Abstract
High oncogenic risk types of human papillomaviruses are mainly transmitted via sexual contact and are the main cause of cervical cancer in females in developing countries. Molecular detection of HPV infection enables early cancer detection; however, it is not widely used in low-income countries due to resource constraints. The aim of this study was to assess economical yet sensitive HPV detection and genotyping assays for both physician and self-collected cervical samples in a resource limited diagnostic setting. A previously reported polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) based HPV detection and genotyping protocol was verified using direct DNA sequencing to accurately identify the HPV 16 and 18 genotypes in a routine-diagnostic set-up. Then the HPV prevalence in a cohort of 433 clinically normal females was performed using PCR–RFLP diagnostic tool. Finally, the performance of the PCR–RFLP HPV screening tool was further evaluated against self-collected samples. HPV 16 and 18 genotyping with the PCR–RFLP consistently agreed with the sequencing data. The HPV prevalence in the screening cohort was 5.8%. HPV 16 and 18 were the most common high-risk HPV genotypes detected in the study cohort. Self-sampling vs physician collected samples from the same subject resulted in an overall concordance of 93% for HPV detection. The PCR–RFLP protocol can be used effectively under low resource settings for HPV 16/18 diagnosis and genotyping. The self-sampling approach can be recommended to increase HPV screening among women in Sri Lanka.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human papillomavirus (HPV) infection is the most common sexually transmitted disease in the world, primarily causing cervical cancer (CC) through persistent infections in the cervix [10]. Over 100 known HPV types exist, with 13 classified as high-risk genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 68, and 59) [20]. The prevalence of HPV is high among women diagnosed with invasive cervical cancers, with HPV 16 and HPV 18 being the predominant genotypes among CC cases in Sri Lanka (80.6%) [15]. Therefore, it is recommended that even women who are clinically healthy and have normal cervical cytology, but test positive for HPV 16/18, undergo additional evaluations to prevent CC [6, 28].
The HPV cannot be grown easily in tissue culture systems or detected with serology based methods. Therefore, most diagnostic methods of HPV rely on detection of viral nucleic acids. Molecular testing for oncogenic HPV infections using commercial kits is the common early screening method for CC in high-resource settings [7, 11, 26, 32]. However, this is not feasible in resource-limited healthcare systems in developing countries due to high costs and a lack of skilled personnel. Polymerase chain reaction (PCR) based HPV detection is highly sensitive and can detect a broad spectrum of mucosal HPV genotypes in a single PCR using only a small amount of specimen [5, 21]. It is widely used in epidemiological studies and can be adopted for clinical setting where resources are limited and inputs are low. HPV DNA amplified in PCR can then be further analyzed with multiple HPV genotyping methods, such as sequencing [31], hybridization with HPV specific probes [12], microarray platforms [3], multiplexed PCR using matrix assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) [24] or restriction fragment length polymorphism (RFLP) [5, 21]. RFLP analysis following PCR amplification is a simple, inexpensive, robust, reproducible, and rapid method of HPV genotyping. It is based on digestion of the PCR products using one or several specific restriction enzymes. Previously, a study has described a PCR–RFLP analysis method to distinguish five clinically relevant HPV genotypes in clinical samples, HPV 16, 18, 11, 6, and 33, solely using the RsaI restriction enzyme [21]. Nevertheless, the study was constrained by its limited sample size and lack of confirmation through direct sequencing. In the present study, we intended to use the same PCR–RFLP protocol, but results will be verified through sequencing and by testing a substantially larger number of clinical specimens. For the first time we evaluated suitability of the above PCR–RFLP method for routine HPV screening.
Cytology-based HPV screening is the norm in government clinical settings for monitoring HPV or CC in Sri Lanka due to limited access to molecular diagnostics. Regular prevalence monitoring for HPV or cervical cancer is lacking due to unawareness and unavailability as well. Self-collected sampling and urine based less-invasive sampling have emerged as alternatives to clinician collected samples for HPV screening with increased acceptability and increased rate of compliance among women with social stigma for screening in remote and low resource setting [1, 4, 16, 18, 23]. However, the validity of such a sampling technique for HPV screening has not yet been evaluated in Sri Lanka.
