Abstract
The dorsal root ganglion (DRG) and its anatomically and functionally associated spinal nerve and ventral and dorsal roots are important components of the peripheral sensory-motor system in mammals. The cells within these structures use a number of peptides as intercellular signaling molecules. We performed a variety of mass spectrometry (MS)-based characterizations of peptides contained within and secreted from these structures, and from isolated and cultured DRG cells. Liquid chromatography-Fourier transform MS was utilized in DRG and nerve peptidome analysis. In total, 2724 peptides from 296 proteins were identified in tissue extracts. Neuropeptides are among those detected, including calcitonin gene-related peptide I, little SAAS, and known hemoglobin-derived peptides. Solid phase extraction combined with direct matrix-assisted laser desorption/ionization time-of-flight MS was employed to investigate the secretome of these structures. A number of peptides were detected in the releasate from semi-intact preparations of DRGs and associated nerves, including neurofilament- and myelin basic protein-related peptides. A smaller set of analytes was observed in releasates from cultured DRG neurons. The peptide signals observed in the releasates have been mass-matched to those characterized and identified in homogenates of entire DRGs and associated nerves. This data aids our understanding of the chemical composition of the mammalian peripheral sensory-motor system, which is involved in key physiological functions such as nociception, thermoreception, itch sensation, and proprioception.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A large amount of sensory information within the mammalian nervous system is conveyed from the organism’s periphery to the spinal cord and brain stem via sensory neurons residing in the dorsal root ganglion (DRG), which consists of the neuron cell bodies and support cells [1, 2]. A sensory neuron has a single axon extending from the cell body that bifurcates, sending one process to the dorsal region of the spinal cord, while the other process innervates a variety of targets [1, 3, 4]. DRG neurons innervate a number of target organs, including skin, muscles, and joints [5, 6], and are involved in mechanoreception, limb proprioception, thermoreception, and nociception. A number of neuropeptides and other cell-to-cell signaling peptides have been identified within DRGs, including Substance P and calcitonin gene-related peptide I (CGRP-1) [7]. Many common pain medications (e.g., opioids) target this neuronal network. DRGs are structurally associated with the spinal nerve (SN) and its dorsal root (DR) and ventral root (VR). The DR and SN contain neurites from the sensory neurons. The VR possesses mostly terminals of spinal motoneurons and presents an opportunity for a comparative analysis of motor and sensory components of the nervous system.
Both normal and pathologic functions of the peripheral sensory-motor system depend on the overall chemical composition of corresponding cells and their extracellular environments. Important studies have been performed to better understand the metabolite, lipid, and protein content of DRGs and their surrounding structures [8–14]. However, the cell-to-cell signaling molecules in these areas and their releasates have not been completely characterized. Measurements of activity-dependent release are challenging for several reasons. First, cells release a broad range of physiologically active compounds over a wide concentration range. Second, only small amounts of the compounds within the tissue are released, making measurement detection limits important. Moreover, peptide signaling can be terminated by internalization of the signal or efficient enzymatic extracellular degradation. Therefore, effective capture and accumulation of released peptides is important for successful measurements. Lastly, the collection of the sample should minimally perturb the system and allow for a controlled stimulation of release and efficient analyte collection.
Methods that combine analytical techniques, such as liquid chromatography (LC)-MS, have become mainstream approaches used in proteomic investigations of tissues, organs, and cell cultures [15–23]. In contrast to LC-MS, direct assay of tissues or cells using matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) MS is also effective, either via mass spectrometry imaging [24–26] or by characterization of the isolated cells, nerves, and ganglia deposited on a conductive sample plate [27, 28]. Both LC-electrospray ionization (ESI)-MS and direct MALDI MS can be used to investigate peptide release. Direct MS is well suited for the smallest volume samples and can accommodate larger numbers of samples as a separation is not required. However, most released analytes are present at low concentrations and require preconcentration before the MS analysis. Also, the extracellular media contains high inorganic salt levels that are not compatible with direct MS measurements. Therefore, effective methods for collecting released material and conditioning/concentrating the peptides are required. For example, solid phase extraction (SPE) using collection capillaries or probes placed at appropriate locations in close proximity to tissue regions, cell populations in culture, or individual cells enables the measurement of compounds released upon electrical or chemical stimulation [29–31].
Microfluidics-based cell culturing systems can also be used to provide a wealth of information pertaining to tissue and cell biochemistry, growth, and morphology [32, 33]. We previously reported a microfluidic device that permits the culture and maintenance of neurons, temporal application of selective chemical stimuli, and collection of peptide release, with the releasate analyzed using MALDI-TOF MS [34]. The advantages of interfacing microfluidics to MALDI include its small sample volume requirements, wide dynamic range for peptide characterization [35, 36], and ease of making a large number of measurements, although tandem MS (MS/MS)-based identification of released compounds using direct MALDI MS tends not to be as effective as LC-ESI-MS. Therefore, the separation and preconcentration of peptides on an LC system aids in the identification of endogenous peptides and proteins. Well-known benefits of having LC on the front end of MS include minimized suppression effects from co-eluting ion species and increased dynamic range for better detection of low abundance peptides. The combination of direct tissue measurements with LC-Fourier transform (FT)-ESI-MS and cell release measurements using MALDI-TOF MS is particularly effective in examining peptide release, thereby enabling the assignment of signals detected in release by mass matching to peptides characterized in the larger samples [30, 37].
One of our long-term goals is to recreate different functional networks of well-defined DRG cells within engineered microfluidic devices [38–40] so that we can characterize the biochemical responses, including cellular release, of these networks to different stimuli and changing microenvironments. However, in order to establish and validate our working model and perform the required measurements, it is important to have an inventory of the compounds present within and released from DRGs and their associated structures. Here we present a series of LC-ESI-FTMS/MS investigations of the peptide content of the rat peripheral sensory-motor system, including the DR, VR, SN, and DRG, as well as isolated DRG cells. In parallel, SPE-assisted MALDI MS and LC-ESI-FTMS were used to characterize the release from semi-intact preparations of DRGs with attached nerves, a robust DRG cell culture was established in miniaturized wells, and a number of peptides were identified as being released from the DRG cells and nerves. The data obtained on the peripheral sensory-motor system peptidome and secretome will be used to enable follow-up studies on the function of this complex neuronal system, and on the growth and formation of defined DRG networks in engineered microfluidic devices.
