Abstract
Alkylamines are widely used as ion-pairing agents during LC-MS of oligonucleotides. In addition to a better chromatographic separation, they also assist with the desorption of oligonucleotide ions into the gas phase, cause charge state reduction, and decrease cation adduction. However, the choice of such ion-pairing agents has considerable influence on the MS signal intensity of oligonucleotides as they can also cause significant ion suppression. Interestingly, optimal ion-pairing agents should be selected on a case by case basis as their choice is strongly influenced by the sequence of the oligonucleotide under investigation. Despite imposing major practical difficulties to analytical method development, such a highly variable system that responds very strongly to the nuances of the electrospray composition provides an excellent opportunity for a fundamental study of the electrospray ionization process. Our investigations using this system quantitatively revealed the major factors that influenced the ESI ionization efficiency of oligonucleotides. Parameters such as boiling point, proton affinity, partition coefficient, water solubility, and Henry’s law constants for the ion-pairing reagents and the hydrophobic thymine content of the oligonucleotides were found to be the most significant contributors. Identification of these parameters also allowed for the development of a statistical predictive algorithm that can assist with the choice of an optimum IP agent for each particular oligonucleotide sequence. We believe that research in the field of oligonucleotide bioanalysis will significantly benefit from this algorithm (included in Supplementary Material) as it advocates for the use of lesser-known but more suitable ion-pair alternatives to TEA for many oligonucleotide sequences.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mass spectrometry is one of the most widely used analytical techniques for biomedical applications [1]. This has only become possible as electrospray ionization (ESI) [2] and matrix-assisted laser desorption/ionization (MALDI) [3] provided the necessary means for generating gaseous ions from biological macromolecules [4–6]. Electrospray ionization is particularly well suited for biochemical analysis because of its ease of coupling liquid chromatography with mass spectrometry, as well as its ability to preserve specific solution phase interactions between molecules into the gas phase [7–11]. Nevertheless, despite the widespread expansion of applications involving ESI-MS, there is still significant debate on the mechanisms by which gaseous ions are formed through electrospray ionization [12, 13].
According to the charged residue model (CRM) [14, 15], as the droplet size decreases by evaporation of solvent, it will reach the Rayleigh limit and break up into a number of smaller droplets. These second-generation droplets would continue to subdivide into still smaller droplets. A succession of such Coulombic fissions would eventually lead to the formation of ultimate droplets so small that each of them would only contain one analyte molecule. Conversely, the ion evaporation model (IEM) argues that before a charged droplet becomes small enough to contain only one solute molecule, the charge density on its surface would become so high that the resulting electric field pushes one or more of those surface ions into the gas phase [16]. The general agreement used to be that small molecules are primarily ionized via IEM, whereas the formation of large macromolecular ions is better explained by CRM [17]. However, early nanospray experiments revealed that shorter chain-like proteins (such as insulin) can leave the ESI droplet when it is still quite large and contains more than one molecule of the analyte [18]. The more recently proposed chain ejection model (CEM) applies to such conditions. Based on this model, unlike globular natively folded proteins, unfolded proteins and disordered polymer chains immediately migrate to the surface when placed in a Rayleigh-charged nanodroplet in order to minimize contact between their exposed hydrophobic moieties and the aqueous microenvironment within the droplet interior. One chain terminus then gets expelled into the vapor phase, followed by stepwise sequential ejection of the remaining protein and separation from the droplet [19, 20]. Despite its success in explaining the ionization of unfolded protein chains, the CEM is unlikely to apply to nucleic acid chains because their hydrophobic and hydrophilic moieties are homogenously distributed along the chain [21].
An attempt for generating a predictive model for the MS signal intensity of small ions based on the IEM has been successful. Under this model [22, 23], solvation energy and surface activity of the ions are the major determinants of their MS response. In contrast, the main focus of the theoretical models of CRM has been to predict the charge state of ionized proteins. Based on these models, the maximum charge of a protein in the gas phase does not depend on the analyte charge in solution. It is rather governed by the Rayleigh charge limit of the ultimate electrospray droplet that contains only one protein molecule. This value is only dependent on the size of a protein, which can either be represented via the protein radius [24] or its surface area [25]. However, no general theoretical framework for the prediction of ion intensity of large analytes that are produced by the CRM can be found.
The equilibrium partitioning model of Cech and Enke [26] takes a distinct approach for predicting ESI ion intensities that could be applicable to both IEM and CRM. The basic postulation of this model is that because the excess charge resides on the surface of ESI droplets, only the analytes that can migrate to the surface will be charged and eventually detected by a mass spectrometer [27]. Therefore, the variability in the MS response of various compounds can be explained by their surface activity. Several parameters, including nonpolar surface area, Gibbs free energy of transfer from nonpolar to polar solutions, and reversed phase HPLC retention factors have been used as surrogates of surface activity in this model with varying degrees of success [28].