Therefore, this study aimed to assess the feasibility and performance of a previously described PCR–RFLP based HPV screening workflow to detect HPV in low resource routine diagnostic settings with further emphasis on self-sampling HPV detection to enhance the CC prevention in Sri Lanka.
Materials and methods
Control cell lines, HPV recombinant plasmid vectors and method validation
HPV screening and genotyping workflow was established in-house using a model system consisting of 4 HPV infected cancer cell lines (DNA from CasKi, SiHa, HeLa, C4 cells) and with plasmids containing full length HPV genomes of HPV type 16 and HPV 18, kindly gifted by Dr. Stephanie Liu, Department of Obstetrics and Gynaecology, Faculty of Medicine, University of Hong Kong, Hong Kong SAR [19]. In addition, HPV genotyping based on PCR–RFLP was validated using 20 known HPV positive samples (positive with CareHPV™ kit) collected at the National Cancer Control Program, Colombo, Sri Lanka.
Primers and PCR protocols for HPV detection and genotyping
All DNA samples from HPV infected cancer cell lines and clinical material collected in this study were evaluated by amplification of the human β-globin gene using primers PCO4 (5′-CAACTTCATCCACGTTCACC-3′), and GH20 (5′-GAAGAGCCAAGGACAGGTAC-3′) as an internal control. The general primer GP5 + (5′-TTTGTTACTGTGGTAGATACTAC-3′), and GP6 + (5′-TTTGTTACTGTGGTAGATACTAC-3′), which produces a 150 bp PCR product were used for primary screening of HPV DNA in all clinical samples as described previously (GP-PCR) [13]. Further genotyping of HPV positive cases was performed using PCR–RFLP and the method was verified for use in routine diagnostic set-up by sequencing 20 known HPV positive cases.
HPV genotyping analysis by restriction fragment length polymorphism (RFLP)
Twenty HPV positive cases were selected for HPV genotyping with RFLP analysis. HPV DNA was first amplified with degenerate consensus primers MY09 (5′-CGTCCMARRGGAWACTGATC-3′) and MY11 (5′-GCMCAGGGWCATAAYAATGG-3′) which amplified a 450 bp region in the viral L1 gene region (MY-PCR) [9]. RFLP analysis of clinical samples positive for HPV was performed using the RsaI restriction enzyme as described earlier [21]. A 10 µL of the MY-PCR products were digested in a total volume of 20 µL with 10 units of RsaI at 37 °C for 2 h. Digested products were then separated electrophoretically in 10% polyacrylamide gels and stained with ethedium bromide. The banding pattern in the restriction profile of each sample was evaluated against the standard banding pattern in controls for HPV 16 and HPV 18 (HPV 16 and HPV 18 plasmid control vectors).
Confirmation of genotyping by direct sequencing
To confirm the results obtained by PCR–RFLP analysis, HPV positive GP-PCR products from 20 selected cases were sequenced. The sequence data obtained were compared with all sequences available in GeneBank database (www.ncbi.nlm.nih.gov) using the BLAST tool.
Detection of HPV prevalence in clinically normal female cohort
Ethical clearance was obtained from the Ethical Review Committee Faculty of Medicine, University of Peradeniya, Peradeniya, Sri Lanka (2017/EC/20). Cervical cytology samples for the standard Papanicolaou test (Pap smear), and Cytobrush (EDM, USA) sampling for screening for HPV infections were collected from the clinically normal females after obtaining informed written consent. Cervical brush samples were collected in 10 mL ice-cold sterile Phosphate buffered saline (PBS) (Sigma-Aldrich, USA) and DNA was extracted and purified from the cellular specimen by proteinase K digestion followed by a column clean up method (ReliaPrepTM gDNA Tissue Miniprep System, Promega, USA).
In total, 433 samples were collected from a clinically normal female cohort between 20 and 65 years of age attending CC screening in Teaching Hospitals Peradeniya and Kandy after obtaining informed written consent from 2017 to 2019. Demographic data, history for exposure to risk factors of CC, and clinical assessment of the cervix were collected from the participants. Data on age, occupation, monthly family income, parity, number of sexual partners, previous exposure to HPV vaccination, and methods of contraception were also collected. All the samples were screened using blinded cervical cytology. HPV prevalence was determined in the entire cohort of clinically normal females using the established PCR–RFLP method (Fig. 1).
