Abstract
PlK1 has a significant role in the development of breast cancer. Thus, silencing of PlK1 gene may arrest the growth of breast cancer. However, the in vivo stability of PlK1 siRNA after injection remains a challenge to target the specific site. The delivery of siPlK1 RNA via viral vector and amine group–terminated dendrimer is associated with immune reaction and cellular cytotoxicity. Thus, in the present study, hyaluronic acid–functionalized and –thiolated polycaprolactone nanoparticles (SH-HPP NPs) were developed for enhancing the targeting capabilities of siRNA towards human breast cancer cells. NPs displayed size in the range of 180–217 nm, and with sustain and pH-dependent release of siRNA up to 120 h. The in vitro treatments with siRNA-containing NPs showed the high number of necrotic cells and the cell cycle arrest at the G2/M phase. The gene expression analysis depicts the decrease of endogenous PLK1 siRNA expression on MCF-7 cells upon PLK1 NPs treatment. In vitro cytotoxicity experiments demonstrated effective anticancer properties against MCF-7. Finally, in vivo results showed that substantial tumor inhibition was achieved with PLK1 siRNA–containing SH-HPP NPs in comparison of the control group. Hence, HPP NPs have enormous potential for the selective delivery of siRNA, i.e., breast cancer cells.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) has become a major health concern globally and is accountable for 12% of all new annual cancer cases worldwide. According to the World Health Organization (WHO), 2.3 million women were diagnosed with BC, and 685,000 deaths reported across the globe in 2020. Various factors, i.e., lifestyle, genetic predisposition, and the previous history of BC in the family, increase the risk of BC in females [1].
Polo-like kinase 1 (PLK1) has a crucial role in cell division and DNA damage. The presence of PLK1 was detected in actively dividing cancerous cells [2]. In the case of breast cancer, the elevated level of PLK1 is responsible for poor clinical outcomes [3]. Inhibition of PLK1 demonstrated the significant cell death of both tumor-initiating and cancerous cells in triple-negative breast cancer [4].
To reduce the expression of target gene protein at a post-transcriptional level, the specific siRNA-mediated selective down-regulation of a complementary mRNA is needed. However, the poor cellular activity of siRNA is a major problem due to its biodegradability [5]. Interestingly, several attempts in the field of oncology have been made for the effective in vivo delivery of siRNA. But, no one is effective for the targeted in vivo delivery of siRNA. Hence, the development of a novel carrier system is required to prevent degradation, prolong circulation time, and target the desired site. The nanocarriers may prevent nuclease-mediated degradation and allow the entry of siRNA into the cytoplasm followed by gene silencing of targeted genes. Thus, nanocarriers could play a crucial role to enhance the delivery of siRNA [6].
However, several nanosystems failed or showed less efficacy in delivering siRNA at the target site. The problems could be related to preparing the required copolymers, the inadequate expression of functional groups, minimum entrapment efficiency, and a decrease in targeting potential with time. Among several biomaterial-based siRNA delivery vehicles, biodegradable and biocompatible materials have gained much attention due to many flexible features [7, 8].
Polycaprolactone (PCL), a biodegradable, biocompatible, and non-toxic polymer, is promising for the development of nanoparticles. However, the hydrophobic characteristic of PCL leads to fast in vivo removal as well as poor aqueous stability of PCL nanoparticles. Hence, surface modification may be considered an excellent strategy to avoid fast in vivo removal [9]. Functionalization of nanocarriers with polyethylene glycol (PEGylation) can modulate the protein corona, increase the circulation time, and improve biodistribution. Also, PEGylation reduces immunogenicity and toxicity [10].
Furthermore, hyaluronic acid (HA) has extensive and critical role in the development of cancer, progression, migration, embryonic development, and regulation of inflammation. Thus, HA owns its potential to target cancer cells due to overexpression of HA receptors in several cancerous tissues, including BC [11]. Hence, HA can be used to increase in cellular uptake of siRNA [12].
In this study, we developed pegylated and HA functionalized PCL NPs for enhanced and site-specific delivery of PLK1 siRNA to tumor with CD44 receptors, and characterization and therapeutic efficacy of developed NPs were evaluated and summarized in this present work.
Experimental section
Materials
Polycaprolactone, hyaluronic acid, and poly (ethylene glycol) diamine were procured from Sigma-Aldrich. PLK1 siRNA (10 µm) was purchased from Santacruz Biotechnology. Pluronic F-68, EDAC.HCl (1-Ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride), and DMSO were supplied by HiMedia Lab, Mumbai, India. Acetone was procured from Qualigen Fine Chemical Ltd, Mumbai, India. Dichloromethane and RNase and DNase-free distilled water were purchased from Fisher Scientific, Mumbai India. Lawesson’s reagent was obtained from Central Drug House. Mumbai, India. All the other chemicals and reagents were used as received.