There have also been empirical studies of the fact.ors that influence ESI signal intensity of small organic compounds. Some parameters identified in various studies were: the degree of conjugation [29], charge delocalization [30], partition coefficient [31–33], gas-phase basicity [34], hydrogen to carbon (H/C) ratio [35] and molecular volume [32]. Nevertheless, in the particular case of oligonucleotides, very few investigations have been performed. It has been reported that solution pH [36], organic solvent percentage [37], and analyte hydrophobicity [38] can affect the signal intensity of oligonucleotides from pure solutions. But since LC-MS of oligonucleotides is almost always performed in the presence of alkylamine ion-pairing (IP) agents, the following studies that have included the use of IP agents are more relevant.
Gaus et al. [39] compared the signal intensity for phosphorothioates when using seven different alkylamines as IP agents and observed the highest ion intensity with tripropylamine (TPA). Erb and Oberacher [40] compared triethylamine (TEA) with dimethylcyclohexylamine (DMCHA) and concluded that TEA generates a stronger signal for a 27-mer DNA. Sharma et al. [41] used six different alkylamines (including DMCHA, TPA, and TEA) with a 17-mer DNA strand and observed the best MS sensitivity with dimethylbutylamine (DMBA). In an attempt to explain the effect of IP agents on oligonucleotide ion intensity, Chen et al. [42] carefully examined the signal intensity of a 24-mer DNA oligonucleotide in the presence of seven different IP agents and found a very strong relationship between the Henry’s law constant of the alkylamines and MS signal intensity of the oligonucleotide. The IP agents with a lower Henry’s law constant generated stronger MS signals for the oligonucleotide [42]. Nevertheless, it was later demonstrated that the effect of IP agents on the signal intensity of oligonucleotides is very much sequence-dependent and cannot be solely explained by the Henry’s law constant [43, 44].
In this manuscript, we present a comprehensive study of the IP and oligonucleotide-related parameters that influence the ESI-MS signal intensity using 11 different DNA sequences and 15 different IP agents. Statistical analysis of this set of data reveals important details about the factors that impact ESI efficiency. It will be further discussed in the following sections that identification of these factors provides the necessary means for a more detailed mechanistic understanding of the ESI process. This understanding is particularly important for oligonucleotides – as well as many other macromolecules – because several explanations have been put forward regarding their interactions in the electrospray droplet [26, 37, 38, 42] and each explanation is successful for interpreting some experimental results, indicating that all of them are partially accurate. Therefore, a more quantitative account of such interactions is necessary for generating an integrative model of the oligonucleotide ESI process. Furthermore, as demonstrated in the Results section, distinguishing the major parameters that affect the ESI signal intensity of oligonucleotides allows for the generation of an empirical formula for the prediction of oligonucleotide signal intensity in various mobile phase compositions. The proposed formula has extensive applications for LC-MS method development.
Materials and Methods
Chemicals and Reagents
The ion pairing agents N,N-dimethylbutylamine (DMBA), octylamine (OA), tripropylamine (TPA), N,N-dimethylhexylamine (DMHA), diisopropylamine (DIPA), N-methyldibutylamine (MDBA), propylamine (PA), triethylamine (TEA), hexylamine (HA), tributylamine (TBA), N,N-dimethylcyclohexylamine (DMCHA), N,N-diisopropylethylamine (DIEA), tetramethylethylenediamine (TMEDA), dibutylamine (DBA), and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU), as well as LC-MS grade methanol and water were purchased from Sigma-Aldrich Inc. (St. Louis, MO, USA). DNA Lobind microcentrifuge tubes were purchased from Eppendorf (Hauppauge, NY, USA). The ssDNA strands with the following sequences were purchased from Eurogentec (Seraing, Belgium): AAAAAAAAAAAAAAAAAAAAAAAA, TTTTTTTTTTTTTTTTTTTTTTTT, CCCCCCCCCCCCCCCCCCCCCCCC, GTGTGTGTGTGTGTGTGTGTGTGT, ATTTCTTTGTTTATTTCTTTGTTT, ATTCTTGTTATTCTTGTTATTCTT, ATCTGTATCTGTATCTGTATCTGT, TCGTACTAGTGGTCCTAATCGTAC, ATCGATCGATCGATCGATCGATCG, ACGACGACGTTTACGACGACGACG and CGGAGGAAACCTACGACGAGGAAA. The 24-mer TCGTGCTTTTGTTGTTTTCGCGTT was purchased from Integrated DNA Technologies (Coralville, IA, USA).
Preparation of Working Solutions for Direct Infusion Experiments
Precalculated volumes of various IP agents were added to aliquots of 20 μg/mL solutions of oligonucleotides in 50:50 methanol/water prepared fresh each day during the course of this investigation to the final concentration of 15 mM for all IP agents, except DBU. The final concentration of DBU was 2.5 mM. Importantly, all samples were prepared and analyzed in DNA Lobind tubes in order to eliminate any sample to sample variation due to differential non-specific losses to the tube walls. The pH of all of these oligonucleotide/ion-pair solutions fell in the very narrow range of 8.5 to 9.5 without adding any buffers.