Workflow for detection of HPV prevalence in clinically normal female cohort. n = number of samples collected. Categories for cervical cytology sample evaluation: NILM, negative for Intraepithelial Lesion or Malignancy; ASCUS-L, atypical squamous cells of undetermined significance; ASCUS-H, atypical squamous cells cannot exclude high-grade squamous intraepithelial lesion; LSIL, Low Grade Intraepithelial Neoplasia; HSIL, High Grade Intraepithelial Neoplasia; CC, cervical cancer
Evaluation of self-sampling for HPV testing using PCR–RFLP method
The validity of the self-collected HPV screening compared to physician-collected samples was determined using the in-house developed HPV detection assay in a cohort of females participated in CC screening program (n = 68) (Fig. 2). During their scheduled appointment for cervical smear collection with the physician, the participants were provided with a Qvintip® self-sampling device (Aprovix AB, Uppsala, Sweden) along with proper instructions. On the same day, the physician collected the samples after the participants collected their own samples. Self-collected cervico-vaginal samples were placed in 500 µL of sterile normal saline and the supernatant containing DNA was obtained using a direct lysis buffer. Sensitivity, specificity, negative predictive value, and positive predictive value were calculated using physician collected HPV detection and genotyping as the gold standard method.
Statistical analysis
Statistical analysis was performed using Statistical Software for Social Sciences (SPSS version 20.0®). Mean and standard deviations were calculated for continuous variables. The kappa statistic was calculated to determine the level of chance agreement between different methods. Statistical significance was set at p < 0.05. The overall percentage of agreement between paired samples was calculated as the proportion of concordant sample sets divided by the total number of samples.
Results
Primary HPV screening and genotyping using PCR–RFLP
A successful PCR–RFLP-based strategy for HPV detection and genotyping (for HPV 16 and 18) was implemented, using a model system comprising HPV-infected cancer cell lines and HPV 16 and 18 recombinant plasmids (Fig. 3).
HPV DNA detection using PCR was established using cell culture model system consisting four HPV infected cancer cell lines (Caski, HeLa, SiHa, C4) and HPV recombinant vectors for HPV 16 and HPV 18. a Amplification of the internal control globin gene was used to confirm the integrity of the samples. b, c Amplification of the HPV DNA using GP5+/6+ and MY09/11 primers. d H16 and H18 are showing the banding pattern for HPV 16 and 18 genotyping with RsaI based RFLP. The theoretical sizes of each band of the HPV 16 reference banding pattern are 310 bp, 72 bp and 70 bp. The theoretical sizes of each band of the HPV 18 reference banding pattern are 135 bp, 125 bp, 85 bp, 72 bp and 38 bp. e PCR–RFLP analysis of HPV infected clinical specimen. Lane 1, 2, 5 and 7 are showing HPV 16 genotype isolated from clinical samples. Lane 6 showing HPV 18 isolated from clinical samples. H16: HPV 16 reference banding patter. H18: HPV 18 reference banding pattern
The RsaI enzyme showed high digestion efficiency, and no partial digestions were visualized under the RFLP conditions mentioned above (Fig. 3D).
Comparative genotyping of HPV positive clinical samples with PCR–RFLP and DNA sequencing
Genotyping results of 20 selected HPV positive samples with in-house developed assay were further verified by sequencing of GP-PCR products. There was a 100% concordance for detecting HPV 16/18 genotyping using PCR–RFLP and direct sequencing. However, the overall concordance between the two methods was 90%. Two samples that could not be genotyped using RFLP because of faint bands in PCR were sequenced and confirmed as non-HPV 16/18 types. The exact genotypes could not be determined in multiple infections with either method (Table 1).