Experimental
Animals and Tissue Dissection
All procedures related to animal handling and euthanasia were performed in accordance with local, state, and federal regulations, and approved by the Illinois Institutional Animal Care and Use Committee. DRGs, spinal cords, SNs, VRs, and DRs were surgically dissected from 2.5–3 mo-old Sprague–Dawley outbred male rats (Harlan Laboratories, Inc., Indianapolis, IN, USA), 21–28 d postnatal Sprague–Dawley outbred male rats (Charles River, St. Constant, QC, Canada), or 6–12 wk-old Long-Evans/BluGill male and female rats (University of Illinois at Urbana−Champaign, USA), euthanized by decapitation. To reduce the detrimental metabolic effects of a stopped blood flow, 40–120 mL of ice-cold modified Gey’s balanced salt solution (mGBSS) was injected under the skin above the location of the DRGs of interest (the lumbar area) immediately after decapitation. The spinal canal was surgically opened, the spinal cord removed, and DRGs with adjacent nerves were individually isolated and placed into ice cold mGBSS containing (in mM): 1.5 CaCl2, 4.9 KCl, 0.2 KH2PO4, 11 MgCl2, 0.3 MgSO4, 138 NaCl, 27.7 NaHCO3, and 0.8 Na2HPO4, and 25 HEPES, pH 7.2. In some experiments, other media types were used as described below. Most of the presented work used 2.5–3 mo-old Sprague–Dawley outbred male rats (Harlan Laboratories, Inc.). Animals of other strains, sex, and age investigated in the DRG peptidomics experiments produced similar results to data obtained from the Sprague–Dawley rats.
DRG Cell Isolation
Immediately after dissection, DRGs were sustained in a volume of ice cold Hibernate A media (BrainBits, Springfield, IL, USA) for up to 48 h. For dissociation, approximately 10 DRGs dissected from 2.5–3 mo-old Sprague–Dawley outbred rats were digested in a solution of 0.25% collagenase (Worthington Biochemical Corp, Lakewood, NJ, USA) in DRG cell culture media containing Neurobasal A without phenol red (Life Technologies, Carlsbad, CA, USA), 100 U/mL penicillin/streptomycin (Life Technologies), 0.5 mM GlutaMAX (Life Technologies), 50 ng/mL nerve growth factor (Life Technologies), 50 ng/mL brain-derived neurotrophic factor (BDNF) (Prospec Bio, Rehovot, Israel), and 500 μL B27 Growth Supplement (Life Technologies) for 1.5 h at 37°C. After digestion, the samples were subjected to centrifugation for 3 min at 200 ×g. The supernatant was removed, and the pellet washed with Hank’s Balanced Salt Solution (HBSS) (Life Technologies). The sample was centrifuged, the supernatant removed, and the pellet digested using 0.25% trypsin with ethylenediaminetetraacetic acid (Life Technologies) for 15 min at 37°C. After incubation, the sample was centrifuged, supernatant removed, and 0.5–1 mL DRG cell culture media + 1% fetal bovine serum (Life Technologies) added to inactivate the trypsin. The pellet was mechanically dissociated by trituration with fire-polished pipettes. Following trituration, some of the pellet was allowed to resettle. Once a small pellet formed in the bottom of the microcentrifuge tube, the supernatant was removed and spun for 3 min at 200 ×g. After centrifugation, the supernatant was removed and the pellet washed with HBSS. After a final centrifugation, the cell pellet was resuspended in 1 mL of DRG cell culture media per 10 original DRG.
DRG Cell Culture
Polydimethylsiloxane (PDMS) blocks, punched with 1.5 or 2 mm diameter biopsy punches (Acuderm, Inc., Ft. Lauderdale, FL, USA) to create small restricted-space culture wells, were attached to either glass coverslips or PDMS substrates. Prior to cell plating, glass coverslips were treated with concentrated H2SO4 for 24 h to remove any contaminants and then washed with distilled water. The PDMS was sterilized in an O2 plasma cleaner for 30 s at 100 W. After treatment, the substrates were washed with 70% ethanol and air dried in a sterile cell culture hood for 30 min. To functionalize the substrates for neuronal culture, 0.05 mg/mL poly-D-lysine (PDL) (BD Biosciences, Franklin Lakes, NJ, USA) and 0.1 mg/mL laminin (Sigma Aldrich, St. Louis, MO, USA) were placed on the substrates and incubated for at least 1 h at 37°C. Prior to seeding, the PDL/laminin solution was removed and substrates were washed with HBSS. A 5–10 μL DRG cell suspension in DRG cell culture media obtained from 2.5–3-mo-old Sprague–Dawley rat ganglia was seeded into the restricted space wells. To regulate evaporation, about 200 mL of HBSS was added to the dish outside of the wells. The seeded cells were cultured at 37°C with 5% CO2 for 7–10 d before stimulations. DRG cell culture media was added every 2–3 d, or more frequently if levels appeared low due to evaporation.
Release Stimulation and Analysis
Regional Whole DRG Release Sampling
Left and right L4 DRGs, with the roots and spinal nerve attached, were collected from 2.5–3-mo-old Sprague–Dawley outbred rats after euthanasia by decapitation. Immediately after decapitation, cold mGBSS (120 mL, 4°C) was injected into the subepidermal areas neighboring the SNs and their corresponding DRG. Rat trunks were maintained on ice during the surgical dissection procedure. The collected semi-intact preparations of DRGs with attached nerves were sequentially incubated in ice cold Na+-free artificial cerebrospinal fluid (aCSF-1) composed of (in mM): KCl 2.5, NaH2PO4 1.25, CaCl2 0.5, MgCl2 3.5, NaHCO3 26, glucose 10 and sucrose 233, and HEPES 15 for approximately 3 h, followed by ice cold aCSF-1 supplemented with 63 mM NaCl for 20 min. This series of incubations reduced analyte release induced by surgical isolation. Next, the DRGs and nerves were moved into standard oxygenated artificial cerebrospinal fluid (aCSF-2) (in mM): NaCl 115, KCl 2.5, NaH2PO4 1.25, CaCl2 0.5, MgCl2 3.5, NaHCO3 26, HEPES 15, and incubated at 36°C for 20–30 min. The structures were placed into an incubation chamber with multiple SPE probes [C18 ZipTip pipette tips (Millipore, Billerica, MA, USA)] mounted near specific areas of the peripheral sensory-motor system (Figure S1). Outward flow of the aCSF-2 through the pipette tips was maintained during the process. Release from the semi-intact preparation was stimulated by replacing aCSF-2 with a high K+ aCSF-3 (100 mM KCl, with a corresponding reduction in Na+). Solution flow inside the pipette tips was reversed to inflow, and incubation and analyte collection continued for 20 min at 36°C. Four experiments were performed using four animals and eight L4 ganglia. Analytes retained on the pipette tips were eluted onto a metal MALDI sample plate and mixed with 2.5 μL of 2,5-dihydroxybenzoic acid (DHB) MALDI matrix solution [10 mg/mL DHB in acetonitrile (ACN)/water (50:50)].
