Abstract
Supramolecular mass spectrometry has emerged in the last decade as an orthogonal method to access, at the molecular level, the structures of noncovalent complexes extracted from the condensed phase to the rarefied gas phase using electrospray ionization. It is often considered that the soft nature of the ESI source confers to the method the capability to generate structural data comparable to those in the condensed phase. In the present paper, using the ammonium ion/cucurbituril combination as a model system, we investigate using ion mobility and computational chemistry the influence of the instrumental parameters on the topology, i.e., internal versus external association, of gaseous host/guest complex ions. MS and theoretical data are confronted to condensed phase data derived from nuclear magnetic resonance spectroscopy to assess whether the instrumental parameters can play an insidious role when trying to derive condensed phase data from mass spectrometry results.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucurbit[n]urils are macrocyclic receptors constructed by the association of n glycoluril repeat units [1]. These pumpkin-shaped molecules present a hydrophobic inner cavity and two identical carbonyl portals, making them suitable for encapsulation of hydrophobic molecules or the hydrophobic part of molecules in aqueous media [2,3,4,5,6]. The main drawback of cucurbiturils is their low solubility in water requiring low pH or high ionic strength to ensure their dissolution upon protonation of the carbonyl portal or cationization [2,3,4,5,6]. On the other hand, the cucurbit[n]uril (CB[n]) family of molecular containers has attracted a huge interest due to their outstanding recognition properties, exceptional strength of their interaction with various guests, and the numerous applications offered by the encapsulation propensity of the cucurbituril family members [7, 8]. Cucurbiturils form stable inclusion complexes with various protonated alkyl- and aryl(di)amines [1,2,3,4,5,6,7,8]. Nuclear magnetic resonance (NMR) experiments are most of the time conducted to investigate the host–guest complexes of cucurbiturils. The interior of the cucurbituril cavity represents a 1H NMR shielding region, with upfield shifts of more than 1 ppm being common [9]. By NMR integration, 1:1 complexes associating alkyl- or aryl-ammonium ions are most of the time detected with CB[6], the common cucurbituril receptor. Apparently, the exchange between bound and free alkylammonium ions is really slow on the NMR time scale, making the signals from both species observable when an excess of guest is present. Counter-examples are reported when, for instance, n-propylammonium ions are engaged in complexation studies with CB[6]. In such a case, only an averaged NMR spectrum is observed [9]. The association of protonated arylamines and CB[6] represents a typical example of such a behavior. Indeed, CB[6] is able to accommodate the benzene ring within the hydrophobic cavity, even if benzene has a van der Waals diameter larger than the calculated internal cavity of CB[6]. Mock et al. suggested that the benzene ring exceeds the strain-free binding capacity of CB[6], with the cage structure getting distorted into an ellipsoid shape upon benzene encapsulation. A decrease by more than 0.4 Å of the diameter of the CB[6] molecule in a direction orthogonal to the guest ring has been measured from the crystal structure of the p-xylylenediamine complex of CB[6] [2,3,4,5,6,7,8]. Therefore, the relatively low binding affinities of the arylamine guests with CB[6] must be understood as a balanced compensation between favorable noncovalent interactions and the stress-strain relationship between the host and the guest [1]. As a typical example, the dissociative constants for the host–guest complexes associating ethanamine and aniline, respectively, amount to 10–2 M and 10-4 M [9]. It was also demonstrated that when the inner cavity of CB[6] is occupied by a benzene ring, there is no extra room for additional ortho- and meta substituents. For the para isomers, both substituents are able to extend toward both carbonyl portals, making the complexation of substituted anilines specific to the para isomers [1]. In addition, CB[6] usually presents slow kinetic of guest encapsulation since a significant deformation of the portals must occur to let the guest molecule enter the CB[6] cavity. The complexation mechanisms have also been a extensively studied, and Nau et al. proposed that the inclusion step corresponds to a flip-flop process, with the exclusion complexes considered as intermediate in the pre-equilibrium step [10]. Beside solution-phase methods, mass spectrometry has also been abundantly used to study the complexation propensity of several cucurbituril congeners. For instance, Dearden et al. reported elegant papers revealing the efficiency of MS-based methods, such as collision-induced dissociation (CID) and ion mobility spectrometry (IMS), to define the topology of gas-phase inclusion/exclusion complex ions [11,12,13,14,15,16,17]. Nau and coworkers also investigated the gas-phase chemistry of cucurbituril complexes with a special attention to the inner-phase chemical reactions undergone by the encapsulated guest molecules upon CID activation [18]. However, to the best of our knowledge, none of these MS-based investigations focused on the instrumental (ion optics) and experimental (solvent, concentrations, time in solution) parameters. In a recent paper [19], our research groups depicted the outstanding capabilities of the association between mass spectrometry methods, ion mobility spectroscopy, and computational chemistry to monitor the slow encapsulation process of the guest para-phenylenediamine (DAPHEN) within the CB[6] host. The use of the ESI source allowed us to study the noncovalent associations present in solution within the rarefied gas phase. The binary complex formed between CB[6] and DAPHEN has been used as a model system to demonstrate, by means of ion mobility and collision-induced dissociation measurements, that the inclusion/exclusion topology ratio of gas-phase ions [CB[6] + DAPHEN + 2H]2+ (m/z 553) varies as a function of the equilibration time in solution prior to the Electrospray process [19].
In direct continuity with this previous study, we report here a detailed study of the complex formation between CB[6] and numerous mono- and diamino benzenic compounds, paying special attention to the in-solution equilibration time, i.e., the time between the preparation of the solution and the mass spectrometry analysis. The selected guests are presented in Scheme 1. These guests all possess an aromatic ring – to be entangled within the hydrophobic cavity – and at least one amino group – to be associated with the carbonyl rim. Aniline has been previously demonstrated to be entangled within the CB[6] cavity with a binding constant of up to 104 M–1 [9]. As such, aniline corresponds to the simplest guest to be included in the present investigation. The halogeno-compounds (ortho- and para-) have been chosen to create a (increasing) steric hindrance on the way to the encapsulation. The para-xylylenediamine compound (PXD) basically presents the same structural features as DAPHEN (two amine moieties, one aromatic ring) but with a different backbone and in particular nonconjugated and more flexible amino groups, due to the presence of the methylene bridges. As in our reference article [19], the assignment of the topology of the ions separated by ion mobility will be achieved on the basis of energy-resolved collision-induced dissociation experiments (ER-CID) and further corroborated by calculating the collisional cross-sections from topologies generated with computational chemistry methods using the density functional theory at the B97D level of theory with a 6-31G(d,p) basis set. A special emphasis on the determination of the relative energies of the inclusion/exclusion structures has been set up to confirm our experimental observations. CB[6] and different guests have been chosen to better understand the influence of the guest and host sizes on the equilibration time in solution prior to the Electrospray process.