HPV prevalence among clinically normal female cohort
The mean age of the 433 participants in primary HPV screening was 40.65 ± 9.5 years. The mean age at first sexual intercourse was 25.4 ± 4.8 years. All were non-smokers. Of these women, 18.3% were nulliparous, and 64.7% had no contraceptive use. None of the women had been previously vaccinated against HPV. Of the 433 women, 64.7% had no any contraceptive use, 12.8% had an intrauterine contraceptive device (IUCD), 14.3% were on oestro-progestins, and 8.1% had undergone tubal ligation. About 42% of the women were housewives, and 48.2% were employed (Table 3).
Only 426 samples were considered satisfactory for evaluation using the HPV DNA test (426/433), and the overall prevalence of HPV was 5.8% among clinically normal women (Table 2). The highly oncogenic HPV 16/18 genotypes constituted 52% of high-risk HPV infections in the study cohort. Two cases of multiple infections were reported. Two samples could not be genotyped using RFLP because of faint band in the MY PCR. However, they were confirmed to be non-HPV 16/18 genotypes using sequencing. Normal Pap cytology was observed in 80.5% of the cohort; however, minor cytological abnormalities like 4% of ASCUS, 1.5%, ASC-H and 1.1% LSIL were observed. Only 1.8% of the women had glandular cell atypia and 11.4% of the PAP samples were unsatisfactory for evaluation.
The prevalence of HPV was highest in the 20–29 age group. The HPV prevalence decreased with increasing age from 30 to 49 yrs. The HPV prevalence again seen higher in the 50–60 age group. There was no statistically significant relationship between HPV prevalence and age, employability, parity, or ethnicity (p > 0.05). However, high HPV prevalence rates have been reported among employed women and women of high parity. A High HPV prevalence was also reported among females who had their first sexual exposure below the age of 19 yrs (Table 3). The HPV DNA test developed in-house detected all high-grade lesions (high grade squamous cell abnormality (HSIL)/cervical intraepithelial neoplasia 2 (CIN2) or worse) with 100% sensitivity and 90% specificity.
Comparison of self-sampling vs physician collected samples
The HPV screening assay was further evaluated for 68 self-collected samples. The mean age of study cohort was 45 ± 7 years. The mean age at the first sexual intercourse was 25 ± 4 yrs. The peak age of respondents was 40 years. Normal Pap cytology was reported in 75% of the cohort; however minor cytological abnormalities of 10.3% of ASCUS, 1.5% of LSIL were observed. A small number (7.4%) of the samples were unsatisfactory for further evaluation. Two females had HSIL/CC in the studied cohort.
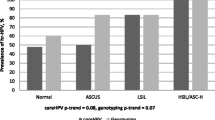
All the females who were invited to collect a self-sample for HPV testing accepted to perform self-sampling. In total 68 females provided both physicians collected cervical brush sample and self-collected cervico-vaginal fluid sample for HR-HPV testing. Eight of the self-sampled specimens were excluded from the analysis, as the specimens tested negative for β-globin. The HPV prevalence reported in self and physician collected sample types was 8.3% and 5.9% respectively (Supplementary Table 1). The overall concordance between self-collected and the physician collected samples was 93%. However, only a moderate agreement was reported between self and physician collected sample HPV detection with a κ (Kappa) = 0.47, 95% (CI 0.434–0.492). The overall sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for HPV detection in self-samples were 75%, 96%, 60%, and 98%, respectively. The sensitivity and specificity, PPV and NPV for detecting CIN2+ with self-sample HPV testing were 100%, 90%, 40% and 100%, respectively. Therefore, no underlying cervical high-grade lesions were missed. Out of five samples HPV positive in self-collected samples, four were HPV 16/18 infected. Among the four physician-collected samples, only two were HPV 16/18 infected. There was an overall 60% concordance between HPV genotyping results of self and physician collected samples. However, self-sampling HPV testing and genotyping had 100% concordance in detecting HSIL or worse, compared to physician collected sample HPV testing and genotyping (Supplementary Fig. 1).
Discussion
There is a limited provision for HPV screening in many developing countries worldwide, including Sri Lanka and the current screening rates remain unsatisfactory. Therefore, there is a need to introduce simpler sampling methods and to implement rapid and cost-effective molecular diagnostics for HPV detection. PCR-based RFLP analysis, as tested in this study, seems to be a straightforward and economical tool for characterizing clinically important HPV subtypes, such as HPV 16 and 18, in genital HPV infections. Notably, this method doesn’t demand sophisticated or expensive equipment, making it applicable to settings with limited financial resources.