Sampling of Release from DRG Cell Cultures
To chemically stimulate DRG cell release, a high K+ DRG cell culture media bath (60 mM K+) was used on the restricted-space cultures 7–10 d after seeding. Details on the fluid exchanges used to stimulate release were as follows: first, a control sample consisting of DRG cell culture media in which the cells had been incubating was collected. At this point, fresh media was placed on the cells and they were incubated for 30 min. That media was then removed, creating a pre-stimulation sample. Next, the high K+ DRG cell culture media was added to the cells and removed after 30 min, creating a stimulation sample. A final bath of DRG cell culture media was added to the cells and collected as the post-stimulation sample after 30 min.
Depending on the experiment and numbers of wells with viable cultures, samples from 1–10 culture wells per dish were combined by sample type before further processing for MALDI MS and/or LC-ESI-FTMS measurements. An initial exploratory release experiment used cells cultured from a single rat; samples obtained from individual cell culture wells were subjected to MALDI analysis. Another experiment, also with a single animal, pooled samples from multiple wells for both MALDI and FTMS analysis. Next, we performed six separate cell culture experiments (using six different animals, with samples collected on six separate days). We collected control, pre-stimulation, stimulation, and post-stimulation samples from each viable well (for a total of 143 wells containing viable cultures), and then combined the samples from individual wells together by dish (18 dishes total). For the combined samples, the analytes were desalted and concentrated using SPE probes [C18 ZipTip pipette tips (Millipore)]. The bound sample was washed with 5% methanol/0.1% TFA in deionized water and eluted, first with 50% ACN/0.1% TFA and then with 75% ACN/0.1% TFA solutions. The eluted samples were mixed 1:1 with the DHB MALDI matrix [10 mg/mL DHB in ACN/water (50:50)] on a metal sample plate. Mass spectrometric profiling was performed using an UltrafleXtreme MALDI-TOF MS workstation (Bruker Daltonics, Billerica, MA, USA) in the reflectron mode operating at positive polarity. Individual mass spectra were analyzed with flexAnalysis (ver. 3.3, Bruker Daltonics). External mass calibration was performed using peptide calibration standard II (Bruker Daltonics). Differential comparisons between multiple experiments were done using the ClinProTools (Bruker Daltonics) software. In addition to the MALDI MS measurements, release samples were analyzed with LC-ESI-FTMS; for this experiment, the analytes collected from 34 culture wells from several of the experiments were preconcentrated and desalted using SPE, pooled, and then subjected to LC-ESI-FTMS (as described below).
Peptide Identification in Biological Samples
Six groups of DRGs and associated nerve tissues were subjected to peptide extraction and analyzed with LC-ESI-FTMS. To extract the peptides, DRGs were immersed in ice-cold acidified acetone (acetone/water/HCl, v/v/v, 40/6/1) immediately after isolation from the animals until all dissections were finished. For groups 1 and 2, the collected DRGs were transferred into 12 mM of ice cold HCl, homogenized, and placed in an ice bath for 1 h. Supernatant was collected and combined with the acidified acetone mentioned above after centrifugation at 20,000 × g for 15 min. For groups 3–6, the DRGs were placed into an 80°C water bath for 10 min to stop enzymatic protein degradation. The heat-denatured DRGs were homogenized, placed in an ice bath for 1 h, and centrifuged as described above. The supernatant was saved and tissue pellets were resuspended in 600 μL of 0.25% acetic acid solution and placed on ice for a 1 h peptide extraction. After centrifugation, the saved supernatant was combined with the acidified acetone. The combined peptide extracts from all experiments were cleaned using C18 spin columns (Thermo Fisher Scientific, Waltham, MA, USA), dried down with a Speedvac concentrator, and reconstituted in H2O/ACN (95:5).
Each extracted peptide sample was separated on a nanocapillary column (10 cm × 75 μm inner diameter) containing ProteoPep II media (C18, 300 Å, 5 μm, New Objective, Inc., Woburn, MA, USA) using an Eksigent nanoLC 1D Plus system (SCIEX, Framingham, MA, USA) and analyzed with an 11 Tesla FT mass spectrometer (LTQ-FT Ultra, Thermo Fisher Scientific). Separation conditions were as follows: (1) buffer A, 95% water, 4.8% ACN, and 0.2% formic acid; (2) buffer B, 95% ACN, 4.8% water, and 0.2% formic acid; (3) gradient conditions were: 0–80 min, 0%–30% B; 80–105 min, 30%–45% B; 105–120 min, 45%–60% B; 120–125 min, 60%–85%; 125–130 min, 85%–85%; 130–145 min, 85%–0%; (4) the operating flow rate was 300 nL/min.
Data acquisition on the LTQ-FT mass spectrometer consisted of a full scan event (m/z 300–2000 at 50 K resolving power) and data-dependent collision-induced dissociation FTMS/MS scans of the five most abundant peaks from the previous full scans. MS/MS settings were as follows: isolation width = m/z 10; minimum signal threshold = 5000 counts; normalized collision energy = 35%; activation Q = 0.25; activation time = 50 ms. The RAW files were searched against a lab-built rat proteome database using PEAKS software (ver. 7.0, Bioinformatics Solutions Inc., ON, Canada) and ProSightPC (Thermo Fisher Scientific). For database search with PEAKS, 15 ppm and 0.1 Da mass tolerances were used for MS and MS/MS modes, respectively. Identified peptides with score (−logP > 15) were kept. For ProSightPC, 81.1 Da and 10 ppm mass tolerance were used for MS and MS/MS modes. Identified peptides with P value < 10−4 were kept. A large mass tolerance was used here because of the ProsightPC biomarker search mode, which involves matching an observed mass to the theoretical masses of possible subsequences in the protein database, and then comparing calculated fragments of those subsequences to observed fragments. A large mass tolerance window allows peptides with post-translational modifications (PTMs) to be matched to the subsequences. The value of 81.1 Da was used here because it can cover the mass shifts related to most common PTMs, including amidation, oxidation, and phosphorylation.