Methods
Synthesis of hyaluronic acid–coupled and –pegylated polycaprolactone (HPP)
First, hyaluronic acid was conjugated to diamine PEG. In brief, 0.574 g of HA (1.5 × 106 Da, 3.83 × 10−4 mMol) was weighed and dissolved in 4 mL of deionized water to obtain the solution with the help of a magnetic stirrer. Next, 20 mg of EDAC.HCl (191.7 g/mol, 0.10 mMol) was added dropwise to the solution of hyaluronic acid for the activation of the –COOH group present in HA, and allowed to stir for 4 h. Afterward, the diamine PEG (3400 Da, 340mMol) solution in deionized water was added to the EDAC. HCl activated the HA solution and was allowed to stir overnight at room temperature. The pure HA conjugated PEG was obtained by dialyzing the product against 200 mL of deionized water using a dialysis membrane (molecular weight cut-off, 6000 Da) for 48 h at room temperature. In order to obtain the pure product, the water was changed at every 12 h. After dialysis, the product was lyophilized (Labconco, Freezone 2.5, Kansas, Missouri) and kept in an airtight container at room temperature.
For the synthesis of HA-PEG-PCL, 2.0 g of ℇ-caprolactone was solubilized in toluene, then HA-PEG was added to the solution of ℇ-caprolactone and allowed to react in the presence of stannous octoate at 110 ℃ for 5 h. After 5 h, the reaction mixture was allowed to cool at room temperature, followed by precipitation in cold diethyl ether. The precipitate was collected and washed with cold diethyl ether, dried under vacuum, and stored in the desiccator to protect from moisture [8, 11].
Thiolation of HPP
The thiolation of HPP was carried out with Lawesson’s reagent. 1.0 g of HPP was dissolved in DCM, and then 100 mg of Lawesson’s reagent was added to the solution of HPP. The mixture was kept in the microwave (LG 20L solo Microwave Oven MS2043DB) for 10 min at 100 °C. The synthesis was confirmed using the FTIR and 1H-NMR spectroscopic methods.
FTIR and 1H-NMR analyses
The synthesis of HPP and SH-HPP was confirmed using FTIR spectra (Bruker ALPHA-T, Bruker, Germany). The samples were placed directly on the sample plate and scanned against FTIR spectra ranges 400 to 4000 cm−1 range. Similarly, 1H-NMR spectra (JEOL Peabody, MA, USA) were used to confirm the conjugation process for the development of HPP and SH-HPP NPs.
Preparation of nanoparticles formulations
HPP NPs and thiolated HPP (SH-HPP) NPs were prepared as per the earlier described process with slight modifications. Both, HPP and SH-HPP (10 mg), were added in 10 mL of DCM, vortexed for 10 min. The above organic solution was added very slowly to the 20 mL of 0.1% w/v Pluronic F-68 solutions ( DCM: water = 1:2) while stirring at 2000 rpm overnight, and DCM was evaporated with the help of rotary evaporator (Buchi rotavapor R-300). After the complete removal of solvent, the HPP and SH-HPP nanoparticle formulations were centrifuged, washed, lyophilized, and stored at 8 ℃ [11]. After that, the obtained HPP and thiolated HPP NPs were dispersed in 20 mL of RNA-free water, separately. Twenty microliters of PLK1 siRNA was added to the aqueous dispersion of HPP and thiolated HPP (SH-HPP) NPs under constant stirring with the help of a magnetic stirrer.
Particle size, zeta potential, and polydispersity index (PDI)
The z-average diameter, zeta potential, and PDI of siRNA-loaded thiolated and nonthiolated NPs were measured by using SZ-100 Zetasizer (Horiba Scientific, Kyoto, Japan). To ensure the zeta potential of siRNA-loaded thiolated and nonthiolated NPs, the zeta sizer (Malvern, UK) was used. In brief, aliquots of NPs formulations (thiolated and nonthiolated) of 10 mL were made up to 100 mL by adding double-distilled water. The resulting samples were mixed well and kept for 1 min in a polyethylene (non-glass) bottle followed by ultra-sonication. Then, the samples were transferred to a polystyrene cuvette, and analysis was carried out in triplicate.
Morphological assessment
To evaluate the morphological characteristics of developed NPs formulations, atomic force microscopy (AFM) (Alpha 300RA AFM, WITec, Germany) was used. A small drop of NPs solution was employed on a carbon-coated copper grid having 300 mesh, and the grid was air-dried. AFM studies of siRNA-HPP NPs and siRNA-SH-HPP NPs were carried out at different magnifications using glass substrate in AC mode at RT. The extra sample was evacuated by blotching with channel paper.
Percent encapsulation efficiency (% EE)
The percent encapsulated efficiency of PLK1 siRNAs loaded to HPP and SH-HPP NPs was measured using Eq. (1), and UV–VIS spectrophotometer (Thermo Scientific Evolution 160) at 260 nm. Briefly, the formulation was dispersed in double-distilled water and subjected to the ultracentrifuge (Rotor NV 70.1 in Optima L-100 XP Ultracentrifuge, Beckman-Coulter, USA), After, ultracentrifugation of the supernatant was collected and the amount of released siRNA was measured with the help of dual-beam UV–Vis spectrophotometer (UV-1601, Shimadzu, Japan), at 260 nm wavelength [13].
where Csample is the concentration of sample siRNA in formulation and Csupernatant is the concentration of siRNA in the supernatant phase.