Instrumental Conditions
Samples were directly infused to a Waters (Milford, MA, USA) Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer in the negative ion electrospray ionization mode via the instrument’s built-in fluidics system. The TOF-MS tuning parameters were as follows: capillary voltage −2.0 kV, cone voltage 25 V, extraction cone voltage 2 V, source temperature 125 °C, desolvation temperature 450 °C, cone gas 0 L/h, and desolvation gas (nitrogen) 1000 L/h. The infusion flow rate was set to 50 μL/min and the data were collected in continuum full-scan MS mode with a 1 s scan time over the mass range from 500–3000 m/z. All measurements were performed in triplicate and base peak signal intensities were measured by combining 50 scans.
Calculation of the Normalized Signal Intensity of the Oligonucleotide/Ion-Pair Solutions
It is a well-known phenomenon that absolute ESI-MS signal intensities are arbitrary and quite variable from experiment to experiment. Therefore, the establishment of a reliable electrospray ionization efficiency indicator that can be used to allow for comparisons among results obtained over several days of these experiments was an absolute necessity. It could be argued that the summation of all charge states detected for an oligonucleotide is necessary in order to represent its ionization efficiency. Nevertheless, since the charge state distribution of an oligonucleotide does not change dramatically with the use of different alkylamines, the ion count of the most intense charge state (the base peak) changes proportionally to the total current of all charge states. Therefore, for the sake of simplicity, we selected to use the MS signal intensity of the base analyte peak instead of the total ion current for calculating the ionization efficiency of oligonucleotides as described below. An inspection of the Supplementary Figure S1 clearly indicates that calculations based on these two parameters (base peak intensity versus sum of all charge states) do not generate any significant differences in the final results.
To deal with the highly varying base peak ESI-MS signal intensities for the same solution from day to day, we decided to use the oligonucleotide solutions in TMEDA as the reference. Therefore, the ionization efficiency of oligonucleotides in different solutions was reported as their normalized signal intensity, which was calculated as the ratio of their base peak intensity in any particular solution to the MS signal intensity of the base peak for the same oligonucleotide in the TMEDA solution:
The logic behind this method and other merits of this approach are further discussed in the Results section.
Databases and Computer Software
The MS operation and data acquisition were performed using Waters (Milford, MA, USA) MassLynx 4.1. Statistical analyses and graphs were created using GraphPad Software (La Jolla, CA, USA) Prism 6 and SAS JMP Pro 12 (Cary, NC, USA). Henry’s law constants were computed using HENRYWIN module of the US Environmental Protection Agency’s EPI suite ver. 4.11 (Washington, DC, USA). Proton affinity and gas-phase basicity values were obtained from the NIST database (http://webbook.nist.gov/chemistry/). The remaining physicochemical parameters were obtained from the CAS REGISTRY of the American Chemical Society (https://scifinder.cas.org/).
Prediction Formula
The signal intensity prediction formula is as follows. We have also supplemented a Microsoft Excel sheet containing this formula with the online version of this manuscript for the convenience of our readers. Please note that Content A, T, C, and G need to be entered as decimals.
Results and Discussion
In order to determine the extent to which different parameters influence the ESI ion intensity of oligonucleotides, we chose 11 DNA sequences of varying compositions and tested each one alongside 15 different ion-pairing agents with distinct physicochemical properties. Tables 1 and 2 list the oligonucleotide sequences and IP agents utilized for this study, respectively. Because previous studies in our laboratory [42, 44] had demonstrated that alkylamine IP agents produce the highest oligonucleotide MS signal intensity when used at concentrations around 15 mM, we used all alkylamine IP agents at this concentration. The only exception was DBU, which was used at 2.5 mM in order to maximize its signal intensity as suggested by Sharma et al. [41] and confirmed through our own observations. Furthermore, an oligonucleotide concentration of 20 μg/mL was chosen because we were able to acquire robust mass spectra at this concentration, which made data interpretation easier. We should also mention that the performance of ion-pairing reagents was not dependent on the concentration of the oligonucleotides; i.e., the ion-pairing reagent that generated the strongest MS signal intensity at higher oligonucleotide concentrations also had the best performance at lower oligonucleotide concentrations. Therefore, the concentration of the studied oligonucleotides was kept constant throughout this investigation.
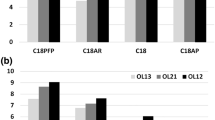
Figure 1 shows the representative data acquired from our experiment with two different DNA strands. It is clear that in the absence of IP agents, the oligonucleotide MS signal was weaker than when IP agents were being utilized. This observation might initially seem contradictive considering that IP agents are highly surface active and it is generally accepted that surface active additives cause ion suppression [45, 46]. However, the model suggested by Chen et al. [42] can explain this phenomenon perfectly. Based on their model, the interaction between oligonucleotides and highly surface active IP agents helps oligonucleotides to reach the surface of the electrospray droplet. This effect is further amplified due to the alkylamine concentration gradient that exists from the center to the surface of the electrospray droplet because of the continuous ion-pair evaporation from the surface. Nevertheless, this explanation is only true when the ion-pair concentrations are not high enough to hinder oligo access to the surface of the droplet. That is why the concentrations of the ion-pairing reagents need to be chosen very carefully. The concentration of 15 mM (or 2.5 mM for DBU) completely satisfies this need.