Experimental
Materials
Cucurbit[6]uril (CB[6]) and all guest molecules are commercially available (Sigma Aldrich, Overijse, Belgium) and are used without further purification. The host stock solution (10–3M) is prepared in formic acid/water (50:50). The guest stock solutions (10–3M) are prepared in methanol/water (80:20) with 1% HCOOH. HPLC-grade solvents are used for all solutions.
Mass Spectrometry Investigations
For the MS experiments, the stock solutions are mixed to reach final concentrations at 10–5 M. The solvent for dilution is methanol/water (80:20) with 1% HCOOH.
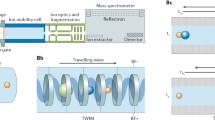
Measurements are performed using a hybrid quadrupole (Q)-traveling wave (T-wave) ion mobility (TWIMS)-time-of-flight (TOF) mass spectrometer (Synapt G2-Si, Waters, UK) equipped with an Electrospray (ESI) ionization source. Typical ion source conditions are: capillary voltage, 3.1 kV; sampling cone, 40 V; source offset, 80 V; source temperature, 150 °C; and desolvation temperature, 300 °C. This mass spectrometer is used for the recording of ESI full scan mass spectrum, for the (energy-resolved) collision-induced dissociation (CID), as well as for the ion mobility experiments. Briefly, the core of the instrument comprised of the so-called TriWave setup of three successive T-wave elements, named the trap cell, the IMS cell, and the transfer cell, in which the wave speed and amplitude are user-tunable. The trap and transfer cells are filled with argon, whereas the IMS cell is filled with nitrogen. A small rf-only cell filled with helium is fitted between the trap and the IMS cells. Collision energy may be applied to the trap cell (trap CE) and to the transfer cell (transfer CE) to fragment ions, respectively. before and after the ion mobility separation. For the energy-resolved CID experiments, mass-selected ions are subjected to increased collision energies in the trap cell. For the energy-resolved CID experiments on ions separated by ion mobility, the ions are decomposed within the transfer cell.
Ion Mobility Spectrometry (IMS): Data Analysis
Ion mobility mass spectra are analyzed using Waters Masslynx. Collisional cross-sections are obtained following a calibration protocol using polyalanine as the calibrant [20] and taking into account the APEX of the arrival time distribution (ATD), named the arrival time (tA). Peak deconvolution is performed using Origin 8.9’s Multiple Peak Fit tool. Each ATD is deconvoluted into symmetric Gaussian functions with constant signal width (full width half maximum - FWHM). Ion mobility resolutions, say R = CCS/Δ CCS where ΔCCS represents the full width of the mobility peak at half height, are determined using Origin 8.9 software by converting the complete arrival time distributions into CCS distributions using the polyalanine calibration.
NMR Measurements
The 1H-NMR spectra are recorded at room temperature on an AVANCEII 500 spectrometer working at 500 MHz (Bruker, Karlsruhe, Germany). All the samples are prepared in 50% DCOOD and 50% D2O/CD3OD 80/20 at a concentration of 5.10–3 M. All spectra are recorded with 64 scans.
DFT Calculations
All molecular structures reported herein are optimized at the DFT level using the B97D functional including dispersion corrections and a 6-31G(d,p) basis set [21]. All DFT calculations are performed within the Gaussian09 package [22]. The optimized structures are then injected in the MOBCAL program using the trajectory method [23] to derive the collisional cross-sections (CCS) to be readily compared with the data generated by the ion mobility experiments. The ε0 and r0 parameters for bromine, chlorine, and iodine have been set to 1.35 10-3 eV and 3.5 Å, respectively, in the MOBCAL program since they were lacking in the original version.
Results and Discussion
In a previous report [19], we investigated by mass spectrometry and theoretical chemistry the complexes associating para-phenylenediamine, DAPHEN, with cucurbit[6]uril. By monitoring the time evolution of the ATD derived from ion mobility experiments, we demonstrated that the full encapsulation of the guest molecule within the host cavity is hindered by the size of the carbonyl portals, leading to slow kinetics for ingression [19]. DAPHEN presents two major structural features: two amine moieties and one aromatic ring that makes this molecule suitable for complexation within the cucurbituril cavity. For the present study, we investigate the influence of the nature and position of substituents on the phenyl ring on the kinetic of ingression. As a starting point, the pristine aniline molecule has been selected since aniline possesses only one amine function and no other substitution in ortho or para position of the amino group, which makes it the simplest guest with a structure close enough to DAPHEN. The aniline phenyl ring is then progressively decorated by halogen atoms (chlorine, bromine and iodine) in ortho and/or para positions.
Aniline•CB[6] or Aniline@CB[6] as Exclusion or Inclusion Complexes
Figure 1 presents the global MS-analysis obtained when a fresh equimolar solution (10–5 M) of aniline and CB[6] is submitted to Electrospray ionization. Beside the doubly protonated naked CB[6] at m/z 499, the binary complex associating one aniline guest to one CB[6] host is also detected with two different charge states. M/z 1090 ions correspond to the singly charged complex ions, [CB[6]+aniline+H]+, whereas m/z 545 ions are readily identified as the doubly charged species, [CB[6]+aniline+2H]2+. The signal at m/z 592 corresponds to the doubly charged ternary complexes, associating two aniline molecules to a macrocyclic receptor, affording the [CB[6]+2aniline+2H]2+ ions.