In vitro release study
The release behavior of PLK1 siRNA from NPs formulations was assessed by tube dialysis method by using cellulose acetate membrane (MWCO 12,400) at three different pH, i.e., 4.5 (0.01 M acetate buffer), 5.5 (0.01 M acetate buffer), and 7.4 (0.01 M phosphate buffer). The aliquots were withdrawn at predefined intervals (1, 2, 4, 8, 24, 48, 72, 96, and 120 h) and replaced with the same amount of respective buffer solutions. The aliquots were centrifuged at 13,000 rpm for 10 min at 25℃. The supernatant was pipetted out up, and by UV–VIS spectrophotometer (Thermo Scientific Evolution 160 UV–VIS v8.01 Spectrophotometer) at 260 nm to detect the amount of released siRNA [13, 14].
In vitro apoptosis study
FRET technique was used to detect caspase activation. Caspases are cysteine proteases that play a crucial role in apoptosis. Upon activation, caspases slice multiple proteins leading to apoptosis. MCF-7 cells were transfected with Plasmid ECFP-DEVD-EYFP, using Lipofectamine LTX plus reagent as the protocol of the manufacturer. The fluorescent protein ECFP acts as the FRET donor and EYFP as the acceptor, and tetrapeptide sequence DEVD served as the linker. Caspase 3 (Effector), upon activation, identifies the tetrapeptide DEVD and cleaves it, and causes loss of fluorescence in caspase-activated cells [12, 13]. Hence, the loss of fluorescence could be an indication for activation of apoptosis.
Treatment with siRNA-HPP NPs and siRNA-SH-HPP NPs
The stock of dispersion of NPs (5% w/v) was prepared and dilutions were made by using mixture of RPMI1640/DMEM cell media. Then, media containing the NPs was added to the plates having a single layer of cells (5 × 103 cells/well). The plates and wells were labelled according to the concentration of NPs added to respective cells. The plates were incubated for 24 h and images were acquired at the mentioned time points. Cells were imaged using BD pathway™ 435 Bioimager (BD biosciences, USA) using the Attovision™ software. For imaging, the excitation single 438 ± 12 nm wavelength was used, and emissions were collected at two wavelengths, 483 ± 15 nm for ECFP and 542 ± 27 nm for EYFP. Photographs were taken using a 20X dry objective with an NA of 0.75. Cells displaying FRET reduction were considered caspase-activated cells. [15, 16].
Effect of PLK1 siRNA–loaded HPP NPs and SH-HPP NPs on cell cycle
Quantification of DNA content was determined using DNA binding dye and with flow cytometry. As the cell cycle progresses from the G1 phase to the S phase, the DNA content of the cell increases due to the synthesis of DNA, while at the G2 phase, the DNA content of the cell is expected to approximately double that the of G1 phase. This increase in DNA content can be visualized as increased fluorescence more brightly and can be quantified accordingly in a histogram of fluorescence intensity. The cell cycle analysis was carried out using the DNA binding dye Hoechst 33,342. Briefly, the cells were trypsinized and incubated in media containing 5 µg/mL Hoechst 33,342 for 15 min at 37 ℃. The cells were then rinsed with PBS, and doublet was excluded by passing cells through 40 µm cell strainer and the cells were analyzed using flow cytometry [15, 16].
Autophagy study
The MCF-7 LC3 EGFP cells were incubated with two different concentrations of siRNA-containing HPP and SH-HPP NPs formulations (100 and 200 µg/mL) for 24 and 48 h at 37 ℃. The LC3 green emission was collected with FITC filter sets using a Nikon Ti-Eclipse microscope. The images were obtained with EMCCD camera (AndoriXON 897) equipped with NIS element software (Nikon) were used.
In vitro transfection with PLK1 siRNAs and PLK1 siRNA–loaded NPs
The oligofectamine protocol (Invitrogen) was followed to transfect the MCF-7 cells with siRNAs. For the transfection, the cells were grown using 25 × 25 cm2 flasks. Briefly, 1 day previous to transfection, cancer cells were seeded without antibiotics. The cells were treated with a 50 nM concentration of siRNAs plus oligofectamine and siRNA-SH-HPP NPs in Opti-MEM at 37 ℃ for 4 h. Next, the fresh culture medium was added and incubated for another 48 h. Cells were harvested for gene expression (mRNA) and protein expression analysis. The experiment was repeated three times to assure reproducibility of results.
RNA isolation and qRT-PCR
TRIzol method (Invitrogen) was used for the isolation of total RNA. Using an oligo (dT) primer, further synthesis of cDNA was performed by reverse transcription. On the other hand, green real-time PCR amplification of SYBR was done with the CFX real-time PCR detection system (Bio-Rad). Human PLK1 sense: 5′- GGCAACCTTTTCCTGAATGA-3′, antisense: 5′-AATGGACCACACATCCACCT-3′ and GAPDH sense: 5′-AGATCCACAACGGATACATT-3′, antisense: 5′-TCCCTCAAGATTGTCAGCAA-3′ were exploited as primer. With the help of Applied Biosystem Step one, SYBR Premix examination of PLK1 gene expressions was done in “Applied Biosystem Step one” under given PCR conditions. Initially, denaturation at 95 ℃ for 1 min and then denaturation at 95 ℃ for 30 s (approx. 40 cycles), toughening with 60 ℃ for 45 s and extension at 72 ℃ for 45 s, were carried out. GAPDH was used as an internal control; the assay was performed in duplicates with 20 µL reaction. Q-PCR data were quantitatively analyzed by using the 2-ΔΔCt method.