Based on their model, Chen et al. also suggested that IP agents with a lower Henry’s law constant would evaporate faster and create a more steep concentration gradient from the droplet center to the surface leading to stronger oligonucleotide MS signal intensity [42]. Examining Figure 1 reveals that this scheme can only explain the extreme cases. For example, OA has the smallest Henry’s law constant and it is often among the top performing IP agents, whereas TMEDA with a very large Henry’s law constant often performs poorly. Nevertheless, although HA has a much larger Henry’s law constant than MDBA, it has performed better in both cases. Yet, perhaps the most interesting observation was the different performance patterns of IP agents based on the oligonocleotide sequence. These observations indicated the need for a comprehensive set of experiments to understand the nuances that can influence the ESI ion intensity of oligonucleotides.
It is very well known that the absolute ESI-MS signal intensities are quite variable for an experiment performed at different times, even at similar experimental conditions. Therefore, a major prerequisite for performing this investigation was the establishment of a reliable ESI-MS intensity indicator that can be used to allow for comparisons among results obtained over several days of these experiments. To this end, we monitored the signal intensity of oligonucleotide solutions in the presence of various alkylamine ion-pairing agents over the course of a few days. Our results indicated that by choosing one of the oligo/IP solutions as a reference and reporting the ratio of MS signal intensity for other mixtures of the same oligonucleotide to the reference solution, a much more dependable measure of the MS signal intensity could be generated with minimal variability between the experiments. This approach was consistent with the efforts of Leito and colleagues in generating an electrospray ionization efficiency scale [47–49]. Since oligonucleotide solutions usually had the lowest signal intensities in TMEDA solutions, the oligonucleotide/TMEDA solutions were chosen as references and all other intensities were reported as their ratios to these reference solutions:
Table 3 shows the absolute and relative intensities of similar oligonucleotide solutions that were prepared and measured on two different days. It is seen that while the difference in absolute intensity from 1 d to another can easily exceed 30%, normalized intensities were not more than 15% different. Note that while only a representative number of measurements is shown here, these trends are consistent across much larger data sets.
Having a standardized response (normalized intensity) as well as variables related to the ion pair (physicochemical properties of Table 2) were important initial considerations toward modeling the ESI process for oligonucleotides. However, in order to proceed with statistical analysis, we still needed to construct a set of descriptors for the oligonucleotides. Studies have shown that the oligonucleotide hydrophobicity has the strongest correlation with its electrospray ionization efficiency [38, 44]. Furthermore, it has been demonstrated that the hydrophobicity of an oligonucleotide can be calculated solely from the percentages of its nucleobases with very good accuracy [38, 50]. Therefore, we decided to use nucleotide proportions (%A, %T, %C, %G) of each oligonucleotide as the additional variables of our statistical model.
A partitioning (or decision tree-based) method was used for our analysis. Partitioning is a way to describe the relationship between a response and set of factors without a mathematical model [51]. The goal is to divide the data into groups, which differ maximally with respect to MS response. Partitioning is an iterative process, the visualization of which resembles a tree – hence the term “decision tree”. The bootstrap forest (or random forest) [52] averages the results of many trees. For each of these trees, only a random sample of the observations is considered; then for each split, only a random subset of the candidate variables is considered. In this way, it is highly probable that all of the variables useful in predicting the response will eventually be chosen as splitting variables. Figure 2 shows the output of the bootstrap forest for our data. Not surprisingly, some familiar liquid–gas parameters such as boiling point and vapor pressure have been revealed to substantially contribute in MS response determination. The Henry’s law constant was also shown to have a considerable contribution.
The significant role of partition coefficient and water solubility of IP agents can be easily understood in light of the previously mentioned model of Chen et al. [42]. Based on their model, the surface activity of alkylamine IP agents plays a vital role in the increased ion intensity of oligonucleotides. Water solubility and partition coefficient are both very closely related to surface activity. The effect of gas-phase basicity and proton affinity is most probably brought forward during the dissociation process of oligonucleotides from ion-pairs in the gas phase, and a hypothetical mechanism suggested by Muddiman et al. [37] regarding the role of piperidine and imidazole in charge state reduction of oligonucleotides can help to explain this effect. It is reasonable to assume that alkylamines can hydrogen bond to the phosphate backbone of oligonucleotides. Through this hydrogen bonding, they can displace cations, which results in reduced cation adduction and subsequently increases signal intensity. But for this to happen, the proton that originally resided on the alkylamine in solution should be transferred to the phosphate backbone in the gas phase so that the hydrogen bound ion-pair can be released from the oligo. Therefore, it is expected for the gas-phase proton affinity of the top-performing IP agents to be lower than the oligonucleotide phosphodiester backbone. The proton affinity of the phosphodiester group is estimated at 315 Kcal/mol [53], whereas all utilized alkylamines have proton affinities in the range of 220–240 Kcal/mol, which is well below the estimated value for the phosphodiester backbone. One important observation in support of this proposal was the excellent capability of OA in suppressing cation adduction. In comparison, DBA with very closely related structure and physicochemical properties to OA that often resulted in similar effects on MS signal intensity of oligonucleotides was not very effective in reducing cation adduction. Examination of Table 2 reveals that the proton affinity of OA is about 10 Kcal/mol lower than DBA.
