. Biomass Convers Biorefin:1–14. https://doi.org/10.1007/s13399-022-03062-3
Cui H, Feng Y, Yin Z, Qu K, Wang L, Li J et al (2023) Organic carbon release, denitrification performance and microbial community of solid-phase denitrification reactors using the blends of agricultural wastes and artificial polymers for the treatment of mariculture wastewater. Ecotoxicol Environ Saf 255:114791. https://doi.org/10.1016/j.ecoenv.2023.114791
Bhattacharyya P, Bhaduri D, Adak T, Munda S, Satapathy BS, Dash PK et al (2020) Characterization of rice straw from major cultivars for best alternative industrial uses to cutoff the menace of straw burning. Ind Crop Prod 143:111919. https://doi.org/10.1016/j.indcrop.2019.111919
Shen Z, Wang J (2011) Biological denitrification using cross-linked starch/PCL blends as solid carbon source and biofilm carrier. Bioresour Technol 102(19):8835–8838. https://doi.org/10.1016/j.biortech.2011.06.090
Pan X, Sano Y (2005) Fractionation of wheat straw by atmospheric acetic acid process. Bioresour Technol 96(11):1256–1263. https://doi.org/10.1016/j.biortech.2004.10.018
Manikandan V, Balasubramanian B, Bharti B, Velmurugan P, Elango D, Baskaran R et al (2022) Development of ZnO/MOGAC nanocomposites for enhanced photocatalytic removal of PO43− and NO3-ions from wastewater under various light irradiations. Biomass Convers Biorefin pp 1–18. https://doi.org/10.1007/s13399-021-02173-7
Chen X, Yu J, Zhang Z, Lu C (2011) Study on structure and thermal stability properties of cellulose fibers from rice straw. Carbohydr Polym 85(1):245–250. https://doi.org/10.1016/j.carbpol.2011.02.022
Yusefi, M., Ali, R. B. R., Abdullah, E. C., & Shameli, K. 2020 Analysis on physiochemical properties of cellulose fiber from rice straw waste. In IOP Conference Series: Materials Science and Engineering (Vol. 808, No. 1, p. 012038). IOP Publishing. https://doi.org/10.1088/1757-899X/808/1/012038
Nandiyanto, A. B. D., Rahman, T., Fadhlulloh, M. A., Abdullah, A. G., Hamidah, I., & Mulyanti, B. 2016 Synthesis of silica particles from rice straw waste using a simple extraction method. In IOP Conference Series: Materials Science and Engineering (Vol. 128, No. 1, p.012040). IOP Publishing. https://doi.org/10.1088/1757-899x/128/1/012040
Rout PR, Dash RR, Bhunia P (2016) Effectiveness of aegle shell (AS) and groundnut shell (GS) as carbon sources for bio-denitrification of nitrate rich aqueous solution. Adv Sci Lett 22(2):354–358. https://doi.org/10.1166/asl.2016.6858
Park S, Baker JO, Himmel ME, Parilla PA, Johnson DK (2010) Cellulose crystallinity index: measurement techniques and their impact on interpreting cellulase performance. Biotechnol Biofuels 3:1–10. https://doi.org/10.1186/1754-6834-3-10
Shelly Y, Kuc ME, Iasur-Kruh L, Azerrad S, Kurzbaum E (2020) A new acinetobacter isolate is an extremely efficient biofilm-formative denitrifying bacterium. Front Environ Sci 8:556226. https://doi.org/10.3389/fenvs.2020.556226
Pradhan S, Prabhakar MR, Karthika Parvathy KR, Dey B, Jayaraman S, Behera B, Paramasivan B (2023) Metagenomic and physicochemical analysis of Kombucha beverage produced from tea waste. J Food Sci Technol 60(3):1088–1096. https://doi.org/10.1007/s13197-022-05476-3
Rout PR, Bhunia P, Dash RR (2017) Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour Technol 244:484–495. https://doi.org/10.1016/j.biortech.2017.07.186
Kundu P, Pramanik A, Dasgupta A, Mukherjee S, Mukherjee J (2014) Simultaneous heterotrophic nitrification and aerobic denitrification by Chryseobacterium sp. R31 isolated from abattoir wastewater. BioMed Res Int 2014. https://doi.org/10.1155/2014/436056
Acknowledgements
The authors are thankful to Dr B R Ambedkar National Institute of Technology, Jalandhar and Thapar Institute of Engineering and Technology, Patiala for supporting this work.
Availability of data and materials
Not applicable
Author information
Authors and Affiliations
Contributions
Mansi Achhoda—investigation; methodology; formal analysis; validation; writing—original draft, Nirmalya Halder—writing—formal analysis; validation; writing—original draft, Neha Thakur—investigation; methodology; formal analysis; validation, Dwarikanath Ratha—reviewing and editing, Bunushree Behera—conceptualization; supervision; writing—review and editing, Prangya Ranjan Rout—conceptualization; supervision; writing—review and editing
Corresponding authors
Ethics declarations
Ethical approval
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Achhoda, M., Halder, N., Thakur, N. et al. Assessing the efficacy of waste organic solids as carbon sources in denitrification bio-filters for abating nitrate toxicity from wastewater. Biomass Conv. Bioref. (2024). https://doi.org/10.1007/s13399-024-05418-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13399-024-05418-3