The GP-PCR assay method utilized for primary HPV screening demonstrated the capability to detect a wide range of both known and unknown HPV genotypes [29]. In the present study, the results of the GP-PCR were in good agreement with the cytological diagnosis of HSIL/CC, reporting a clinical sensitivity of 100% and specificity of 90% for detecting CIN 2+. Importantly, prior research has already established the non-inferiority of this method compared to commercial HPV detection platforms [30]. This study evaluated an RsaI based PCR–RFLP to distinguish the two most common high oncogenic risk HPV types, HPV 16 and 18, from other high-risk types as described previously [21], a capability often not achievable even with many proprietary kit-based HPV detection methods. The patterns generated were clearly distinguishable in common, inexpensive polyacrylamide gels, making it comparable with direct sequencing of PCR products or commercial qPCR methods. All HPV 16/18 genotypes identified using the PCR–RFLP method were consistent with the DNA sequencing results. Several previous studies have performed PCR–RFLP analysis to distinguish high-risk genotypes [21, 22, 27]. However, many of these studies used multiple restriction enzymes which complicated the analysis for routine diagnostic application [27]. In the present study, the use of a single restriction enzyme reduced the complexity of the analysis. The banding patterns were not shared with any other known high-risk types and hence were distinguishable among known and common HR-HPV genotypes [22]. Similar to our study, the PCR–RFLP method using the RsaI restriction enzyme was employed in a study in Brazil to identify high risk HPV genotypes in cervical fluid samples. It also showed that this technique is highly sensitive and can be used to discriminate the presence of HR-HPV types such as 16, 31, 39, 51, 58, 68, and LR-HPV types 6b, 11, 44, and 55 [2]. Thus, the RsaI based PCR–RFLP can be used for accurate genotyping the HPV 16 and 18 types in a limited-resource setting, without the need for additional costly sequencing or hybridization procedures. According to the sequencing results, HPV genotyping results can also be used to identify other genotypes such as HPV 33, HPV 58 and HPV 90 (Table 1). However, it is necessary to maintain stringent quality control measures to avoid any risk of DNA carry over contamination while performing this technique. We used sterilization methods such as ethanol disinfection and UV radiation of biosafety cabinets before and after sample processing. For each batch of samples, DNA extraction and PCR negative controls were included to monitor for any contamination. Furthermore, samples were processed in small batches, typically comprising around 10 samples per batch, to enhance control over the process and minimize the likelihood of contamination. These rigorous procedures were essential components of our methodology to ensure the reliability of the results. The crude prevalence of cervico-vaginal HPV infection among the studied cohort consisting of married women 21–65 years of age was 5.8%, and the HPV prevalence among females with normal cytology was 3.2%. Interestingly, there are no existing reports on population-based HPV prevalence in Sri Lanka or on the prevalence of HPV among females with normal cytology. A community-based study conducted in Gampaha district, Sri Lanka in 2012, involving 2000 females, reported an HPV prevalence of 3.3% among married women aged 20–59. This study remains the sole investigation into HPV prevalence in Sri Lanka to date [8]. The current study revealed a 3.2% prevalence of HPV 16/18 types in clinically normal females, while the study in Gampaha district reported a 1.2% prevalence. Among the reported HPV infections, HPV 16 and 18 were the most common genotypes identified, both of which are vaccine-preventable. We also detected significant number of non HPV 16/18 genotypes in the clinically normal screening cohort. Therefore it will be interesting to study the contribution of these genotypes to occurrence of cervical lesions or cancer among Sri Lankan females in a future study. Nevertheless, both studies agreed to the fact that the highest HPV prevalence is in the 19–29 age group.