We stress here that our principal objective is to study the evolution of the gas-phase ion topology as a function of equilibration time in solution. The ESI mass spectrum of the same aniline/CB[6] solution recorded 24 h after the preparation of the equimolar solution is therefore presented in Supplementary Figure SI1 and is superimposable with the ESI mass spectrum recorded after 5 min in solution, raising the question of the kinetic dependence of the encapsulation process. To gain structural information about the detected gas-phase ions, ion mobility spectroscopy experiments in association with mass spectrometry (IMMS) are carried out after a mixing time close to 0 (t = 5 min) and after 24 h of agitation at 25 °C. For the present investigation, we focus only on the doubly charged complex ions that are by far the most abundant detected ions, without considering that the charge state distribution of the complex ions within the solution phase is more than likely not reproduced by the relative signal intensities within the ESI spectrum of Figure 1.
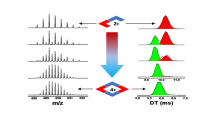
Figure 2a presents the IMMS plots obtained from the full mass spectrum, without any precursor mass selection prior to the ion mobility separation, for the m/z 545 ions for t = 5 min and t = 24 h, namely the [CB[6]+aniline+2H]2+ doubly charged complexes. For both equilibration times, two different drift times are recorded at 4.80 and 5.22 ms, highlighting the presence of two different non-interconverting populations of ions on the time-scale of the ion mobility experiments. Obviously, these two families correspond to ions possessing different topologies. It is worth noting that these two families are still detected after 24 h in solution with almost no modification in their relative proportions (Figure 2a). This already contrasts with the DAPHEN case [19], where only one structure assigned to an inclusion topology is detected after 24 h of equilibration time in solution at room temperature. The identification of the topologies that correspond to these different drift times is made below based on the comparison of the experimental and theoretical data (Figure 2). The experimental collisional cross-sections (CCSexp) in Figure 2a have been obtained after calibration (see the Experimental section) of the Waters Synapt G2-Si and amounts to 206 and 216 Å2 (tA = 4.80 and 5.22 ms).
ESI-IMS-MS analysis of an equimolar solution (10–5 M) of CB[6] and aniline (CV = 40 V) for two different equilibration time (t = 5 min and 24 h). (a) Arrival time distribution (ATD) (no mass selection prior the IMS separation) of the [CB[6]+aniline+2H]2+ ions (m/z 545.7) – the CCSexp and the relative abundances are obtained using, respectively, the calibration and the deconvolution procedures described in the Experimental section. (b) Optimized topologies for the [CB[6]+aniline+2H]2+ ions obtained at the DFT level using the B97D functional and the 6-31G(d,p) basis set – the CCSth are calculated by the trajectory method in the MOBCAL software starting from DFT-optimized geometries
Figure 2b summarizes the data obtained by the DFT calculations to generate the ion structures and calculate their relative energies and MOBCAL (trajectory method) software [23] to calculate the theoretical collisional cross-sections (CCSth). The inclusion topology is calculated to be more stable than the exclusion complexes by about 15 kcal/mol, see Table 1. For the exclusion topology, we identify two different structures, see Table 1 and Figure 2b. In one of their reports, Dearden et al. proposed that for the 2+ exclusion complex ions associating para-phenylenediamine (DAPHEN) to CB[6], the doubly protonated guest molecule is lying on the carbonyl rim with both ammonium groups in close interaction with the carbonyl oxygen [14]. In the aniline case, at the B97D/6-31G(d,p) level of theory, we also obtain two stable exclusion topologies, one presenting such a spatial arrangement with the benzene ring lying parallel to the rim (17 kcal/mol above the inclusion topology) and the other with a perpendicular layout (15 kcal/mol less stable than the inclusion topology), see Figure 2b. The theoretical CCS values obtained using the trajectory method in Mobcal amount at, respectively, 190, 204, and 208 Å2, as presented in Figure 2b for the inclusion and both the exclusion topologies. A quick comparison between the experimental (206 and 216 Å2) and theoretical values (190, 204, and 208 Å2) does not allow a straightforward structure assignment. Nevertheless, the ion mobility resolution is limited on a Waters Synapt G2 instrument to around 40 (R = CCS/ΔCCS – where ΔCCS represents the full width of the mobility peak at half height) [24]. We measure the resolution of both signals (see Experimental section for the procedure) and we obtain, respectively. R = 26 and 21 for the 4.8 and 5.22 ms signals. Theoretically, the temporal separation of the exclusion complexes, with theoretical CCS at 204 and 208 Å2, would require an ion mobility resolution around 50 that is not reached in the present measurements, allowing to propose that the ion mobility separated ions are, respectively, the internal and (a mixture of both) the exclusion complexes. Furthermore, since both ion families are separated upon ion mobility, we can state that the inclusion and exclusion complexes do not interconvert during the ion mobility experiment. As a third set of data, the E50 values have been obtained by recording the survival yield curves (see Supplementary Figure SI2 for a typical example) obtained from the energy-resolved CID experiments (ER-CID) carried on after the mobility cell (in the transfer cell) to study each topology separately. The E50 data are converted into center-of-mass energies to allow comparing the different host–guest systems. As presented in Table 1, the measured E50 values (ca. 0.9 eV) are significantly reduced in comparison to that measured previously for the DAPHEN@CB[6] inclusion complexes (≈2 eV) [19]. We believe that without the second amino group in para compared to DAPHEN, the aniline guest might probably easily flip out of the cavity generating the exclusion topology. Further evidence for the presence of the exclusion topology at tA = 5.22 ms comes from the following sequence of ion manipulations. We submit to collision-induced dissociation (CID) in the trap cell of the TriWave setup the mass-selected doubly charged ternary complexes at m/z 592 (see Figure 1 for the ESI mass spectrum), the [CB[6]+2aniline+2H]2+ ions, in order to generate the m/z 545 binary complexes that are likely to correspond to exclusion complexes. Indeed, the ternary complexes are expected to present both aniline molecules pointing outside the CB[6] cavity in a full exclusion structure. This is partially confirmed by measuring the ATD of the m/z 592 ions, see Supplementary Figure SI3. The ATD signal is at least comprised of two overlapping traces that correspond, upon deconvolution (see Experimental), to ion families characterized by CCSexp = 206 and 230 Å2 for, respectively, tA = 5.1 and 5.6 ms (see Supplementary Figure SI3). By comparing those CCSexp with the CCSth of two different candidates topologies, one with both aniline guests pointing outside the CB[6] cavity (CCSth = 225 Å2 ) and the other with one aniline outside and one aniline inside the cavity (CCSth = 209 Å2) (see Supplementary Figure SI3), we observe that both topologies are present in a 56%–44% relative abundances. We first check whether, upon CID, these m/z 592 ions generate the m/z 545 ions by loss of a neutral aniline molecule, see Supplementary Figure SI3 for the CID spectrum of the m/z 592 ions. The so-produced m/z 545 ions are consecutively subjected to ion mobility and the recorded ATD is now characterized by a quasi-unique signal at tA = 5.22 ms, confirming the exclusion topology of those complex ions (see Supplementary Figure SI3).