Calculation of Peptide Signal Intensity Fold Changes across Cell Culture Stimulations
Comparisons of the peptide profiles were performed on the MALDI MS data from the six cell culture experiments in their original format using averaged peak statistics functions (Kruskal-Wallis test for not normally distributed peak signal intensities as determined by the Anderson-Darling normality test) in ClinProTools (ver. 3.0, Bruker Daltonics). Representative spectra acquired from each culture dish from each of the six experiments were combined into control, pre-stimulation, stimulation, and post-stimulation sample data sets and imported into ClinProTools as separate classes. Each set consisted of 18 spectra. The spectra preparation settings were: a resolution of 800; top hat baseline subtraction of 10% minimal baseline width; two cycles of Savitzky-Golay smoothing with a width of 2.0 m/z; and a data reduction factor of 3. Peak picking was done on the class average spectrum with a signal-to-noise threshold of 3.00. The Benjamini-Hochberg procedure was automatically applied to correct for the multiple testing hypothesis issue commonly associated with MS data. Peak statistics were calculated by ClinProTools after manual removal of signals originating from the physiological media, and addition of signals that were not automatically picked. Peak statistic tables were generated and manually analyzed for trends in the change of average peak intensity over the four class types.
Results and Discussion
Peptidomics of the DRG Tissues
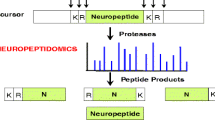
Our first goal was to characterize the endogenous peptides present within the DRG and nerves, and to determine those that were secreted. Accordingly, our initial studies involved characterization of the peptides present within the DRG. Unlike standard bottom up proteomics experiments, we did not digest the proteins and characterize the resulting peptides but, instead, worked with the peptides naturally present within the samples. Here, six groups of DRG tissues were extracted and analyzed with LC-ESI-FTMS, with 2290 peptides from 296 proteins identified after de novo sequencing and database search using the PEAKS software (Table S1). The database search was performed again using a different search tool, ProsightPC (Thermo Fisher Scientific). The ProsightPC search identified 1650 peptides, 434 of which were unique peptide identifications to the total readout (Table S2). In summary, 1216 peptides were identified by both PEAKS and ProsightPC; 1074 peptides were exclusively identified by PEAKS and 434 by ProsightPC. Two and four groups of DRG tissues were processed with the two different peptide extraction methods described above. As a result, 145 proteins were observed with both extraction procedures, and 84 and 67 proteins were unique for just one of the extraction procedures, respectively. Our results suggest that peptide and protein coverage is increased by using different sample processing approaches. Identified peptides were categorized into functional classes, as shown in Figure 1. The neuronal category represents 40% of all of the peptides detected and consists of peptides related specifically to neuron structure or processes involved in neuron formation, function, and maintenance. Among them, several peptides originating from proteins involved in neurotransmission and implicated in neurodegenerative diseases were identified, such as complexin-1 [41] and gamma-synuclein [42], as well as those involved in axon growth and myelination, including vimentin [43] and periaxin [44].
Endogenous cell–cell signaling peptides act as neurotransmitters and neuromodulators. Among the results from six experiments, 18 neuropeptides derived from five prohormones were detected. Figure 2 shows examples of MS/MS spectra of neuropeptides of various lengths, including LVV-hemophin-7, (mass 1323.10 Da), calcitonin gene-related peptide I 19–37 (CGRP-1 19–37, mass 1921.95 Da), and secretogranin-2 derived peptide (mass 3679.80 Da). Another prohormone described most commonly in the DRG is protachykinin-1. Short forms of neuropeptide K cleaved at dibasic sites and a C-flanking peptide were also detected.
Representative MS/MS sequencing of peptides obtained from DRG tissue and release experiments. (a) MS/MS fragmentation of LVV-hemorphin (m/z 662.86, 2+); (b) MS/MS fragmentation of CGRP-1 19–37 (m/z 961.98, 2+); (c) MS/MS fragmentation of secretogranin-2 derived peptide (m/z 1217.61, 3+). The amino acid sequence of each peptide is given above the spectrum
The presence of CGRP-1 and its mRNA in rat DRGs has been previously reported in immunohistochemistry and in situ hybridization studies [45, 46]; CGRP-1 has been shown to be essential for pain signaling and increases innervation in DRGs during inflammation [46]. Here, full length CGRP-1 (1–37) was detected and confirmed with MS/MS. In addition, three truncated forms of CGRP-1 (1–37) were identified from the DRG tissues, including CGRP-1 1–17, 18–37, and 19–37. The cleavage site had one arginine, a monobasic site that is often involved in neuropeptide enzymatic processing. Saghatelian and coworkers [47] discovered that the CGRP-1 1–17, 18–37, and 19–37 are produced enzymatically by peptidases. Therefore, those short forms identified in this work are likely endogenous signaling molecules and not products of degradation of CGRP-1 during sample preparation.
The majority of bioactive peptides are generated from larger precursor proteins. However, in recent years it has been shown that bioactive peptides can also be generated from the cleavage of cytosolic proteins [48]. As a notable example, hemoglobin can be processed to yield multiple bioactive peptides, including the hemorphins [49]. Hemoglobin-derived peptides have been found in a number of cell types within the body, from lens and alveolar cells to macrophages, and have also been found within neurons and oligodendrocytes, as well as in the sciatic nerve [49]. We observed several hemoglobin-derived peptides within the tissue samples, including N-terminus extended LVV-hemorphin-3, lengthened forms of neokyotorphin, and LVV-hemorphin-7. Hemorphins are a group of endogenous opioid peptides derived from the β-chain of hemoglobin. They work as signaling molecules by binding to opioid receptors and play roles in pain and inflammation [50, 51]. LVV-H7 attenuates hyperalgesia effects at the spinal level. Perhaps the hemorphins detected in these systems participate in the regulation of hyperalgesia given the projection of DRGs toward the spinal cord.
Peptide Profiling of Release
Our next goal was to determine what molecules are released from the DRG and associated nerves, as well as from the cells within the DRG. We used two distinct means to probe release: a semi-intact preparation that consisted of the DRG and nerves, and a separate set of experiments using a miniaturized microfabricated chamber to allow us to culture isolated DRG cells, exchange the culturing fluid to stimulate release, and then efficiently collect releasate for characterization.