The HPV screening and genotyping workflow was extended to include HPV detection in the self-collected samples. In this study, a higher level of agreement was observed between the self-collected cervico-vaginal specimens tested for HPV DNA and physician-collected cervical samples tested for HSIL or worse (CIN2+). These findings suggest that a self-sampling HPV DNA test could serve as a primary CC screening method, especially in settings with limited resources or where there is reluctance for physician-directed screening due to social taboos. Self-collected samples HPV analysis also circumvents the need for DNA extraction, offering a more streamlined approach to HPV genotyping utilizing PCR–RFLP, further eliminating the laboratory risk of DNA carry over. Some self-samples tested negative for beta globin (12%, 8/68) relative to physician-collected samples, possibly because they were not familiar with the procedure, as none had used the device before. Therefore, strategies to enhance awareness of the sampling procedure among Sri Lankan females are crucial before introducing self-sample-based HPV screening into routine clinical setups. There are no prior reports on the validity of self-sampling HPV detection compared to physician-collected sample HPV detection among a cohort of Sri Lankan females. Notably, the cervical cancer screening coverage remains around 10% among high risk groups in Sri Lanka [14]. One main reason for such low coverage is due to the social stigma for cervical cancer screening with physician collected samples. Therefore, the findings of this study show that self-sampling presents an intriguing alternative for cervical cancer screening, particularly in enhancing screening coverage. This approach could potentially reach underserved populations and contribute to improving overall cervical cancer detection rates in Sri Lanka.The HPV prevalence reported among the self-samples was higher than physician collected samples, and the results were consistent those of previous studies that reported higher HPV prevalence among self-collected samples using the Cobas 4800 PCR based assay [16]. The higher prevalence of HPV among self-sampling compared to physician-collected samples may be attributed to self-samples containing clinical material from the vaginal epithelium, which can include many low-risk HPV types, subsequently reducing the specificity of self-sampling HPV testing [16, 18]. False-positive HPV results with self-collected samples may lead to increased referral rates of HPV-positive but cytology-normal females to colposcopy. Secondary tests, such as triaging with HPV mRNA expression or other molecular markers with higher performance that assess not only the presence of HPV infection but also the risk for oncogenesis, could help reduce the number of such colposcopies [25].
According to the results, self-sampling based HPV detection using GP5+/6+ PCR followed by RFLP is non-inferior to physician collected samples in detecting HSIL or worse. The self-sampling HPV detection showed an overall concordance of 93% with a moderate agreement of Kappa value of 0.47 with physician collected sample HPV detection. According to the literature, this is among the few studies that used a clinically validated GP-PCR based RFLP method for HPV screening and genotyping among self-collected samples in low-resource clinical settings. Interestingly, a study conducted in South India among women with histologically confirmed CC using PCR based screening showed similar agreement values (93%) between physician and self-collected sample [17]. The self-sampling has shown increased acceptance among non-attendees around the world. However, conducting a large-scale population-based study for HPV screening using self-collected cervico-vaginal samples is recommended to gain more insight into employing self-sampling HPV testing in resource-limited settings as a primary CC screening method.
Data availability
All relevant data are within the paper and its Supporting Information (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9683586/#sec015) file. Any additional information are available from the corresponding author on reasonable request.
References
Adler DH, Laher F, Lazarus E, Grzesik K, Gray GE, Williamson A. A viable and simple self-sampling method for human papillomavirus detection among South African adolescents. J Immunol Tech Infect Dis. 2013. https://doi.org/10.4172/2329-9541.1000113.
Alves Melo IM, Pereira Viana MR, Pupin B, Bhattacharjee TT, Canevari RDA. PCR-RFLP and FTIR-based detection of high-risk human papilloma virus for cervical cancer screening and prevention. Biochem Biophys Rep. 2021;26:100993. https://doi.org/10.1016/j.bbrep.2021.100993.
Arron ST, Skewes-Cox P, Do PH, Dybbro E, Da Costa M, Palefsky JM, et al. Validation of a diagnostic microarray for human papillomavirus: coverage of 102 genotypes. J Nucleic Acids. 2011;2011:1–6. https://doi.org/10.4061/2011/756905.
Bottari F, Igidbashian S, Boveri S, Tricca A, Gulmini C, Sesia M, et al. HPV self-sampling in CIN2+ detection: Sensitivity and specificity of different RLU cut-off of HC2 in specimens from 786 women. J Clin Pathol. 2017;70:327–30. https://doi.org/10.1136/jclinpath-2016-204044.