At this point of the discussion, the origin of both the inclusion and exclusion ion families is to be debated. Embarrassing is also the fact that the ratio between both ATD signals does not evolve from t = 5 min to t = 24 h (Figure 2). To further discuss this point, we need to consider the solution phase situation. Mock et al. used NMR spectroscopy to characterize the selectivity of cucurbiturils toward several guest compounds [9]. In particular, they demonstrated that aniline is entangled in the CB[6] cavity with a dissociation constant that amounts to 10–4 M in a 1:1 HCOOH/H2O solution [9]. To confirm these data, we repeat the 1H-NMR measurements on the aniline/CB[6] system using solution conditions similar to the Electrospray analysis, say 50% DCOOD and 50% D2O/CD3OD 80/20 (see Experimental), except that the final concentrations are, respectively, around 10–3 and 10–5 M for the NMR and MS analyses. We first record the NMR spectra of pure aniline and pure CB[6] (see Supplementary Figure SI4a, b). The aniline protons are observed between 6.2 and 6.3 ppm, whereas the CB[6] protons are well resolved at 3.05 ppm (doublet), 4.3 ppm (singlet), and 4.55 ppm (doublet) (see Supplementary Figure SI4b for assignment). This nicely correlates with the literature data and in particular the 1H-NMR spectrum of CB[6] presented in the literature [25]. We then prepare different host/guest solutions taking care to exactly conserve the same solvent composition. We record the NMR spectra for three different stoichiometries, i.e., 2:1, 1:1, and 1:2 for aniline:CB[6] (see Supplementary Figure SI4). After 24 h in solution and whatever the starting stoichiometry, signals corresponding to free aniline and CB[6] are still observed in the NMR spectra. Nevertheless, additional signals are detected and correspond to the aniline@CB[6] complex. The resonance of the aniline protons are shifted upfield and are now detected in the 5.3–5.8 ppm region, indicating that aniline is encapsulated within the CB[6] cavity. Indeed, as reported by Mock et al. in 1988 and reminded in a recent paper [26, 27], the interior of cucurbituril is a proton-shielding region. The resonances of the CB[6] receptor also undergo chemical shifts compared with the free receptor [26, 27]. In our measurements, we observe the most significant displacement for the 4.3 ppm band that is displaced around 4.4 ppm (Supplementary Figure SI4 for assignment). The detection of resonances of both the free and bound molecules, either aniline and CB[6], means that the IN-to-OUT exchange process is really slow in the solution phase (slow on the typical NMR time scale). By integrating the free and bound aniline proton resonance signals, we can estimate that 33% of the aniline molecules are entangled within the CB[6] cavity for the aniline/CB]6] equimolar solution after 24 h in solution. We also evaluate the overtime evolution of the NMR spectra of the 1:1 host:guest solution and we determine that for the equimolar aniline/CB[6] solution after 5 min, 24 h, and 48 h in solution, 3%, 33%, and 32% out of the total aniline molecules are entangled within the cucurbituril cavity. Marquez and Nau demonstrated that the ingress (kingress) and egress rate constants (kegress) as well as the binding constants for the production of the inclusion complex between CB[6] and several primary amines is highly dependent on the pH. They showed that the complexes are destabilized at low pH values (<2) at which protonation of CB[6] occurs, and thereby reduces the driving force for complexation of organic ammonium salts [9]. In our MS and NMR experiments, we are using acidic conditions limiting the production of inclusion complexes, as observed upon NMR. We must then consider as the starting point of our MS investigations that in the aniline/CB[6] solution, inclusion complexes are present in solution but that a significant proportion of aniline and CB[6] molecules remained unassociated for the equimolar aniline/CB[6] solution, both after 5 min (3% bound aniline molecules) and 24 h (33%) in solution. Upon Electrospray ionization, we then propose that the inclusion complexes are transferred to the gas phase of the mass spectrometer and that, given the acidic conditions, any nonspecific exclusion complex ion is likely to be generated.
It is then tempting to deduce from all those experimental/theoretical results that decompositions of the ternary [CB[6]+2aniline+2H]2+ complex ions (m/z 592 – see Figure 1 for ESI mass spectrum) either in the ion source or somewhere else during the flight of the ions from the source to the TWIG cell can be responsible for the production of the m/z 545 binary complex ions presenting the exclusion topology observed in Figure 2a. Basically, the m/z 592 ions can decompose into m/z 545 at mainly two specific places during their travel from the source to the ion mobility cell (see Supplementary Figure SI5). First, before the quadrupole, the ions are accelerated between the stepwave and the first traveling wave ion guide (TWIG), and the applied voltage is discussed as the sample cone voltage in the tune page, see Supplementary Figure SI5. This corresponds to a voltage applied to increase sensitivity by declustering the ion adducts. Secondly, after the quadrupole, the ions are stored in the so-called trap cell that is floated at a voltage – the trap DC bias – that is the voltage applied between the trap and the helium cells allowing the ion injection into the mobility cell. The trap cell can also be used as a CID cell by modifying the Trap CE (trap collision energy).
First, with regard to these three parameters, we focus on the in-source events by mass-selecting the m/z 545 ions by the quadrupole analyzer and monitoring the evolution of the IN/OUT ratio as a function of the sample cone parameter.