Western blot
In western blot analysis, the treated HMECs cells were washed with phosphate buffer saline and lysed using SDS buffer having 4% SDS, 20% glycerol, 0.12 M, and Tris pH 6.8 (Remi, Mumbai, India) containing protease inhibitor mixture. Analysis of protein concentration was performed then for 10 min, and lysates were boiled. Twelve percent SDS–polyacrylamide gel was used to separate 100 µg of total protein and then placed in Immobilon™-FL PVDF transfer membranes with 25 mmol/L Tris, 192 mmol/L glycines, 20% v/v methanol, and pH 8.3 transfer buffer. Membranes were incubated for 1 h with monoclonal antibodies against PLK1 (1:2000 for PLK1 and 1:5000 for β-actin) in 5% non-fat dry milk, further in 5% non-fat dry milk for 30 min with goat anti-mouse serum (1:2000) and determined with LAS-3000 (Fujifilm), initiated by HRP-substrate luminol reagent, and HRP-substrate peroxide solution (Millipore) (ChemiDoc™ XRS + Imaging System, Bio-Rad, California, USA). PLK1 and β-actin expressions were quantified with the help of western blotting techniques (Bio-Rad gel documentation system). Further quantitative comparison of PLK1 and β-actin expressions was performed after the integration of signal intensities from scanned images [17,18,19,20].
Determination of cell viability
MTT (3‐(4,5‐dimethyl thiazol‐2yl)‐2,5‐diphenyl tetrazolium bromide) assay was carried out to evaluate cytotoxicity of PLK1 siRNA–containing NPs. The 1 × 104 cells per well-seeded plates well plate. Percentage of proliferating cells was ascertained by the numbers of siRNA-treated, NPs treated, and control cells. After 48-h incubation with the NPs formulation, MTT was added to the cells and left for 3 h at 37 °C. The contents of all wells were removed and 100 µL of SDS solution in DMSO was added to dissolve the formazan crystals. The absorbance of each sample was recorded with the help of JBMP-8 microplate reader (Jayagen Biologics, Chennai, India) at 540 nm [21].
In vivo antitumor study
Twenty mice (5-week-old) were selected and housed under specific ventilated pathogen-free micro isolator cages. In a tumor-bearing model of mice, 5 × 106 MDA-MB 231 cells in 0.1 mL of Matrigel (BD Biosciences) were administered via interscapular route to mice. When the average tumor volume reached ~ 150 mm3, mice were divided into three groups: (A) control group (without treatment), (B) positive control groups (treated with Adriamycin (ADR) 2.5 mg/kg once a week for 4 weeks), and (C) third groups, animals were treated with 100 µl NPs formulation (siRNA-SH-HPP NPs). NPs formulation that was suspended in 0.9% NaCl was administered at a dose of 100 µL/mice (20 g bodyweight) once a week for 4 weeks by i.v. injection. Tumor growth and tumor volume were measured using digital Vernier caliper (Simplex bearing & tools, Mumbai MH). Animals were monitored at routine interludes for a duration of approximately 1 month for tumor volume, body weight, and mortality.
Relative tumor volume was determined with the equation (mentioned below)
In the above equation, W1 and W2 represent the smallest and largest diameter in centimeter, respectively.
The antitumor efficacy of formulation was determined against T/C ratio that can be used to represent tumor regression. The percentage treatment/control (T/C) is determined with the help of the following equation [22].
T—mean tumor volume of the treated group with formulation.
RTV represents the mean tumor volume of formulation-treated group after completion of study – mean tumor volume group on initial day of dosing.
C denotes mean tumor volume of control group.
All protocols for animal studies were approved by Institutional Animal Ethics Committee (Registration Number: 65/GO/ReBiBt/S/99/CPCSEA), ACTREC, Tata Memorial Centre, Navi Mumbai.
Statistical analysis
Experiment data are presented as the mean ± standard deviation. Treated groups were examined with standard groups. Results were determined statistically with Student’s T-test. The information was found significant at p < 0.01.
Result and discussion
Nanoparticles synthesis
FTIR and NMR analyses of siRNA-loaded NPs
Synthesis of HPP and SH-HPP was confirmed using FTIR and NMR analyses and depicted in Fig. 1a, b. The formation of HPP conjugate was confirmed by the signal present due to -C = O stretching at 1729 cm−1, the peak at 1628 cm−1 represents -C = O due to –CONH present in hyaluronic acid, and the—and N–H stretching at 3500 cm−1 Fig. 1a(i). In thiolated HPP (SH-HPP), the peak of SH was found at 2400 cm−1, shown in Fig. 1a(ii). The synthesized copolymer of the 1H-NMR spectrum is represented in Fig. 1b(i) and b(ii). All the characteristic peaks of polycaprolactone, PEG, and hyaluronic acid (HA) were found to be present in the 1H-NMR spectra. The PEG peak can be identified at ~ 3.63 ppm in both HPP and SH-HPP. The HA peaks were identified at ~ 2.8 to 3.43 ppm, and ~ 0.8 ppm. However, after thiolation with Lawesson’s reagent, the shift was observed in the HA region, i.e., 3.22 to 3.43, and 3.43 to 3.68 ppm. The shift can be observed due to difference of electronegativity of oxygen and sulfur. Thus, 1H-NMR NMR spectra confirm the synthesis of HPP and SH-HPP.