Coser J, da Rocha Boeira T, Kazantzi Fonseca AS, Ikuta N, Lunge VR. Human papillomavirus detection and typing using a nested-PCR-RFLP assay. Braz J Infect Dis. 2011;15:467–72. https://doi.org/10.1016/S1413-8670(11)70229-X.
Elfgren K, Elfström KM, Naucler P, Arnheim-Dahlström L, Dillner J. Management of women with human papillomavirus persistence: long-term follow-up of a randomized clinical trial. Am J Obstet Gynecol. 2017;216:264.e1-264.e7. https://doi.org/10.1016/j.ajog.2016.10.042.
FDA. FDA Executive summary new approaches in the evaluation for high-risk human papillomavirus nucleic acid detection. 2019;1–34.
Gamage D, Rajapaksa L, Abeysinghe MR, De Silva A. Prevalence of carcinogenic human papilloma virus infection and burden of cervical cancer attributable to it in the District of Gampaha, Sri Lanka; 2012;UNFPA, Epidemiology Unit, ISBN 978-955-8375-06-8
Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Hildesheim A, Schiffman MH, et al. Improved amplification of genital human papillomaviruses improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–61. https://doi.org/10.1128/JCM.38.1.357-361.2000.
Hausen H. Papillomaviruses in the causation of human cancers—a brief historical account. Virology. 2009;384:260–5. https://doi.org/10.1016/j.virol.2008.11.046.
Heideman DAM, Hesselink AT, Berkhof J, Van Kemenade F, Melchers WJG, Daalmeijer NF, et al. Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol. 2011;49:3983–5. https://doi.org/10.1128/JCM.05552-11.
Hubbard RA. Human papillomavirus testing methods. Arch Pathol Lab Med. 2003;127:940–5. https://doi.org/10.5858/2003-127-940-HPTM.
Husnjak K, Grce M, Magdić L, Pavelić K. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J Virol Methods. 2000;88:125–34. https://doi.org/10.1016/S0166-0934(00)00194-4.
Iqbal A, Joseph N. Cervical cancer in Sri Lanka, South Asian. J Cancer. 2023;12:39–40. https://doi.org/10.1055/s-0043-1764236.
Karunaratne K, Ihalagama H, Rohitha S, Molijn A, Gopala K, Schmidt JE, et al. Human papillomavirus prevalence and type-distribution in women with cervical lesions: a cross-sectional study in Sri Lanka. BMC Cancer. 2014;14:1–6. https://doi.org/10.1186/1471-2407-14-116.
Ketelaars PJW, Bosgraaf RP, Siebers AG, Massuger LFAG, van der Linden JC, Wauters CAP, et al. High-risk human papillomavirus detection in self-sampling compared to physician-taken smear in a responder population of the Dutch cervical screening: results of the VERA study. Prev Med. 2017;101:96–101. https://doi.org/10.1016/j.ypmed.2017.05.021.
Kuriakose S, Sabeena S, Binesh D, Abdulmajeed J, Ravishankar N, Ramachandran A, et al. Diagnostic accuracy of self-collected vaginal samples for HPV DNA detection in women from South India. Int J Gynaecol Obs. 2020. https://doi.org/10.1002/ijgo.13116.
Lam JUH, Elfström KM, Ejegod DM, Pedersen H, Rygaard C, Rebolj M, et al. High-grade cervical intraepithelial neoplasia in human papillomavirus self-sampling of screening non-attenders. Br J Cancer. 2018;118:138–44. https://doi.org/10.1038/bjc.2017.371.
Liu SS, Leung RCY, Chan KKL, Cheung ANY, Ngan HYS. Evaluation of a newly developed GenoArray human papillomavirus (HPV) genotyping assay and comparison with the Roche linear array HPV genotyping assay. J Clin Microbiol. 2010;48:758–64. https://doi.org/10.1128/JCM.00989-09.
Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27.
Naqvi SH, Wajid S, Mitra AB. Restriction fragment length polymorphism of L1 amplicon using Rsa 1 detects five different human papillomavirus types and their co-infections among women attending a gynaecological outpatient department. J Virol Methods. 2004;117:91–5. https://doi.org/10.1016/j.jviromet.2003.12.002.