As presented in Figure 3a, even at the lowest tested sample cone value (10 V), ions presenting the exclusion topology are already detected with a relative abundance of about 10%. The stepwise increase of the sample cone voltage from 10 to 70 V clearly induces the modification of the relative proportions between the IN and OUT topologies with the exclusion complex ions becoming more abundant above ca. 50 V (Figure 3a). Figure 3b presents the sample cone voltage dependence of the relative abundances of three different key ions, namely [CB[6]+2aniline+2H]2+ (m/z 592), [CB[6]+aniline+2H]2+ (m/z 545), and [CB[6]+2H]2+ (m/z 499). From Figure 3b, we can conclude that in the sample cone voltage range 10–70 V, no extensive decomposition of the [CB[6]+2aniline+2H]2+ (m/z 592) or the [CB[6]+aniline+2H]2+ (m/z 545) ions is observed. Therefore, we can deduce that the IN/OUT evolution is mainly caused, in the sample cone voltage range 10–70 V, by the collision-induced isomerization – not decomposition – of the m/z 545 inclusion complex ions toward the less stable and less compact exclusion complexes (Figure 2b).
MS analysis of an equimolar solution (10–5 M) of CB[6] and aniline. (a) Influence of the sample cone voltage on the IN/OUT ratio determined by IMS on mass-selected m/z 545 ions (trap bias 30 V – trap CE 0.5 V). (b) Influence of the sample cone voltage on the relative intensities of the m/z 499, 454, and 592 ions within the ESI-ToF analysis (trap bias 30 V – trap CE 0.5 V). (c) Influence of the trap DC bias on the IN/OUT ratio determined by IMS on mass-selected m/z 545 ions (sample cone voltage 40 V – trap CE 0.5 V). (d) Survival yield of the mass-selected m/z 545 ions with CID in the trap and influence of the trap DC on the IN/OUT ratio determined by IMS (sample cone voltage 40 V – trap bias 30 V)
We then assess the influence of the trap DC bias on the IN/OUT ratio by again mass-selecting the m/z 545 ions by the quadrupole analyzer and monitoring the evolution of the IN/OUT ratio as a function of the trap DC bias voltage, taking care to avoid the m/z 545 ion decomposition. The data presented in Figure 3c again suggest that the increase in the trap DC bias clearly induces the IN-to-OUT isomerization.
As a final experiment, we probe the influence of the trap collision energy (trap CE) on the survival yield (SY) and the IN/OUT ratio of the mass-selected m/z 545 ions. The SY corresponds, in a CID mass spectrum, to the ratio between the abundance of the non-fragmented parent ions and the abundances of all ions (parent plus fragments) going out of the collision cell. The IN/OUT ratio is measured by injecting the m/z 545 emerging from the trap cell into the ion mobility cell. The results of such an experiment are presented in Figure 3d. Basically, the parent ions start to decompose at a trap CE above 10 V. Nevertheless, already at a value of 5 V, the IN-to-OUT ratio is in favor of the exclusion topology, indicating that the IN-to-OUT isomerization requires less internal energy to occur than the complex ion dissociation. It is also remarkable to note that the m/z 545 parent ions start to decompose when the proportion of exclusion complex ions reaches a maximum around 12–13 V. This clearly indicates that the collisionally activated m/z 545 inclusion complex ions first undergo the IN-to-OUT isomerization prior to their decomposition. The dominant decomposition reaction corresponds to the loss of neutral aniline and the formation of [CB[6]+2H]2+ at m/z 499.
Adamantylamine•CB[6] and Diaminohexane@CB[6] as Exclusion and Inclusion Model Complexes
Adamantylamine (ADAM) and diaminohexane, see Scheme 2, are introduced in the present investigation as model guests for the ion mobility experiments since the binary complexes associating ADAM and diaminohexane have already been demonstrated to create specifically exclusion and inclusion complexes with CB[6], respectively, [28].
As presented in Table 1, the [CB[6]+diaminohexane+2H]2+ complexes present only one gas phase structure – i.e., a single signal in the ATD – both at t = 5 min and t = 24 h. This structure is clearly identified as an inclusion topology. The nice agreement between theoretical and experimental CCS in addition to the high E50 value (center-of-mass energy) at 1.59 eV confirms the inclusion character of the detected ions. This inclusion topology possesses by far the most stable structure in comparison with the exclusion candidate, with the exclusion structure being 63.5 kcal/mol less stable than the inclusion one. In Supplementary Figure SI6, the CID mass spectrum of the m/z 557 ions is presented. The decomposing complex ions are shown to mostly end up being protonated CB[6] by a formal loss of protonated diaminohexane. The corresponding signal (m/z 117) is not detected amongst the CID fragments while m/z 100 cations, corresponding to NH2-(CH2)5-CH2 + cations, are detected. This suggests that the decomposition process is more complicated than a simple decomplexation reaction. We previously reported that the decomposition of the inclusion complexes created between 1,4-diaminophenylene and CB[6] occurs via a complicated mechanism involving bond breaking within the cucurbituril backbone [19]. The observation of CB[6] fragment ions points to a similar mechanism that will not be discussed further in the present study.
Table 1 also presents the results for the [CB[6]+ADAM+2H]2+ ions. The experimental data clearly point to the exclusive presence of an exclusion topology. The identification of an exclusion complex is mainly based on the nice agreement between theoretical (217 Å2) and experimental (228 Å2) CCS. The CCS calculated for the inclusion and exclusion complexes are, respectively, 196 and 217 Å2; this huge difference is readily associated to the large volume of the adamantyl group. Interestingly, the relative energy between both topologies only amounts to 6 kcal/mol. The exclusive occurrence of the exclusion complexes finds its origin in a very low kinetic constant of ingress for the guest encapsulation process [10]. The huge transition barrier for ingress is clearly associated with the requirement of significant deformations of the rigid portals of the CB[6] receiver to allow the adamantly group entering the cavity. When exposed to energy-resolved CID experiments, those exclusion complexes are characterized by a E50 of about 0.9 eV. This E50 energy around 0.9–1 eV is associated with the dissociation of the H-bonds present between the ammonium group of the guest and the carbonyl oxygen atoms of the receiver portal. In the case of the adamantylamine complexes, based on the high proton affinity of the molecule (PA = 226.8 kcal/mol versus 210.9 kJ/mol for aniline [29]), protonated adamantylamine plus protonated CB[6] are the dominating CID fragments (see Supplementary Figure SI7).