As expected, the composition of oligonucleotides also plays a major role in determining the ESI response in the form of a very large contribution for %T in the sequence of the oligo. A more significant contribution from thymine compared with other nucleotides was not unexpected. It has been shown that T is the most hydrophobic DNA base (T > A > G > C) [50, 54, 55], and since hydrophobicity significantly affects signal intensity, it is reasonable for T content to be more important than the other nucleobases. The major significance of hydrophobicity for electrospray ionization efficiency of our analytes is also in complete agreement with the previously mentioned equilibrium partitioning model [26, 27].
Creating a predictive model with these data is also very desirable from a practical standpoint. Based on these experiments, it is clear that different DNA sequences generate the highest MS signal intensity with different alkylamine IP agents. Therefore, all different combinations of oligo/IP would need to be investigated to find the optimum ion-pair every time prior to the development of a LC-MS method. This would create an additional burden on the analysis and hinder method development. Therefore, a predictive algorithm that can rapidly assist with the selection of optimal IP agents without the need to perform dozens of experiments would be extremely useful. We managed to generate a partial least squares (PLS) regression model capable of predicting intensity values that are very well aligned with our experimental observations (Figure 3).
To further confirm the prediction power of the resulting PLS model, we used a DNA strand with the sequence 5′-TCG TGC TTT TGT TGT TTT CGC GTT-3′, which had not been examined in the previous experiments. The sequence was run through the algorithm and the expected normalized signal intensities were calculated. Then the experimental values were obtained. As shown in Figure 4, the prediction of our model fit the observed data, well. The only exception was HA, where prediction and observation showed opposite trends. We believe this is due to a complete lack of adenines in this particular sequence. Figure 5 shows the experimental MS signal intensities for two sequences that are both composed of 50% T. However, the remaining 50% is equally distributed among all other nucleotides for Figure 5a in contrast to Figure 5b, which only has T and G nucleotides in its sequence. While the trend in Figure 5a is more similar to the predictions of Figure 4, Figure 5b more closely resembles the experimental results. Considering the particularly superb performance of HA with poly-A sequences (not shown), it seems that the presence of adenines is necessary for maximum HA performance. But neither the sequence in Figure 4 nor the sequence in Figure 5b contained any adenines. Therefore, HA was not among the best IP agents for any of them. However, HA performed well with the DNA sequence shown in Figure 5a even though its T content was not very different from the other two. We believe that this is because it contained a greater number of adenines. This very specific synergy between the adenine content and HA is too complicated for our model to accurately extract and therefore it has overestimated HA performance for the sequence in Figure 4. To further demonstrate this point, we repeated this experiment with a different sequence that contained adenine bases. As expected, the prediction pattern for HA and other IP agents was in line with the observations of this experiment (Figure 6). Therefore, our model seems to predict the HA performance inaccurately for the sequences that are completely void of adenines. Nevertheless, this should not be a problem for most common sequences that contain all four nucleotides. Even for sequences like the one in Figure 4, our model accurately determined the response for all of the other ion-pairs.
Experimental and predicted normalized signal intensities for miR-451 (5′- AAA CCG UUA CCA UUA CUG AGU U −3′) in the presence of various ion pairing reagents. Note that in contrast to Figure 4, both prediction and experiment follow similar patterns regarding HA performance
Another important observation concerns TEA. It is the most widely used ion-pair for LC-MS analysis of oligonucleotides [56]. However, when comparing the performance of TEA to other IP agents in Figure 1a, Figure 4, and Figure 5a, the results indicate that using TEA with any of these sequences would cause a substantial decrease in the sensitivity of the resulting LC-MS method. Therefore, it is evident that the use of alternative alkylamine IP agents is necessary for improving the sensitivity of LC-MS methods for oligonucleotides. Of course, other considerations should also be taken into account when using IP agents for oligonucleotide analysis. One such important consideration is the ion suppression caused by these alkylamines when switching from negative to positive ESI. In our experience, all of the IP agents mentioned in this manuscript can cause such an effect. The way we have mitigated this issue is to dedicate separate LC channels to positive and negative ESI buffers. In this way, we have managed to keep the overlap of mobile phases components at a minimum and successfully switch between positive and negative ESI routinely. We would also like to add that from the point of cost and ease of use, there are no dramatic differences among the alkylamines we have used for this study. Therefore, switching from TEA to an alternative ion-pair would not add any undue burden to the bioanalytical methods. In contrast, it provides an easy way for increasing the sensitivity of oligonucleotide quantitation by LC-MS, which represents a potentially quite important outcome. Above all, it is only via increased sensitivity that mass spectrometry can satisfy the many emerging needs of oligonucleotide researchers.