Particle size and zeta potential measurement
Optimized process for the synthesis demonstrated hydrodynamic sizes of the SH-HPP and HPP NPs of 181 ± 2 nm and 217 ± 1 nm, respectively. The zeta potentials were found to be − 8.44 ± 1 mV and − + 4.81 ± 1 mV, respectively. The size of NPs was also affected by the concentration of polymer, surfactant, and siRNA [8, 20] (please refer to Tables 1 and 2). The drug loading efficiency was also found to be decreased, while increasing the concentration of polymer (refer to Table 3 for the particle size, zeta potential, PDI, and % drug loading (%DL) and also refer to supplementary Figs. S1, S2 and S3a for the DLS analysis).
The value of PDI may vary from 0.1 to 0.5 to 0.7, and PDI with more than 0.7 value indicates broad particle size distribution of the formulation [23].
Morphological assessment
AFM (atomic force microscopy) study indicated that the size and shape of NPs were found to be uniform, spherical without aggregation, and in the nanometer range. The size distribution with a small polydispersity index indicated the uniformity of size. The nanoparticle distribution pattern can be observed with a photomicrograph, which offers NPs distribution at similar heights and in diameter (nm) (refer to supplementary Fig. S3b(i) and b(ii).
Entrapment efficiency and in vitro PLK1 siRNA release
The entrapment efficiency of the PLK1 siRNA in nanoparticles is crucial for effective therapeutic efficacy [24]. The percentage entrapment efficiencies of siRNA-SH-HPP NPs and the HPP NPs were found to be 94.6 ± 2% and 80.1 ± 2%, respectively. The cumulative percentage release of PLK1 siRNA (siRNA) was found to be 73.3 ± 2% and 54.5 ± 1% for PLK1 siRNA–HPP, and 43.7 ± 2% for siRNA–SH-HPP NPs for 120 h. Moreover, the release of PLK1 siRNA was found to be pH-dependent (Fig. S3c, supplementary). The less release of PLK1 siRNA was observed at pH 7.4 in comparison with pH 5.4. The more release occurred at low pH might be due to the dissociation of siRNA from NPs. The pH of the tumor microenvironment is slightly acidic; hence. more release of siRNA could have occurred at the site of tumor growth. Another reason for the high release PLK1 siRNA is that tumor microenvironment is acidic and the breakdown of the amide bond between PEG and HA may occur. Thus, breakdown of cationic delivery system in acidic environment of endosomes after intercellularization may cause release of siRNA inside the tumor cells [25].
Caspase 3 activation
The detection of caspase 3 activations proved to be a good marker for early apoptosis [26]. The enhancement of apoptosis after the treatment with siRNA-loaded NPs for 48 h was observed at two different concentrations of NPs formulation (i.e., 100 and 200 µg/mL), and found to be concentration-dependent. Interestingly, the increase in the apoptotic activity was observed after 24 h of treatment with the low concentration of NPs, i.e., 100 µg/mL PLK1 siRNA–loaded NPs. A remarkable increase in caspase 3 activity (18.5 to 20.2%) was observed after 48 h of treatment with 100 µg/mL of NPs. The assay results can be shown in “Caspase activation imaging” Fig. 2a and flow cytometric analysis for caspase activation was done after NPs treatment of the cells was expressed as “Caspase activation FACS” Fig. 2b [27].
Internalization study
To find the internalization of NPs formulations, cells were incubated for 24 to 48 h. At various fluorescence channels, the treated cells were imaged, and as the fluorescent intensity was increased to high levels, a faint signal in the GFP channel was observed. These images were taken using a fluorescent microscope TIE and RetigaExi Camera (Nikon) [28]. The prepared NPs system concentration was increased as 100 and 200 μg/mL at a time interval of 24 and 48 h. The data shows promising results, which leads to the internalization of drugs present in the NPs system into the cell Fig. 3a.
Cell cycle arrest by flow cytometry analysis
The effect PLK1 siRNA loaded NPs on the G0/G1, S, and G2/M cell population was carried out with the help of flow cytometry. The increased population of cells in G1 was confirmed in a time-dependent fashion. Additionally, the ratio of cells present in G1 phase to the NPs formulation was found higher [29]. In HPP NPs, the G2/M phase cell population after 24 h increased from 1.6 to 6.2%, and after 48 h, it further increased to 19.5%, while in the case of SH-HPP after exposure to NPs, NPs show 22.4% after 24 h and 25.2% after 48 h. Hence, more profound effects of SH-HPP were observed clearly at the G2/M phase. The obtained results indicated that the thiolated NPs formulation containing PLK1 siRNA is more efficacious and demonstrated enhanced potential anticancer property and the findings are similar to previously reported work explained by Namvar [30]. FACS and cell distribution were assessed in each phase of cell cycle for 48 h Fig. 3b (also refer to Table 4).