When comparing the fragment ions for the aniline and adamantylamine complex ions, we observe that depending on the relative PA values, the proton remains on the guest molecule (adamantylamine case) or on the receptor macrocycle (aniline case). Given the close similarity between the E50 values whatever the nature of the fragment ions, we conclude that the proton transfer reaction (aniline case) is less energy-demanding than the overall dissociation of the H-bonds. At variance, for the diamino guest, the E50 value is higher (1.59 eV) in relation to the presence of both the ammonium groups creating H-bonds on both rims of the receptor. This was already observed in our previous report (data from reference [19] also added to Table 1) dealing with 1,4-diaminophenylene. However, for the diaminohexane guest, the inclusion complexes are detected immediately after mixing the guest and the host in solution, whereas for the benzenoid counterpart, 24 h are required for full encapsulation [19].
Halogeno-Aniline•CB[6] or Halogeno-Aniline@CB[6] as Exclusion or Inclusion Complexes
The data acquired for aniline, adamantylamine, and diaminohexane will support the discussion of the experimental/theoretical results for the halogeno-aniline guests (see Scheme 1). Briefly, the anilinium guest is enclosed within the cavity of the receptor in the solution phase, but is progressively expelled outside the cavity upon ion activation during the CID experiments but also due to the transfer optics. The E50 amounts to about 0.9 eV for both topologies, demonstrating that the rate determining step on the way to the complex ion dissociation is the H-bond breaking. This is confirmed when analyzing the adamantylamine guest with a E50 of about 0.9 eV.
All experimental and theoretical data obtained for the systems associating CB[6] to 2-chloroaniline (2-Cl), 2-bromoaniline (2-Br), 4-chloroaniline (4-Cl), 4-bromoaniline (4-Br) and 4-iodoaniline (4-I) are presented in Table 1. First, no influence of the equilibration time over the ion mobility data has been monitored pointing to either no or fast encapsulation of the guest within the cavity.
For the ortho-substituted anilines, no complex ions are detected for 2-Br guest, whereas only one arrival time (tA) is measured at 5.63 ms for the 2-Cl counterpart. The associated CCS amounts to 218 Å2 points to the presence of an exclusion topology. This is confirmed by: (1) the E50 value determined at 0.69 eV and; (2) by the theoretical data with a CCSth of 208/211 Å2 for the exclusion complex ions. Similarly to the aniline case described previously, the ion mobility resolution of the Waters Synapt G2-Si mass spectrometer does not allow discriminating between both the exclusion complex topologies. Also the unique observation of the slightly less stable exclusion complexes, being about 5 kcal/mol less stable than the inclusion complexes, is probably associated with a high energy barrier for the ingress of the chlorine ortho-substituted guest within the host cavity through the carbonyl portals. The absence of exclusion complex ions for the bromine-containing guest is probably related to the steric hindrance created around the bromine atom in the ortho position of the anilinium group.
For the para isomers, two traces are observed in the case of 4-Cl and 4-Br guests, whereas only one topology is detected for the 4-I molecule. For both the 4-Cl and 4-Br guests, two arrival time distributions are then measured at 5.08/5.63 and 5.08/5.76 ms, corresponding to CCSexp at 207/218 and 207/224 Å2, respectively (see Table 1). When compared with the theoretical CCS, we can state that we successfully separate the inclusion and exclusion complexes for both guests. However, the weak ion mobility resolution of the Synapt mass spectrometer does not allow establishing the real topology (or mixture of topologies) of the detected 2+ exclusion complexes associating 4-chloro(bromo)aniline to CB[6], see Table 1. The calculated relative energies in Table 1 reveal that the inclusion complexes remain more stable than the exclusion ones, whatever the halogen atoms. It also appears, based on the E50 determination (Table 1), that the presence of the chlorine/bromine atom does not hinder the IN-to-OUT process since the E50 data are all around 0.8 eV; this correlates with the theoretical atom sizes and portal sizes in Supplementary Figure SI8. Thus, in the case of 4-chloro(bromo)aniline, we are probably facing the same behavior as for aniline with partial IN-to-OUT isomerization upon ion activation.
For the 4-I/CB[6] system, only a single arrival time is measured at 5.69 ms corresponding to a CCS of 229 Å2 and a E50 of 0.88 eV (Table 1). The direct comparison with the theoretical CCS suggests the presence of the exclusion complexes, even if the inclusion complexes remain the most stable association, see Table 1. We again use NMR spectroscopy to access the solution topology of the 4-iodoaniline/CB[6] association. As revealed in Supplementary Figure SI9, all H-nuclei NMR signals are slightly deshielded indicating no complexation or external association with H-bond creation between the ammonium hydrogen atoms and the oxygen atoms of the carbonyl groups. The 4-iodoaniline situation is then straightforward to describe with the sole observation of gas phase exclusion complexes explained by the absence of inclusion complexes in the condensed phase. It is likely that the ingress of the bulky 4-iodoaniline guest within the CB[6] cavity is kinetically prohibited, even in solution [10].
PXD•CB[6] or PXD@CB[6] as Exclusion or Inclusion Complexes
As a final guest for the present investigation, para-xylylenediamine – PXD – has been selected based on the structural and functional compatibility with CB[6]. The PXD/CB[6] system was studied in solution and an association constant of 5.102 M–1 has been determined by NMR spectroscopy [28]. As presented in Table 1, two topologies are detected with arrival times and CCS at, respectively, 5.08 and 5.63 ms and 202 and 219 Å2. The complex ions are characterized by E50 of 1.98 and 0.89 eV, respectively. This unambiguously points to the coexistence of inclusion and exclusion complex ions, namely PXD@CB[6] and PXD•CB[6]. Moreover, there is a time dependence of the relative abundances of both topologies, as featured in Figure 4. We previously reported that the quantitative encapsulation of 1,4-diaminophenylene within CB[6] (starting from equimolar solution) requires 24 h at room temperature. Here, almost 6 d are required to reach 90% encapsulation. The extreme E50 value (1.98 eV) indicates that the complex ions are protected from IN-to-OUT isomerization upon ion activation. This value is also clearly associated with the need of covalent bond breaking on the way to complex ion dissociations. This is evidenced by the occurrence of a m/z 120 fragment ion in the CID spectrum of the PXD@CB[6] ions (see Supplementary Figure SI10), instead of the m/z 136 ions for intact (mono protonated) para-xylylenediamine. This behavior mimics the CID reactions of 1,4-diaminophenylene (Ka=1.8 103 M–1 [28]) and 1,6-diaminohexane (4 108 M–1 [28]) containing complex ions.