Conclusion
We have performed a comprehensive set of experiments to help us better understand the physicochemical properties of analytes and mobile phase additives that govern electrospray ionization efficiency. Many of the factors identified through this process have been previously suggested to influence the ESI process by different mechanisms outlined in various models. Therefore, our study provides a unifying platform for several proposed mechanisms in addition to a more quantitative perspective regarding the relative contribution of factors suggested by one model when compared to others. This would make it possible to assess the relative importance of each proposed mechanism toward the overall ESI process and combine those events in the right order to give rise to a generalized model with diverse applications.
We have also generated a PLS regression model with very good predictive power that can help with choosing the optimum IP agent based on the oligonucleotide composition for performing LC-MS analysis. This is based on the fact that most of these alkylamine IP agents have been utilized previously in a few different investigations and all of them have shown acceptable chromatographic performance. Chen and Bartlett have used DIEA [57]. Gong has utilized DBA, DMBA, HA and TPA [43, 58]. Oberacher and colleagues have used DMCHA [40]. McGinnis et al. have separated several modified and unmodified DNA and RNA strands using DIPA [44], and Sharma et al. [41] have reported using DBU among other IP agents. In all of mentioned studies, TEA has also been present as a reference point and the chromatographic performance of these alternative IP agents has been similar to or slightly better than TEA. More importantly, in two of these studies [41, 58], several IP agents have been compared simultaneously, and a close examination of the oligonucleotide retention factors further reveals that the difference in chromatographic performance between various alkylamines is minimal. Furthermore, the concentrations of IP agents in these studies have been in the general range of 10–15 mM, which is in line with our experimental design. The only exceptions were DMBA and DBU, which have been used at the concentrations of 5 and 2.5 mM, respectively.
The study performed by Gong and McCullagh [58] is particularly important in supporting our proposition that the overall performance of oligonucleotide LC-MS methods is primarily governed by the ESI efficiency, and the chromatographic performance of different IP agents is more or less the same. They have used mobile phases containing six different alkylamine IP agents, including DBA, DIEA, DMBA, HA, TEA, and TPA, in order to separate different poly-T sequences (T10, T15, T25, and T40). The obtained resolutions for these oligonucleotides have been largely similar regardless of the choice of ion-pair. Nevertheless, the overall method sensitivity has followed the same pattern as we have shown in Figure 1a with DBA > HA > TPA > DMBA > DIEA > TEA. This data clearly indicates that the alkylamine IP agents that generate the highest MS signal intensity for a particular oligonucleotide are usually the best choice for LC-MS method development as they will most likely have acceptable chromatographic performance. Therefore, we can confidently suggest our PLS predictive algorithm as an efficient tool for ion-pair selection for LC-MS analysis of oligonucleotides although we have not fully studied the chromatographic performance of all IP agents discussed in this manuscript.
References
Woods, A.G., Darie, C.C.: Advancements of Mass Spectrometry in Biomedical Research. Springer International Publishing, Cham (2014)
Fenn, J.B.: Electrospray wings for molecular elephants (Nobel lecture). Angew. Chem. Int. Ed. Engl. 42, 3871–3894 (2003)
Karas, M., Hillenkamp, F.: Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60, 2299–2301 (1988)
Berkenkamp, S., Kirpekar, F., Hillenkamp, F.: Infrared MALDI mass spectrometry of large nucleic acids. Science 281, 260–262 (1998)
Tanaka, K., Waki, H., Ido, Y., Akita, S., Yoshida, Y., Yoshida, T., Matsuo, T.: Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2, 151–153 (1988)
Fenn, J., Mann, M., Meng, C., Wong, S., Whitehouse, C.: Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 (1989)
Heck, A.J.R., van den Heuvel, R.H.H.: Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 23, 368–389 (2004)
Nilsson, T., Mann, M., Aebersold, R., Yates, J.R., Bairoch, A., Bergeron, J.J.M.: Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Method 7, 681–685 (2010)
Percy, A.J., Rey, M., Burns, K.M., Schriemer, D.C.: Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal. Chim. Acta 721, 7–21 (2012)
Kitova, E.N., El-Hawiet, A., Schnier, P.D., Klassen, J.S.: Reliable determinations of protein-ligand interactions by direct ESI-MS measurements. Are we there yet? J. Am. Soc. Mass Spectrom. 23, 431–441 (2012)