Autophagy
As per the published studies the role of autophagy in the development of cancer can not be ignored. Hence, the autophagic effect of PLK1 siRNA-loaded NPs against MCF-7, LC3 EGFP was determined. Upon treatment, LC3 had translocated to the cytoplasm, forming punctate aggregates and LC3 fluorescence intensity was also increased which is shown by indirect immunofluorescence after 24 and 48 h with the concentration of 100 and 200 µg/mL, respectively. The LC3 EGFP punctae formation (indication of autophagy) is shown in Fig. 3c. LC3-EGFP punctae formation was observed in cells treated with NPs systems after 24 and 48 h. Further results provide that NPs induced MCF-7 cell autophagy through the LC3 pathway.
Gene expression studies with RT-PCR
The ability of siRNA and siRNA-loaded nanoparticles to lower the endogenous concentration of PLK1 mRNA in the MCF-7 cancer cell lines can be determined with the help of RT-PCR method. A remarkable low level of PLK1 mRNA in transfected MCF-7 cells has been detected after 48 h of treatment with PLK1 siRNA, and PLK1 siRNA–loaded HPP NPs, and SH-HPP NPs. Similarly, in Wu et al. (2010), GFP-LC3–transfected SMMC-7721 cells treated with LCP-II NPs bright green fluorescent dots were found in SMMC-7721 cells, indicating autophagy [31, 32]. The study of the RT-PCR assay showed that PLK1 mRNA level in MCF-7cells was increased 5.59-fold for PLK1 siRNA–SH-HPP and 2.715-fold lower than that in MCF-7 cells for PLK1 siRNA (Table 5; Fig. 4a). Results were well-matched with previous research.
Western blot analysis
PLK1 siRNA (50 nM) and PLK1 siRNA–SH-HPP NPs (50 nM) were taken for western blot analysis. In western blot analysis, the level of PLK1 protein was assessed after 48 h of transfection of MCF-7 breast cancer cells. β-actin was used as a control for western blotting. The percentage of levels of PLK1 siRNA in cell transfection depends on the amount of PLK1 protein remaining. The cells incubated with Opti-MEM I alone (no siRNA or oligofectamine) were considered control. The protein expression was decreased in transfected MCF-7 cells after the treatment with PLK1 siRNA and PLK1 siRNA–loaded NPs whereas control cell showed no reduction was seen in the level of protein expression Fig. 4b and c. Furthermore, this result showed the targetability of PLK1 siRNA–SH-HPP NPs. With the help of β-actin expression, the results can be correlated well with the previously published report [33].
Effect of siRNA and siRNA-SH-HPP NPs on cell viability
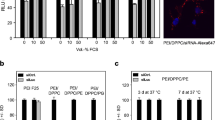
To find out the effect of transfection on the cell viability, the MCF-7 cells were treated with PLK1 si RNA, and PLK1 siRNA loaded (siRNA-SH-HPP) NPs for 48 hrs. In MCF-7 cells, transfection with PLK1 siRNA and siRNA-SH-HPP NPs strategically affects cell viability compared to control cells. Furthermore, treatment with PLK1 siRNA and PLK1 siRNA–SH-HPP NPs (having equivalent concentration of PLK1 siRNA equal to 10, 20, 30, 40, and 50 nM) decreased the cell viability in comparison with control Fig. 5a. According to the result, the PLK1 siRNA and PLK1 siRNA–SH-HPP NPs transfected showed a significant decline in viability at higher concentrations than those with control (p < 0.05), which showed that suppression of PLK1 expression plays a crucial role for in vitro growth of tumor cells (Fig. 5b(i) and (ii)).
The concentration-dependent effect on the in vitro growth on the tumor cells was observed. The treatment with PLK1 siRNA–SH-HPP NPs (50 nM) decreased cell viability up to 80%. The results were in good agreement with the previously published report (31).
As shown in Fig. 6, relative tumor volume was substantially decreased in the mice treated with PLK1 siRNA–SH-HPP NPs in comparison with control group Fig. 6a, T/C value from RTV Fig. 6b and c. Remarkably, in the case of mice treated with PLK1 siRNA–SH-HPP NPs, substantial inhibition of tumor growth was observed as compared to control and the mice model at day 1 and after treatment with ADR and nanoparticle (NPs) (30 days) (n = 6 mice per group).
a The growth of implanted tumor was expressed as relative tumor volume (RTV) ± standard error. b Effects of ADR or NPs on tumor growth reduction were evaluated using T/C ratio based on RTV. c Effects of ADR or NPs on survival rate of mice. d Effects of ADR or NPs on average body weight g of mice (n = 6)
Moreover, there was no significant change in body weight; this may suggest that the administration of PLK1 siRNA–SH-HPP NPs did not affect normal health of mice, adversely (Fig. 6(d)). Group of tumor-bearing mice treated with PLK1 siRNA–SH-HPP NPs observed lower tumor size and weights than the group of controls (Fig. 7). A substantial change was also observed among the groups administered with PLK1 siRNA–SH-HPP NPs versus control in comparison with positive control.