Regional Release from DRG and Associated Nerves
SPE analyte collection was performed simultaneously at four anatomically defined regions of the mammalian sensory-motor system: the DR, VR, DRG, and SN (Figure S1). After stimulating release with elevated K+ media and characterizing the releasates using MALDI MS, more than 100 compounds were observed in the releasates, in both metabolite and peptide mass ranges (Figure S2, Table S4). Although MALDI MS profiling of release from biological structures provides the opportunity for multiplexed, semiquantitative, spatially, and temporally resolved assessment of the chemical composition of release, identifying the compounds using MALDI MS alone can be challenging. In this work, we mass-matched the signals observed in releasates with the identities of the compounds characterized using LC-FTMS from similar tissues. Our mass calibration and analyte identity assignment approaches are outlined in the Supporting Information, including a table of calibration masses (Table S3) and the resulting assignments (Table S4).
A number of peptides known to be released from nervous tissue were observed. Thymosin beta-4 was detected, which is consistent with the results of prior release experiments from a larger variety of tissues [30, 52, 53]. In fact, the ubiquitous nature of thymosin beta 4 in release experiments suggests that it can be used as a marker for effective release stimulation and efficient sample collection. The metabolite and peptide profiles of the releasates we observed from different regions of the mammalian sensory-motor system were similar. Further, since all regions had neuronal termini and only the DRG had the neuronal cell bodies, these results suggest that nerve terminal- and glia-related compounds dominate the profiles (Figure S2). Our previous MS imaging study demonstrated considerable chemical heterogeneity in the chemical content between the DRGs, VRs, DRs, and SNs [54], so it is intriguing that we observed similar compounds released from these regions, as we expect that the content and release profiles would be correlated. In agreement with the observations of similar release profiles for all regions, principal component analysis of the corresponding data sets did not allow us to separate the analyte profiles of release from these regions. Our inability to distinguish the release profiles from these distinct regions may be explained by the dominant release of peptides originating from neurofilament L and M proteins, myelin-related proteins, and vimentin. Such highly abundant peptides may be obscuring differences in release of much lower concentration peptides.
Release from Entire DRG and Cell Culture
The experiments described above used semi-intact preparations, and releasates were collected by placing collection pipettes at specific, spatially defined locations. In the next set of experiments, we investigated release from isolated DRGs and groups of cultured DRG cells within restricted-volume culture wells. The collection system used a flow-through design, and either culture media or stimulation media (with elevated K+ added to cause secretion) was flowed through the chamber for the designated time period. First, we optimized the collection procedure by measuring release from the entire DRG within the device, and ensuring that the ganglion released enough material for characterization. Comparisons between release from a ganglion and cultured cells may yield information about the peptide differences observed from in vitro versus in vivo measurements. DRGs contain more intact connections between cells and, thus, should more closely mimic the semi-intact preparation used above and the in vivo environment (Figure S3).
Next, pre- and post-stimulation studies from isolated DRGs were performed with DRG cell culture media additions. Elevated K+ (60 mM) solution was added for two exposure periods: 15 min and 30 min. Signals not observed in the pre-stimulation appeared following stimulation, and the intensities of a number of signals increased in the mass spectra of the stimulation samples versus mass spectra of the pre-control samples. As one example, a signal corresponding to neurofilament light polypeptide became detectable in the stimulation sample. Finally, intensities of several signals were found to change in the post-control samples, including those that correspond to peptides originating from neurofilament medium polypeptide.
Following isolated ganglion stimulation, a low-density DRG cell culture containing ~20 neurons within a 1.5 mm PDMS well was stimulated with elevated K+ solution for 30 min. Peptides were characterized with MALDI-TOF MS. A series of mass spectra in the peptide region are shown in Figure 3a. Similar to ganglion stimulation data, a number of peaks increased in intensity following the application of elevated K+, suggesting a potentiation of release in response to stimulation. The peptides from the cultured DRG cell release experiments were also analyzed with LC-ESI-FTMS. The identification results show the presence of 28 peptides from 11 proteins. Peptides having known functions related to cell to cell signaling, including LVV-hemorphin-7, VV-hemorphin-7, and their C-terminus extended forms, were detected.
Representative mass spectra acquired with MALDI-TOF MS showing relative changes in peak profiles detected within the peptide mass region upon stimulating release from DRG clusters and cultured cells. (a) Series of mass spectra showing signals within the peptide molecular mass region following control and stimulation of an entire DRG. Peaks that increased in intensity or newly appeared are highlighted with an asterisk. (b) Series of mass spectra of release collected from ~20 DRG neurons within a 1.5 mm PDMS well. Pre- and post-controls were performed with DRG extracellular media; 60 mM elevated extracellular K+ was added to induce peptide release
Interestingly, when compared together (Figure 3b), few signals overlap between the ganglion and dispersed cell culture stimulation data. An explanation for this is that the ganglion data contains signals that were released or secreted in abundance within the tissue, obscuring lower-abundance peptides. By having isolated a much smaller subset of cells from the cluster and culturing them, we are able to observe release from fewer cells so that cell-to-cell chemical heterogeneity may have become more obvious.
These results led us to move forward with multiple high-K+ stimulations using DRG cell cultures. In total, extracellular media was sampled from cultures involving DRGs from six rats on six different days, resulting in the preparation of control, pre-stimulation, stimulation, and post-stimulation samples from cells cultured in multiple wells within 18 different cell culture dishes. Analysis of these samples using MALDI-TOF MS revealed 41 signals of interest. Compared against the LC-ESI-FTMS data of the inventory of characterized compounds from the DRGs, the molecular masses matched 26 peptides from 19 different proteins. These proteins include those involved in axonogenesis and neurite extension such as neurofilament heavy, medium, and light polypeptides [55], vimentin [56], and those expressed in peripheral neurons and Schwann cells, such as high mobility group protein B1 [57], gamma synuclein [42, 58], and peptidyl-prolyl cis-trans isomerase FKBP1A [59]. In addition, proteins integral to the function of nervous tissue, such as periaxin and myelin protein P0 [60, 61], were detected within the samples. Interestingly, one potential hemorphin peptide, L.LVVYPWTQRYFDSF.G, was detected in the MALDI-TOF MS analysis of samples from the cell stimulation experiment.
The average intensity of the peptides in each condition class was used to determine changes across classes (control, pre-stimulation, stimulation, post-stimulation, and a DRG cell culture media blank). Eight signals showed an upward trend in their relative intensity between pre-stimulation and stimulation or post-stimulation samples (Table 1). Of the signals showing an intensity increase, three were identified by mass matching to the LC-ESI-FTMS data. Mast cell protease 1-derived peptide signal increased 1.4- and 1.6-fold from pre-stimulation to stimulation, and pre-stimulation to post-stimulation, respectively, whereas myelin protein P0-derived peptide increased 1.8-fold for both conditions. The signal of a peptide derived from histone H2B had a 1.4-fold increase from pre-stimulation to post-stimulation. In addition, 12 signals showed intensity increases between the control and pre-stimulation samples. These include the previously mentioned histone H2B, myelin protein P0, and mast cell protease 1-derived peptides, as well as nine unassigned signals. We do not believe these increases between the pre-stimulation and control are related to K+-stimulated release but rather, they may be related to the action of removing the culture media and/or the addition of fresh media, both of which could have induced mechanical and/or chemical stimulation.