Liuni, P., Jeganathan, A., Wilson, D.J.: Conformer selection and intensified dynamics during catalytic turnover in chymotrypsin. Angew. Chem. Int. Ed. 51, 9666–9669 (2012)
Hogan, C.J., Carroll, J.A., Rohrs, H.W., Biswas, P., Gross, M.L.: Combined charged residue-field emission model of macromolecular electrospray ionization. Anal. Chem. 81, 369–377 (2009)
Nguyen, S., Fenn, J.B.: Gas-phase ions of solute species from charged droplets of solutions. Proc. Natl. Acad. Sci. 104, 1111–1117 (2007)
Dole, M., Mack, L.L., Hines, R.L., Mobley, R.C., Ferguson, L.D., Alice, M.B.: Molecular Beams of macroions. J. Chem. Phys. 49, 2240–2249 (1968)
Mack, L.L., Kralik, P., Rheude, A., Dole, M.: Molecular Beams of macroions. II. J. Chem. Phys. 52, 4977–4986 (1970)
Iribarne, J.V., Thomson, B.A.: On the evaporation of small ions from charged droplets. J. Chem. Phys. 64, 2287–2294 (1976)
Kebarle, P., Verkerk, U.H.: On the mechanism of electrospray ionization mass spectrometry (ESIMS). In: Cole, R.B. (ed.) Electrospray and MALDI mass spectrometry: fundamentals, instrumentation, practicalities, and biological applications, pp. 1–48. Wiley, Hoboken (2010)
Juraschek, R., Dülcks, T., Karas, M.: Nanoelectrospray—more than just a minimized-flow electrospray ionization source. J. Am. Soc. Mass Spectrom. 10, 300–308 (1999)
Konermann, L., Ahadi, E., Rodriguez, A.D., Vahidi, S.: Unraveling the mechanism of electrospray ionization. Anal. Chem. 85, 2–9 (2013)
Yue, X., Vahidi, S., Konermann, L.: Insights into the mechanism of protein electrospray ionization from salt adduction measurements. J. Am. Soc. Mass Spectrom. 25, 1322–1331 (2014)
Abi-Ghanem, J., Gabelica, V.: Nucleic acid ion structures in the gas phase. PCCP 16, 21204–21218 (2014)
Tang, L., Kebarle, P.: Dependence of ion intensity in electrospray mass spectrometry on the concentration of the analytes in the electrosprayed solution. Anal. Chem. 65, 3654–3668 (1993)
Tang, L., Kebarle, P.: Effect of the conductivity of the electrosprayed solution on the electrospray current. Factors determining analyte sensitivity in electrospray mass spectrometry. Anal. Chem. 63, 2709–2715 (1991)
de la Mora, F.J.: Electrospray ionization of large multiply charged species proceeds via Dole’s charged residue mechanism. Anal. Chim. Acta 406, 93–104 (2000)
Kaltashov, I.A., Mohimen, A.: Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 77, 5370–5379 (2005)
Cech, N.B., Enke, C.G.: Relating Electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 72, 2717–2723 (2000)
Cech, N.B., Enke, C.G.: Effect of affinity for droplet surfaces on the fraction of analyte molecules charged during electrospray droplet fission. Anal. Chem. 73, 4632–4639 (2001)
Cech, N.B., Enke, C.G.: Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 20, 362–387 (2001)
Huffman, B.A., Poltash, M.L., Hughey, C.A.: Effect of polar protic and polar aprotic solvents on negative-ion electrospray ionization and chromatographic separation of small acidic molecules. Anal. Chem. 84, 9942–9950 (2012)
Kruve, A., Kaupmees, K., Liigand, J., Leito, I.: Negative electrospray ionization via deprotonation: predicting the ionization efficiency. Anal. Chem. 86, 4822–4830 (2014)
Henriksen, T., Juhler, R.K., Svensmark, B., Cech, N.B.: The relative influences of acidity and polarity on responsiveness of small organic molecules to analysis with negative ion electrospray ionization mass spectrometry (ESI-MS). J. Am. Soc. Mass Spectrom. 16, 446–455 (2005)
Chalcraft, K.R., Lee, R., Mills, C., Britz-McKibbin, P.: Virtual quantification of metabolites by capillary electrophoresis-electrospray ionization-mass spectrometry: predicting ionization efficiency without chemical standards. Anal. Chem. 81, 2506–2515 (2009)
Liigand, J., Kruve, A., Leito, I., Girod, M., Antoine, R.: Effect of mobile phase on electrospray ionization efficiency. J. Am. Soc. Mass Spectrom. 25, 1853–1861 (2014)
Ehrmann, B.M., Henriksen, T., Cech, N.B.: Relative importance of basicity in the gas phase and in solution for determining selectivity in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 719–728 (2008)
Nguyen, T.B., Nizkorodov, S.A., Laskin, A., Laskin, J.: An approach toward quantification of organic compounds in complex environmental samples using high-resolution electrospray ionization mass spectrometry. Anal. Methods 5, 72–80 (2013)
Bleicher, K., Bayer, E.: Various factors influencing the signal intensity of oligonucleotides in electrospray mass spectrometry. Biol. Mass Spectrom. 23, 320–322 (1994)
Muddiman, D.C., Cheng, X., Udseth, H.R., Smith, R.D.: Charge-state reduction with improved signal intensity of oligonucleotides in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 7, 697–706 (1996)
Null, A.P., Nepomuceno, A.I., Muddiman, D.C.: Implications of hydrophobicity and free energy of solvation for characterization of nucleic acids by electrospray ionization mass spectrometry. Anal. Chem. 75, 1331–1339 (2003)