Conclusions
Aryl- and alkylammonium ions are known to strongly interact with the cucurbituril family of macrocyclic receptors. The primary interactions correspond to H-bonds created between the hydrogen atoms of the ammonium moiety and the oxygen atoms of the carbonyl groups. However, encapsulation of the guest within the cavity is often hindered or kinetically limited by steric factors since the guest molecule must enter the cavity through the small size portals. Mass spectrometry, associating Electrospray ionization, collision-induced dissociation, and ion mobility, has already been advantageously used to study in the gas phase the topology of host–guest complexes associating amino compounds to cucurbituril receptors [11,12,13,14,15,16,17, 19, 30, 31]. In the present work, we selected a model system, extensively investigated in the condensed phase, to assess the influence of the instrumental parameters, i.e., the voltages along the ion optics, on the IN-to-OUT ratio starting from the knowledge of the condensed phase topologies. All experiments were performed on the Waters Synapt G2-Si mass spectrometer and the IN-to-OUT ratios were determined based on ion mobility experiments. We demonstrated here that for mono amino guest molecules, the IN-to-OUT isomerization process can be induced upon ion activation at different locations along the flight of the ions from the source to the ion mobility cell, i.e., in the stepwave (sample cone voltage) and in the Triwave setup (trap DC Bias and trap CE). On the other hand, the diammonium guest molecules entangled within the host cavity and H-bonded on both portals do not undergo the IN-to-OUT process. When increasing the internal energy, the complex ion dissociation proceeds through covalent bond breaking of the cucurbituril backbone. These results complete our previous mass spectrometry investigations on the cucurbituril systems that demonstrated the applicability of ESI, CID, and ion mobility to probe the gas-phase structures of complex ions – the so-called flying boxes [30, 31]. We also previously demonstrated the influence of the equilibration time in solution on the IN/OUT nature of the detected gaseous ions [19]. In this paper, we have also highlighted the key role played by the ion optics on the topology of the supramolecular ions transferred from the solution to the gas phase upon electrospray ionization. The model used in our study is obviously a case study. However, the results clearly demonstrate that greater vigilance should be considered when studying noncovalent systems using mass spectrometry techniques. It was already well known that solution- and gas-phase structures may be intrinsically different. We demonstrate here that the ion structures are likely to be modified all along their path within the mass spectrometer due to ion activation.
References
Mock, W.L., Shih, N.-Y.: Dynamics of molecular recognition involving cucurbituril. J. Am. Chem. Soc. 111, 2697–2699 (1989)
Lagona, J., Mukhopadhyay, P., Chakrabarti, S., Isaacs, L.: The cucurbit[n]uril family. Angew. Chem. Int. Ed. 44, 4844–4870 (2005) Angew. Chem. 117, 4922–4949 (2005)
Lee, J.W., Samal, S., Selvapalam, N., Kim, H.J., Kim, K.: Cucurbituril homologues and derivatives: new opportunities in supramolecular chemistry. Acc. Chem. Res. 36, 621–630 (2003)
Kim, K., Selvapalam, N., Ko, Y.H., Park, K.M., Kim, D., Kim, J.: Functionalized cucurbiturils and their applications. Chem. Soc. Rev. 36, 267–279 (2007)
Isaacs, L.: The mechanism of cucurbituril formation. Isr. J. Chem. 51, 578–591 (2011)
Masson, E., Ling, X., Joseph, R., Kyeremeh-Mensah, L., Lu, X.: Cucurbituril chemistry: a tale of supramolecular success. RSC Adv. 2, 1213–1247 (2012)
Assaf, K.I., Nau, W.M.: Cucurbiturils: from synthesis to high-affinity binding and catalysis. Chem. Soc. Rev. 44, 394–418 (2015)
Barrow, S.J., Kasera, S., Rowland, M.J., del Barrio, J., Scherman, O.A.: Cucurbituril-based molecular recognition. Chem. Rev. 115(22), 12320–12406 (2015)
Mock, W.L., Shih, N.-Y.: Structure and selectivity in host–guest complexes of cucurbituril. J. Org. Chem. 51, 4440–4446 (1986)
Marquez, C., Nau, W.M.: Two mechanisms of slow host–guest complexation between cucurbit[6]uril and cyclohexylmethylamine: pH-responsive supramolecular kinetics. Angew. Chem. 40, 3155–3160 (2001)
Kellersberger, K.A., Anderson, J.D., Ward, S.M., Krakowiak, K.E., Dearden, D.V.: Encapsulation of N2, O2, methanol, or acetonitrile by decamethylcucurbit[5]uril(NH4 +)2 complexes in the gas phase: influence of the guest on 'lid' tightness. J. Am. Chem. Soc. 123, 11316–11317 (2001)
Zhang, H., Paulsen, E.S., Walker, K.A., Krakowiak, K.E., Dearden, D.V.: Cucurbit[6]uril pseudorotaxanes: distinctive gas phase dissociation and reactivity. J. Am. Chem. Soc. 125, 9284–9285 (2003)
Zhang, T., Ferrell, A., Asplund, M.C., Dearden, D.V.: Molecular beads on a charged molecular string: α,ω-alkyldiammonium complexes of cucurbit[6]uril in the gas phase. Int. J. Mass Spectrom. 265, 187–196 (2007)
Dearden, D.V., Ferrell, T.A., Asplund, M.C., Zilch, L.W., Julian, R.R., Jarrold, M.F.: One ring to bind them all: shape-selective complexation of phenylenediamine isomers with cucurbit[6]uril in the gas phase. J. Phys. Chem. A. 113, 989–997 (2009)