Nobre RJ, de Almeida LP, Martins TC. Complete genotyping of mucosal human papillomavirus using a restriction fragment length polymorphism analysis and an original typing algorithm. J Clin Virol. 2008;42:13–21. https://doi.org/10.1016/j.jcv.2007.11.021.
Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–21. https://doi.org/10.1002/rmv.593.
Peng J, Gao L, Guo J, Wang T, Wang L, Yao Q, et al. Type-specific detection of 30 oncogenic human papillomaviruses by genotyping both E6 and L1 genes. J Clin Microbiol. 2013;51:402–8. https://doi.org/10.1128/jcm.01170-12.
Persson M, Elfström KM, Wendel SB, Weiderpass E, Andersson S. Triage of HR-HPV positive women with minor cytological abnormalities: a comparison of mRNA testing, HPV DNA testing, and repeat cytology using a 4-year follow-up of a population-based study. PLoS ONE. 2014;9:1–8. https://doi.org/10.1371/journal.pone.0090023.
Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, et al. Aptima HPV E6/E7 mRNA test is as sensitive as hybrid capture 2 assay but more specific at detecting cervical precancer and cancer. J Clin Microbiol. 2011;49:557–64. https://doi.org/10.1128/JCM.02147-10.
Santiago E, Camacho L, Junquera ML, Vázquez F. Full HPV typing by a single restriction enzyme. J Clin Virol. 2006;37:38–46. https://doi.org/10.1016/j.jcv.2006.06.001.
Schiffman M, Boyle S, Raine-bennett T, Katki HA, Gage JC, Wentzensen N, et al. The role of human papillomavirus genotyping in cervical cancer screening: a large-scale evaluation of the Cobas HPV Test. Cancer Epidemiol Biomarkers Prev. 2015;24:1304–10. https://doi.org/10.1158/1055-9965.EPI-14-1353.
Venceslau EM, Bezerra MM, Lopes ACM, Souza ÉV, Onofre ASC, De Melo CM, et al. HPV detection using primers MY09/MY11 and GP5+/GP6+ in patients with cytologic and/or colposcopic changes. J Bras Patol E Med Lab. 2014;50:280–5. https://doi.org/10.5935/1676-2444.20140028.
Villa LL. Laboratory methods for detection of human papillomavirus infection. In: Rosenblatt A, Guidi HGC, editors. Human papillomavirus. Berlin: Springer; 2009. p. 23–30. https://doi.org/10.1007/978-3-540-70974-9_2.
Wagner S, Roberson D, Boland J, Yeager M, Cullen M, Mirabello L, et al. Development of the TypeSeq assay for detection of 51 human papillomavirus genotypes by next-generation sequencing. J Clin Microbiol. 2019;57:1–11. https://doi.org/10.1128/JCM.01794-18.
Williams J, Kostiuk M, Biron VL. Molecular detection methods in HPV-related cancers. Front Oncol. 2022;12:1–16. https://doi.org/10.3389/fonc.2022.864820.
Acknowledgements
We thank Dr. Stephanie Liu, Department of Obstetrics and Gynaecology, Faculty of Medicine, University of Hong Kong, Hong Kong SAR for providing four HPV infected cancer cell lines DNA (CasKi, SiHa, HeLa, C4) and plasmids containing full length HPV genome of HPV type 16 and HPV 18 for method validation purpose, medical staff who helped in collecting clinical samples and all females consented and participated in this study.
Funding
Funding support from the National Science Foundation, Sri Lanka is acknowledged (Grant No. NSF/RPHS/2016/C06).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There is no conflicts of interests to declare.
Ethics approval
The ethical clearance for the study was obtained from the Ethical Review Committee, Faculty of Medicine, University of Peradeniya, Peradeniya, Sri Lanka on 23th of June 2017 (2017/EC/20).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muhandiram, S., Karunarathna, T.K., Siriweera, E.H. et al. Molecular detection of human papillomavirus prevalence in clinically normal females and identification of high-risk HPV 16 and 18 under low resources setting: a cohort study from Sri Lanka. VirusDis. 35, 271–280 (2024). https://doi.org/10.1007/s13337-024-00875-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-024-00875-w