Peptidomic Analysis of Peptides Released from DRG Cell Culture
In order to aid in identifying the compounds released from these cells, we also performed an LC-ESI-FTMS study of the released peptides. Releasate solutions collected after the stimulation experiments were concentrated and subjected to LC-FTMS analysis as described above. Obtaining enough analyte for an LC-ESI-FTMS-based peptidomic study of released materials was challenging. We expected that high femtomoles to low picomoles of material would be needed for efficient MS/MS peptide sequencing. However, by increasing cell density, performing sample collection in small wells, and combining samples, a number of peptides were identified in the release samples.
As before, we detected several hemorphins, now confidently assigned. LVV-hemorphin-7 and VV-hemorphin 7, as well as longer fragments of both peptides, were observed. These observed hemoglobin-derived peptides act on opioid receptors and most have functions relating to antinociception. Thus, from a functional perspective, their presence in release makes sense, although the mechanisms of their synthesis and processing within these cells are not understood. LVV-hemorphin-7 has also been shown to regulate blood pressure and play a role in learning and memory, as well as cholinergic transmission [49]. Further study into the biosynthetic pathways and release of hemoglobin-derived peptides within DRG tissue and cells is important. Besides hemoglobin-derived peptides, several other classes of peptides were also identified, including thymosin-beta 4, neurofibromin, and myelin basic protein. Neuferricin was also detected and is involved in neuronal apoptosis and plays an important role in neurotrophin signaling during neuronal development [62].
Conclusions
The DRG and adjacent nerves play an essential role in transmitting sensory information from the periphery to the central nervous system, and are major targets in investigations of mechanisms of pain, mechanical injury, and regeneration. We studied this structurally well-defined system across levels, from isolated cells to whole tissue, and releasates, to investigate the peripheral sensory-motor system’s peptidome and secretome. This broad range of sample types and the cellular and chemical heterogeneity of the samples created measurement challenges that we partially addressed using a range of analytical protocols and platforms.
We used a variety of sample preparation and conditioning steps, and multiple ionization and mass analyzers during this work. The individual workflows and instrumentation have different figures of merit and were selected according to the requirements of the experimental objective. To identify peptides in a complex cellular sample, we used LC-ESI-FTMS, whereas to characterize many samples containing low levels of peptides from volume-limited and concentration-limited release samples, we used direct MALDI-TOF MS. The advantages of LC-ESI-FTMS for obtaining comprehensive peptide profiles are well established, and direct MALDI MS provides an opportunity to profile large numbers of individual samples without purification in a short period of time. This combined work flow has been shown to be effective in a number of peptidomics experiments [16, 27, 63, 64], and was particularly effective in these experiments. As a result, we generated more complete peptidome and secretome datasets for the peripheral sensory-motor system, providing a better understanding of its chemical composition.
The inventory of peptides and proteins we report here enables a range of follow-up studies using small numbers of cells that form defined networks within engineered structures. This investigation of the peptidomes and secretomes of the peripheral sensory-motor system has revealed new candidates for peptide neuromodulators, hormones, and trophic factors that may have important roles in local and distal intercellular communication. Characterizing the spatiotemporal and chemical parameters of cellular release provides unique functional insights in fundamental and specific mechanisms of peripheral sensory-motor system function.
References
Altelaar, A.F.M., Van Minnen, J., Jiménez, C.R., Heeren, R.M.A., Piersma, S.R.: Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal. Chem. 77, 735–741 (2005)
Priestley, J.V.: In: Squire, L.R. (ed.) Encyclopedia of Neuroscience, pp. 935–943. Academic Press, Oxford (2009)
Sugiura, Y., Zaima, N., Setou, M., Ito, S., Yao, I.: Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1851–1861 (2012)
Strand, F.L., Rose, K.J., Zuccarelli, L.A., Kume, J., Alves, S.E., Antonawich, F.J., Garrett, L.Y.: Neuropeptide hormones as neurotrophic factors. Physiol. Rev. 71, 1017–1046 (1991)
Monroe, E.B., Jurchen, J.C., Lee, J., Rubakhin, S.S., Sweedler, J.V.: Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 127, 12152–12153 (2005)
Su, J., Al-Tamimi, M., Garnier, G.: Engineering paper as a substrate for blood typing bio-diagnostics. Cellulose 19, 1749–1758 (2012)
Strand, F.: In Peptide Transport and Delivery into the Central Nervous System, (ed.) L. Prokai, K. Prokai-Tatrai. Birkhäuser Basel, vol. 61, pp 1–37 (2003)
Patti, G.J., Yanes, O., Shriver, L.P., Courade, J.-P., Tautenhahn, R., Manchester, M., Siuzdak, G.: Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nat. Chem. Biol. 8, 232–234 (2012)
Ding, H., Goldberg, M., Raymer, J., Holmes, J., Stanko, J., Chaney, S.: Determination of platinum in rat dorsal root ganglion using ICP-MS. Biol. Trace Elem. Res. 67, 1–11 (1999)
Cheng, H., Jiang, X., Han, X.: Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: a shotgun lipidomics study. J. Neurochem. 101, 57–76 (2007)
Wong, A., Sagar, D.R., Ortori, C.A., Kendall, D.A., Chapman, V., Barrett, D.A.: Simultaneous tissue profiling of eicosanoid and endocannabinoid lipid families in a rat model of osteoarthritis. J. Lipid Res. 55, 1902–1913 (2014)