Gaus, H.J., Owens, S.R., Winniman, M., Cooper, S., Cummins, L.L.: On-line HPLC electrospray mass spectrometry of phosphorothioate oligonucleotide metabolites. Anal. Chem. 69, 313–319 (1997)
Erb, R., Oberacher, H.: Comparison of mobile-phase systems commonly applied in liquid chromatography-mass spectrometry of nucleic acids. Electrophoresis 35, 1226–1235 (2014)
Sharma, V.K., Glick, J., Vouros, P.: Reversed-phase ion-pair liquid chromatography electrospray ionization tandem mass spectrometry for separation, sequencing and mapping of sites of base modification of isomeric oligonucleotide adducts using monolithic column. J. Chromatogr. 1245, 65–74 (2012)
Chen, B., Mason, S.F., Bartlett, M.G.: The effect of organic modifiers on electrospray ionization charge-state distribution and desorption efficiency for oligonucleotides. J. Am. Soc. Mass Spectrom. 24, 257–264 (2013)
Gong, L.: Comparing ion-pairing reagents and counter anions for ion-pair reversed-phase liquid chromatography/electrospray ionization mass spectrometry analysis of synthetic oligonucleotides. Rapid Commun. Mass Spectrom. 29, 2402–2410 (2015)
McGinnis, A.C., Grubb, E.C., Bartlett, M.G.: Systematic optimization of ion-pairing agents and hexafluoroisopropanol for enhanced electrospray ionization mass spectrometry of oligonucleotides. Rapid Commun. Mass Spectrom. 27, 2655–2664 (2013)
Cech, N.B., Enke, C.G.: Selectivity in electrospray ionization mass spectrometry. In: Cole, R.B. (ed.) Electrospray and MALDI mass spectrometry, pp. 49–73. Wiley, Hoboken (2010)
Annesley, T.M.: Ion suppression in mass spectrometry. Clin. Chem. 49, 1041–1044 (2003)
Oss, M., Kruve, A., Herodes, K., Leito, I.: Electrospray ionization efficiency scale of organic compounds. Anal. Chem. 82, 2865–2872 (2010)
Leito, I., Herodes, K., Huopolainen, M., Virro, K., Künnapas, A., Kruve, A., Tanner, R.: Towards the electrospray ionization mass spectrometry ionization efficiency scale of organic compounds. Rapid Commun. Mass Spectrom. 22, 379–384 (2008)
Liigand, J., Kruve, A., Liigand, P., Laaniste, A., Girod, M., Antoine, R., Leito, I.: Transferability of the electrospray ionization efficiency scale between different instruments. J. Am. Soc. Mass Spectrom. 26, 1923–1930 (2015)
Gilar, M., Fountain, K.J., Budman, Y., Neue, U.D., Yardley, K.R., Rainville, P.D., Russell II, R.J., Gebler, J.C.: Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. J. Chromatogr. 958, 167–182 (2002)
Cook, E.F., Goldman, L.: Empiric comparison of multivariate analytic techniques: advantages and disadvantages of recursive partitioning analysis. J. Chronic Dis. 37, 721–731 (1984)
Ho, T.K.: Random decision forests. Proceedings of 3rd International Conference on Document Analysis and Recognition. vol. 271, 1, 278–282 (1995)
Lum, R.C., Grabowski, J.J.: Trimethyl phosphate: the intrinsic reactivity of carbon versus phosphorus sites with anionic nucleophiles. JACS 114, 8619–8627 (1992)
Huber, C.G., Oefner, P.J., Bonn, G.K.: High-resolution liquid chromatography of oligonucleotides on nonporous alkylated styrene-divinylbenzene copolymers. Anal. Biochem. 212, 351–358 (1993)
Huber, C.G., Oefner, P.J., Bonn, G.K.: High-performance liquid chromatographic separation of detritylated oligonucleotides on highly cross-linked poly-(styrene-divinylbenzene) particles. J. Chromatogr. 599, 113–118 (1992)
McGinnis, A.C., Chen, B., Bartlett, M.G.: Chromatographic methods for the determination of therapeutic oligonucleotides. J. Chromatogr. B 883/884, 76–94 (2012)
Chen, B., Bartlett, M.G.: Evaluation of mobile phase composition for enhancing sensitivity of targeted quantification of oligonucleotides using ultra-high performance liquid chromatography and mass spectrometry: application to phosphorothioate deoxyribonucleic acid. J. Chromatogr. A 1288, 73–81 (2013)
Gong, L., McCullagh, J.S.O.: Comparing ion-pairing reagents and sample dissolution solvents for ion-pairing reversed-phase liquid chromatography/electrospray ionization mass spectrometry analysis of oligonucleotides. Rapid Commun. Mass Spectrom. 28, 339–350 (2014)
Acknowledgements
The experiments described in this manuscript were supported by the National Cancer Institute (CA176653).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure S1
The normalized signal intensity of T 24 in the presence of various IP agents calculated using either the oligonucleotide base peak signal intensities (solid bars) or the total ion counts of all oligonucleotide charge states (checkered bars) (GIF 1178 kb)
ESM 1
(XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Basiri, B., Murph, M.M. & Bartlett, M.G. Assessing the Interplay between the Physicochemical Parameters of Ion-Pairing Reagents and the Analyte Sequence on the Electrospray Desorption Process for Oligonucleotides. J. Am. Soc. Mass Spectrom. 28, 1647–1656 (2017). https://doi.org/10.1007/s13361-017-1671-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1671-6