Zhang, H., Grabenauer, M., Bowers, M.T., Dearden, D.V.: Supramolecular modification of ion chemistry: modulation of peptide charge state and dissociation behavior through complexation with cucurbit[n]uril (n = 5, 6) or α-cyclodextrin. J. Phys. Chem. A. 113, 1508–1517 (2009)
Yang, F., Dearden, D.V.: Guanidinium-capped cucurbit[7]uril molecular cages in the gas phase. Supramol. Chem. 23, 53–58 (2011)
Yang, F., Dearden, D.V.: Gas phase cucurbit[n]uril chemistry. Isr. J. Chem. 51, 551–558 (2011)
Lee, T.C., Kalenius, E., Lazar, A.I., Assaf, K.I., Kuhnert, N., Grün, C.H., Jänis, J., Scherman, O.A., Nau, W.M.: Chemistry inside molecular containers in the gas phase. Nat. Chem. 5, 376–382 (2013)
Carroy, G., Daxhelet, C., Lemaur, V., De Winter, J., De Pauw, E., Cornil, J., Gerbaux, P.: Influence of equilibration time in solution on the inclusion/exclusion topology ratio of host–guest complexes probed by ion mobility and collision-induced dissociation. Chem. Eur. J. 22, 4528–4534 (2016)
Bush, M.F., Campuzano, I.D.G., Robinson, C.V.: Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal. Chem. 84, 7124–7130 (2012)
Grimme, S.: Semi-empirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 27, 1787–1799 (2006)
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H.P., Izmaylov, A.F., Bloino, J., Zheng, G., Sonnenberg, J.L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, J.A. Jr., Peralta, J.E., Ogliaro, F., Bearpark, M., Heyd, J.J., Brothers, E., Kudin, K.N., Staroverov, V.N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Rega, N., Millam, J.M., Klene, M., Knox, J.E., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Martin, R.L., Morokuma, K., Zakrzewski, V.G., Voth, G.A., Salvador, P., Dannenberg, J.J., Dapprich, S., Daniels, A.D., Farkas, O., Foresman, J.B., Ortiz, J.V., Cioslowski J., Fox, D.J.: Gaussian 09, Revision A.02. Gaussian, Inc.: Wallingford, CT (2009)
Mesleh, M.F., Hunter, J.M., Shvartsburg, A.A., Schatz, G.C., Jarrold, M.F.: Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 100, 16082–16086 (1996)
Giles, K., Williams, J.P., Campuzano, I.: Enhancements in traveling wave ion mobility resolution. Rapid Commun. Mass Spectrom. 25, 1559–1566 (2011)
Li, J., Si, C., Sun, H., Zhu, J., Pan, T., Liu, S., Dong, Z., Xu, J., Luo, Q., Liu, J.: Reversible pH-controlled switching of an artificial antioxidant selenoenzyme based on pseudorotaxane formation and dissociation. Chem. Commun. 51, 9987–9990 (2015)
Mock, W.L., Shih, N.-Y.: Organic ligand–receptor interactions between cucurbituril and alkylammonium ions. J. Am. Chem. Soc. 110, 4706–4710 (1988)
Lin, R.-L., Fang, G.-S., Sun, W.-Q., Liu, J.-X.: Aniline-containing guests recognized by α,α′,δ,δ′-tetramethyl-cucurbit[6]uril host. Sci Rep. 6, 39057 (2016)
Liu, S., Ruspic, C., Mukhopadhyay, P., Chakrabarti, S., Zvalij, P.Y., Isaacs, L.: The cucurbit[n]uril family: prime components for self-sorting systems. J. Am. Chem. Soc. 127, 15959–15967 (2005)
Lias, S.G., Bartmess, J.E., Leibman, J.F., Holmes, J.L., Levin, R.D., Mallard, W.G.: Gas-phase ion and neutral thermochemistry. J. Phys. Chem. Ref. Data 17(Suppl.1) (1988)
Lemaur, V., Carroy, G., Poussigue, F., Chirot, F., De Winter, J., Isaacs, L., Dugourd, P., Cornil, J., Gerbaux, P.: Homotropic allosterism: in-depth structural analysis of the gas-phase noncovalent complexes associating a double-cavity cucurbit[n]uril-type host and size-selected protonated amino compounds. ChemPlusChem. 78, 959–969 (2013)
Carroy, G., Lemaur, V., De Winter, J., Isaacs, L., De Pauw, E., Cornil, J., Gerbaux, P.: Energy-resolved collision-induced dissociation of non-covalent ions: charge- and guest-dependence of decomplexation reaction efficiencies. Phys. Chem. Chem. Phys. 18, 12557–12568 (2016)
Acknowledgments
The UMONS MS laboratory acknowledges the Fonds National de la Recherche Scientifique (FRS-FNRS) for its contribution to the acquisition of the Waters SYNAPT G2-Si mass spectrometers and for continuing support. The work in the Laboratory for Chemistry of Novel Materials was supported by the European Commission/Région Wallonne (FEDER – BIORGEL project), the Interuniversity Attraction Pole program of the Belgian Federal Science Policy Office (PAI 7/05), the Programme d’Excellence de la Région Wallonne (OPTI2MAT project), the Consortium des Équipements de Calcul Intensif (CÉCI), funded by the Fonds de la Recherche Scientifique de Belgique (F.R.S.-FNRS) under grant no. 2.5020.11, and FRS-FNRS. G.C. is grateful to the UMONS and the ULg for a PhD grant. J.C. is an FNRS Research Director. S.L. and C.H. thank the Fonds National pour la Recherche Scientifique (F.R.S.-FNRS), the FEDER, the Walloon Region (Holocancer and Gadolymph programs), the COST Actions, the Centre for Microscopy and Molecular Imaging (CMMI) supported by the European Regional Development Fund of the Walloon region, the ARC, Interreg and UIAP programs.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(DOCX 3261 kb)
Rights and permissions
About this article
Cite this article
Carroy, G., Lemaur, V., Henoumont, C. et al. Flying Cages in Traveling Wave Ion Mobility: Influence of the Instrumental Parameters on the Topology of the Host–Guest Complexes. J. Am. Soc. Mass Spectrom. 29, 121–132 (2018). https://doi.org/10.1007/s13361-017-1816-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1816-7