Zhang, X.-J., Leung, F.P., Hsiao, W.W.L., Tan, S., Li, S., Xu, H.-X., Sung, J.J.Y., Bian, Z.-X.: Proteome profiling of spinal cord and dorsal root ganglia in rats with trinitrobenzene sulfonic acid-induced colitis. World J. Gastroenterol. 18, 2914–2928 (2012)
Komori, N., Takemori, N., Kim, H.K., Singh, A., Hwang, S.-H., Foreman, R.D., Chung, K., Chung, J.M., Matsumoto, H.: Proteomics study of neuropathic and nonneuropathic dorsal root ganglia: altered protein regulation following segmental spinal nerve ligation injury. Physiol. Genomics 29, 215–230 (2007)
Riedl, M.S., Braun, P.D., Kitto, K.F., Roiko, S.A., Anderson, L.B., Honda, C.N., Fairbanks, C.A., Vulchanova, L.: Proteomic analysis uncovers novel actions of the neurosecretory protein VGF in nociceptive processing. J. Neurosci. 29, 13377–13388 (2009)
Perry, M., Li, Q., Kennedy, R.T.: Review of recent advances in analytical techniques for the determination of neurotransmitters. Anal. Chim. Acta 653, 1–22 (2009)
Li, L., Sweedler, J.V.: Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. (Palo Alto, Calif) 1, 451–483 (2008)
Fricker, L.D., Lim, J., Pan, H., Che, F.-Y.: Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom. Rev. 25, 327–344 (2006)
Ebner, K., Rjabokon, A., Pape, H.-C., Singewald, N.: Increased in vivo release of neuropeptide S in the amygdala of freely moving rats after local depolarisation and emotional stress. Amino Acids 41, 991–996 (2011)
Malcangio, M., Ramer, M.S., Jones, M.G., McMahon, S.B.: Abnormal Substance P release from the spinal cord following injury to primary sensory neurons. Eur. J. Neurosci. 12, 397–399 (2000)
Mitsuma, T., Hirooka, Y., Mori, Y., Kayama, M., Adachi, K., Rhue, N., Ping, J., Nogimori, T.: Effects of orexin A on thyrotropin-releasing hormone and thyrotropin secretion in rats. Horm. Metab. Res. 31, 606–609 (1999)
Li, L.J., Kelley, W.P., Billimoria, C.P., Christie, A.E., Pulver, S.R., Sweedler, J.V., Marder, E.: Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J. Neurochem. 87, 642–656 (2003)
Rubakhin, S.S., Page, J.S., Monroe, B.R., Sweedler, J.V.: Analysis of cellular release using capillary electrophoresis and matrix assisted laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis 22, 3752–3758 (2001)
Svensson, M., Skold, K., Nilsson, A., Falth, M., Svenningsson, P., Andren, P.E.: Neuropeptidomics: expanding proteomics downwards. Biochem. Soc. Trans. 35, 588–593 (2007)
Hanrieder, J., Malmberg, P., Ewing, A.G.: Spatial neuroproteomics using imaging mass spectrometry. Biochim. Biophys. Acta 1854, 718–731 (2015)
Norris, J.L., Caprioli, R.M.: Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342 (2013)
Lanni, E.J., Rubakhin, S.S., Sweedler, J.V.: Mass spectrometry imaging and profiling of single cells. J. Proteome. 75, 5036–5051 (2012)
Romanova, E.V., Aerts, J.T., Croushore, C.A., Sweedler, J.V.: Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology 39, 50–64 (2014)
Chen, R., Ouyang, C., Xiao, M., Li, L.: In situ identification and mapping of neuropeptides from the stomatogastric nervous system of Cancer borealis. Rapid Commun. Mass Spectrom. 28, 2437–2444 (2014)
Iannacone, J.M., Ren, S., Hatcher, N.G., Sweedler, J.V.: Collecting peptide release from the brain using porous polymer monolith-based solid phase extraction capillaries. Anal. Chem. 81, 5433–5438 (2009)
Hatcher, N.G., Atkins, N., Annangudi, S.P., Forbes, A.J., Kelleher, N.L., Gillette, M.U., Sweedler, J.V.: Mass spectrometry-based discovery of circadian peptides. Proc. Natl. Acad. Sci. U. S. A. 105, 12527–12532 (2008)
Fan, Y., Rubakhin, S.S., Sweedler, J.V.: Collection of peptides released from single neurons with particle-embedded monolithic capillaries followed by detection with matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 83, 9557–9563 (2011)
Lee, C.Y., Romanova, E.V., Sweedler, J.V.: Laminar stream of detergents for subcellular neurite damage in a microfluidic device: a simple tool for the study of neuroregeneration. J. Neural Eng. 10, 036020 (2013)
Wang, J., Ren, L., Li, L., Liu, W., Zhou, J., Yu, W., Tong, D., Chen, S.: Microfluidics: a new cosset for neurobiology. Lab Chip 9, 644–652 (2009)
Croushore, C.A., Supharoek, S.-A., Lee, C.Y., Jakmunee, J., Sweedler, J.V.: Microfluidic device for the selective chemical stimulation of neurons and characterization of peptide release with mass spectrometry. Anal. Chem. 84, 9446–9452 (2012)
Hou, X., Xie, F., Sweedler, J.: Relative quantitation of neuropeptides over a thousand-fold concentration range. J. Am. Soc. Mass Spectrom. 23, 2083–2093 (2012)
Romanova, E.V., Lee, J.E., Kelleher, N.L., Sweedler, J.V., Gulley, J.M.: Comparative peptidomics analysis of neural adaptations in rats repeatedly exposed to amphetamine. J. Neurochem. 123, 276–287 (2012)
Lee, J.E., Atkins, N., Hatcher, N.G., Zamdborg, L., Gillette, M.U., Sweedler, J.V., Kelleher, N.L.: Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry. Mol. Cell. Proteomics 9, 285–297 (2010)
Millet, L.J., Stewart, M.E., Sweedler, J.V., Nuzzo, R.G., Gillette, M.U.: Microfluidic devices for culturing primary mammalian neurons at low densities. Lab Chip 7, 987–994 (2007)
Croushore, C.A., Sweedler, J.V.: Microfluidic systems for studying neurotransmitters and neurotransmission. Lab Chip 13, 1666–1676 (2013)
Wei, H., Li, H., Gao, D., Lin, J.-M.: Multi-channel microfluidic devices combined with electrospray ionization quadrupole time-of-flight mass spectrometry applied to the monitoring of glutamate release from neuronal cells. Analyst 135, 2043–2050 (2010)
Malsam, J., Kreye, S., Sollner, T.H.: Membrane fusion: SNAREs and regulation. Cell. Mol. Life Sci. 65, 2814–2832 (2008)
George, J.: The synucleins. Genome Biol. 3, reviews3002.3001 - reviews3002.3006 (2001)