Conclusion
PLK1 siRNA–loaded HPP and SH-HPP NPs were prepared successfully. The high entrapment efficiency prolonged the release of PLK1 siRNA from HPP and SH-HPP NPs. The thiolated nanoparticles showed better entrapment efficiency, in vitro drug release, and high in vitro and in vivo therapeutic efficacy. Furthermore, high therapeutic efficacy of siRNA-SH-HPP NPs can be explained on the basis of excellent loading of PLK1 siRNA, high intercellularization, and selective targeting of cancer cells due to presence of HA. Overall, all the findings suggest that the siRNA-SH-HPP NPs formulation could be effective, safe, and alternative therapeutic strategy for the treatment of cancer cells.
Data availability
The datasets generated during the study are available from corresponding authors on reasonable request.
References
Dieterich M, Stubert J, Reimer T, Erickson N, Berling A. Influence of lifestyle factors on breast cancer risk. Breast Care (Basel). 2014;9(6):407–14. https://doi.org/10.1159/000369571.
Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24(2):260–6. https://doi.org/10.1038/sj.onc.1208219.
Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16(2):384–9. https://doi.org/10.1158/1078-0432.CCR-09-1380.
Hu K, Law JH, Fotovati A, Dunn SE. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14(1):R22. https://doi.org/10.1186/bcr3107.
Sajid MI, Moazzam M, Kato S, Yeseom Cho K, Tiwari RK. Overcoming barriers for siRNA.
Bjorge JD, Pang A, Fujita DJ. Delivery of gene targeting siRNAs to breast cancer cells using a multifunctional peptide complex that promotes both targeted delivery and endosomal release. PLoS ONE. 2017;12(6): e0180578. https://doi.org/10.1371/journal.pone.0180578.
Serrano-Sevilla I, Artiga Á, Mitchell SG, De Matteis L, de la Fuente JM. Natural polysaccharides for siRNA delivery: nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules. 2019;24(14):2570. https://doi.org/10.3390/molecules24142570.
Prajapati SK, Jain A, Jain A, Jain S. Biodegradable polymers and constructs: a novel approach in drug delivery. Eur Poly J. 2019;120:109191.
Tavares MR, Menezes LR, Filho JC, Cabral LM, Tavares MI. Surface-coated polycaprolactone nanoparticles with pharmaceutical application: structural and molecular mobility evaluation by TD-NMR. Polym Testing. 2017;60:39–48. https://doi.org/10.1016/j.polymertesting.2017.01.032.
Grossen P, Witzigmann D, Sieber S, Huwyler J. PEG-PCL-based nanomedicines: a biodegradable drug delivery system and its application. J Control Release. 2017;260:46–60. https://doi.org/10.1016/j.jconrel.2017.05.028.
Yadav AK, Mishra P, Jain S, Mishra P, Mishra AK, Agrawal GP. Preparation and characterization of HA-PEG-PCL intelligent core-corona nanoparticles for delivery of doxorubicin. J Drug Target. 2008;16(6):464–78. https://doi.org/10.1080/10611860802095494.
Katas H, Raja MA, Lam KL. Development of chitosan nanoparticles as a stable drug delivery system for protein/siRNA. Int J Biomater. 2013;2013: 146320. https://doi.org/10.1155/2013/146320.
Sharifiaghdam M, Shaabani E, Sharifiaghdam Z, et al. Enhanced siRNA delivery and selective apoptosis induction in H1299 cancer cells by layer-by-layer-assembled Se nanocomplexes: toward more efficient cancer therapy. Front Mol Biosci. 2021;8:639184. Published 2021 Apr 20. https://doi.org/10.3389/fmolb.2021.639184.
Joseph J, Seervi M, Sobhan PK, Retnabai ST. High throughput ratio imaging to profile caspase activity: potential application in multiparameter high content apoptosis analysis and drug screening. PLoS ONE. 2011;6(5): e20114. https://doi.org/10.1371/journal.pone.0020114.
Lekshmi A, Varadarajan SN, Lupitha SS, et al. A quantitative real-time approach for discriminating apoptosis and necrosis. Cell Death Discov. 2017;3:16101. https://doi.org/10.1038/cddiscovery.2016.101.
Kundu AK, Iyer SV, Chandra S, Adhikari AS, Iwakuma T, Mandal TK. Novel siRNA formulation to effectively knockdown mutant p53 in osteosarcoma. PLoS One. 2017;12(6):e0179168. Published 2017 Jun 21. https://doi.org/10.1371/journal.pone.0179168.
Mu X., Lu H, Fan, Yan S, Hu K. Efficient delivery of therapeutic siRNA with nanoparticles induces apoptosis in prostate cancer cells. J. Nanomater. 2018, 2018, 1–10. https://doi.org/10.1155/2018/4719790.
Tian G, Pan R, Zhang B, et al. Liver-targeted combination therapy basing on glycyrrhizic acid-modified DSPE-PEG-PEI nanoparticles for co-delivery of doxorubicin and Bcl-2 siRNA. Front Pharmacol. 2019;10:4. https://doi.org/10.3389/fphar.2019.00004.
Tian Y, Pan ZY, J., Lu, N., Wang, S., Lu, G. Gold nanoparticles increase PLK1-specific small interfering RNA transfection and induce apoptosis of drug resistance breast cancer cells. J Nanomat. 2015;2015:1–9. https://doi.org/10.1155/2015/720198.
Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. https://doi.org/10.1016/0022-1759(83)90303-4.
Kholiya F, Chatterjee S, Bhojani G, et al. Seaweed polysaccharide derived bioaldehyde nanocomposite: potential application in anticancer therapeutics. Carbohydr Polym. 2020;240: 116282. https://doi.org/10.1016/j.carbpol.2020.116282.
Jiang G, Park K, Kim J, et al. Hyaluronic acid-polyethyleneimine conjugate for target specific intracellular delivery of siRNA. Biopolymers. 2008;89(7):635–42. https://doi.org/10.1002/bip.20978.
Chitosan-graft-PAMAM–alginate core–shell nanoparticles: a safe and promising oral insulin carrier in an animal model (Suppl.). RSC Adv. 2015:5:93995–94007. https://doi.org/10.1039/C5RA17729D.
Yue PF, Lu XY, Zhang ZZ, et al. The study on the entrapment efficiency and in vitro release of puerarin submicron emulsion. AAPS PharmSciTech. 2009;10(2):376–83. https://doi.org/10.1208/s12249-009-9216-3.
Peng YY, Diaz-Dussan D, Vani J, Hao X, Kumar P, Narain R. Achieving Safe and Highly Efficient Epidermal Growth Factor Receptor Silencing in Cervical Carcinoma by Cationic Degradable Hyperbranched Polymers. ACS Appl Bio Mater. 2018;1(4):961–6. https://doi.org/10.1021/acsabm.8b00371.
Zhang J, Wang X, Cui W, et al. Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nat Commun. 2013;4:2157. https://doi.org/10.1038/ncomms3157.
Ma YS, Yao CN, Liu HC, et al. Quercetin induced apoptosis of human oral cancer SAS cells through mitochondria and endoplasmic reticulum mediated signaling pathways. Oncol Lett. 2018;15(6):9663–72. https://doi.org/10.3892/ol.2018.8584.
Ouahab A, Cheraga N, Onoja V, Shen Y, Tu J. Novel pH-sensitive charge-reversal cell penetrating peptide conjugated PEG-PLA micelles for docetaxel delivery: in vitro study. Int J Pharm. 2014;466(1–2):233–45. https://doi.org/10.1016/j.ijpharm.2014.03.009.
Baek JS, Na YG, Cho CW. Sustained cytotoxicity of wogonin on breast cancer cells by encapsulation in solid lipid nanoparticles. Nanomaterials (Basel). 2018;8(3):159. https://doi.org/10.3390/nano8030159.
Namvar F, Azizi S, Rahman HS, et al. Green synthesis, characterization, and anticancer activity of hyaluronan/zinc oxide nanocomposite. Onco Targets Ther. 2016;9:4549–4559. Published 2016 Jul 26. https://doi.org/10.2147/OTT.S95962.
Patil YB, Swaminathan SK, Sadhukha T, Ma L, Panyam J. The use of nanoparticle-mediated targeted gene silencing and drug delivery to overcome tumor drug resistance. Biomaterials. 2010;31(2):358–65. https://doi.org/10.1016/j.biomaterials.2009.09.048.
Wu Y, Wang W, Chen Y, et al. The investigation of polymer-siRNA nanoparticle for gene therapy of gastric cancer in vitro. Int J Nanomedicine. 2010;5:129–36. https://doi.org/10.2147/ijn.s8503.
Hou L, Song Z, Xu Z, Wu Y, Shi W. Folate-mediated targeted delivery of siPLK1 by leucine-bearing polyethylenimine. Int J Nanomedicine. 2020;15:1397–408. https://doi.org/10.2147/IJN.S227289.
Acknowledgements
The authors are grateful to RGCB, Thiruvananthapuram (India) for providing the autophagy analysis, caspase assay, cell cycle analysis, and FACS-based analysis. The authors express their sincere thanks to MNIT, Jaipur for providing AFM facility. The authors are thankful to Adina Institute for providing an FTIR facility. The authors also express their sincere thanks to Jayagen Biologics Private Limited Chennai, Tamil Nadu, India, for providing western blotting, gene silencing assay, and cell cytotoxicity analysis. The authors are thankful to Banaras Hindu University (BHU) for providing an NMR facility.
Funding
This work was supported by DBT (DBT/6242-P29/RGCB/PMD/DBT/AWKY/2015).
Author information
Authors and Affiliations
Contributions
Dolly Jain carried out all experimental work, data generation, and writing of the manuscript. Dr. Awesh K. Yadav provided concept, design, and material preparation.
Corresponding author
Ethics declarations
Ethical approval
All protocols for animal studies were approved by Institutional Animal Ethics Committee (Registration Number: 65/GO/ReBiBt/S/99/CPCSEA), ACTREC, Tata Memorial Centre, Navi Mumbai.
Consent to participate
Not applicable. No human subject participated.
Consent for publication
Not applicable. No individual person’s data.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, D., Yadav, A.K. Development of hyaluronic acid–anchored polycaprolactone nanoparticles for efficient delivery of PLK1 siRNA to breast cancer. Drug Deliv. and Transl. Res. 13, 1730–1744 (2023). https://doi.org/10.1007/s13346-022-01288-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13346-022-01288-2