Asch, W.S., Schechter, N.: Plasticin, a type III neuronal intermediate filament protein, assembles as an obligate heteropolymer: implications for axonal flexibility. J. Neurochem. 75, 1475–1486 (2000)
Han, H.J., Myllykoski, M., Ruskamo, S., Wang, C.Z., Kursula, P.: Myelin-specific proteins: a structurally diverse group of membrane-interacting molecules. BioFactors 39, 233–241 (2013)
Gibson, S.J., Polak, J.M., Giaid, A., Hamid, Q.A., Kar, S., Jones, P.M., Denny, P., Legon, S., Amara, S.G., Craig, R.K., Bloom, S.R., Penketh, R.J.A., Rodek, C., Ibrahim, N.B.N., Dawson, A.: Calcitonin gene-related peptide messenger RNA is expressed in sensory neurones of the dorsal root ganglia and also in spinal motoneurones in man and rat. Neurosci. Lett. 91, 283–288 (1988)
Xu, P., Van Slambrouck, C., Berti-Mattera, L., Hall, A.K.: Activin induces tactile allodynia and increases calcitonin gene-related peptide after peripheral inflammation. J. Neurosci. 25, 9227–9235 (2005)
Lone, A.M., Kim, Y.-G., Saghatelian, A.: Peptidomics methods for the identification of peptidase-substrate interactions. Curr. Opin. Chem. Biol. 17, 83–89 (2013)
Murillo, L., Piot, J.M., Coitoux, C., Fruitier-Arnaudin, I.: Brain processing of hemorphin-7 peptides in various subcellular fractions from rats. Peptides 27, 3331–3340 (2006)
Gomes, I., Dale, C.S., Casten, K., Geigner, M.A., Gozzo, F.C., Ferro, E.S., Heimann, A.S., Devi, L.A.: Hemoglobin-derived peptides as novel type of bioactive signaling molecules. AAPS J. 12, 658–669 (2010)
Cheng, B.-C., Tao, P.-L., Cheng, Y.-Y., Huang, E.Y.-K.: LVV-hemorphin 7 and angiotensin IV in correlation with antinociception and anti-thermal hyperalgesia in rats. Peptides 36, 9–16 (2012)
Honda, M., Okutsu, H., Matsuura, T., Miyagi, T., Yamamoto, Y., Hazato, T., Ono, H.: Spinorphin, an endogenous inhibitor of enkephalin-degrading enzymes, potentiates leu-enkephalin-induced anti-allodynic and antinociceptive effects in mice. Jpn. J. Pharmacol. 87, 261–267 (2001)
Annangudi, S.P., Luszpak, A.E., Kim, S.H., Ren, S., Hatcher, N.G., Weiler, I.J., Thornley, K.T., Kile, B.M., Wightman, R.M., Greenough, W.T., Sweedler, J.V.: Neuropeptide release is impaired in a mouse model of Fragile X mental retardation syndrome. ACS Chem. Neurosci. 1, 306–314 (2010)
Romanova, E.V., Roth, M.J., Rubakhin, S.S., Jakubowski, J.A., Kelley, W.P., Kirk, M.D., Kelleher, N.L., Sweedler, J.V.: Identification and characterization of homologues of vertebrate β-thymosin in the marine mollusk Aplysia californica. J. Mass Spectrom. 41, 1030–1040 (2006)
Rubakhin, S., Ulanov, A., Sweedler, J.: Mass spectrometry imaging and GC-MS profiling of the mammalian peripheral sensory-motor circuit. J. Am. Soc. Mass Spectrom. 26, 958–966 (2015)
Yuan, A., Rao, M.V., Veeranna, Nixon, R.A.: Neurofilaments at a glance. J. Cell Sci. 125, 3257–3263 (2012)
Rishal, I., Fainzilber, M.: Retrograde signaling in axonal regeneration. Exp. Neurol. 223, 5–10 (2010)
Müller, S., Ronfani, L., Bianchi, M.E.: Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J. Intern. Med. 255, 332–343 (2004)
Vargas, K.J., Makani, S., Davis, T., Westphal, C.H., Castillo, P.E., Chandra, S.S.: Synucleins regulate the kinetics of synaptic vesicle endocytosis. J. Neurosci. 34, 9364–9376 (2014)
Sezen, S., Blackshaw, S., Steiner, J., Burnett, A.: FK506 binding protein 12 is expressed in rat penile innervation and upregulated after cavernous nerve injury. Int. J. Impot. Res. 14, 506–512 (2002)
Gillespie, C.S., Sherman, D.L., Blair, G.E., Brophy, P.J.: Periaxin, a novel protein of myelinating schwann cells with a possible role in axonal ensheathment. Neuron 12, 497–508 (1994)
Martini, R., Schachner, M.: Molecular bases of myelin formation as revealed by investigations on mice deficient in glial cell surface molecules. Glia 19, 298–310 (1997)
Kimura, I., Nakayama, Y., Konishi, M., Kobayashi, T., Mori, M., Ito, M., Hirasawa, A., Tsujimoto, G., Ohta, M., Itoh, N., Fujimoto, M.: Neuferricin, a novel extracellular heme-binding protein, promotes neurogenesis. J. Neurochem. 112, 1156–1167 (2010)
Buchberger, A., Yu, Q., Li, L.: Advances in mass spectrometric tools for probing neuropeptides. Annu. Rev. Anal. Chem. (Palo Alto, Calif) 8, 485–509 (2015)
Romanova, E.V., Sweedler, J.V.: Peptidomics for the discovery and characterization of neuropeptides and hormones. Trends Pharmacol. Sci. 36, 579–586 (2015)
Acknowledgments
The authors congratulate Lingjun Li for her outstanding achievements in the field of mass spectrometry-based peptidomics and for her well-deserved recognition as recipient of the 2014 Biemann Medal. This work was supported by the National Institute of Mental Health under Award no. R21 MH100704A, the National Institute on Drug Abuse under Award no. P30 DA018310, and the National Science Foundation Division of Chemistry under grant Award no. CHE-11-11705 (with co-funding from the Division of Biological Infrastructure). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. Technical support by Xiying Wang is gratefully acknowledged. This study involved collaborative efforts with Anika Jain in Martha Gillette’s laboratory at the University of Illinois at Urbana-Champaign. The method used for DRG isolation and culture was adapted from procedures partially developed by a former laboratory member, Amy Maduram.
Author information
Authors and Affiliations
Corresponding author
Additional information
Emily G. Tillmaand, Ning Yang and Callie A. C. Kindt contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 3220 kb)
Rights and permissions
About this article
Cite this article
Tillmaand, E.G., Yang, N., Kindt, C.A.C. et al. Peptidomics and Secretomics of the Mammalian Peripheral Sensory-Motor System. J. Am. Soc. Mass Spectrom. 26, 2051–2061 (2015). https://doi.org/10.1007/s13361-015-1256-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1256-1