Abstract
Banana bunchy top virus (BBTV) is a serious threat to banana (Musa spp.) production in India. Generally, BBTV isolates within the country share very low genetic diversity. However, in India, relatively greater diversity has been observed between isolates from north-eastern (NE) region (Meghalaya) and rest of India. Tripura is situated in the south-west corner of NE India and shares international border with Bangladesh. During 2014–2015, diagnostic surveys were conducted in seven districts of Tripura and polymerase chain reaction based detection established that BBTD is widely prevalent in all parts of Tripura showing an average incidence of 22.02%. Among the cultivars, maximum BBTV infection (27.03%) was recorded in ‘Chini Champa’, followed by plantain (24.29%). A representative population (31 isolates) of BBTV from Tripura was characterized based on DNA R and DNA S. Phylogenetic analysis based on BBTV DNA R and DNA S generated two broad clusters of Pacific-Indian Oceans (PIO) and south-east Asian groups including all Tripura isolates within PIO cluster. The clustering pattern and genetic diversity of BBTV population from Tripura suggested monophyletic origin of majority of representative isolates from a common ancestor of PIO group. The exchange of vegetative propagules within and in between countries could have contributed to the geographical expansion of PIO isolates in Tripura. However, four variant BBTV isolates has been identified from North Tripura and Khowai districts possessing somewhat unique variability than that of distinct isolate (BBTV-Umiam) reported from NE India (Meghalaya).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana bunchy top disease (BBTD) caused by banana bunchy top virus (BBTV) is the most devastating viral disease in banana, causing severe crop losses in many banana-growing regions outside the Americas [5, 15, 19]. The disease has resulted in reductions in both productivity and in the area planted [5]. The spread of BBTV occur primarily via distribution of infected planting materials such as suckers and corms and tissue cultured plants [6, 25]. This virus is also transmitted by banana aphid (Pentalonia nigronervosa) in a circulative (persistent) non propagative manner [1, 16].
BBTV belongs to the genus Babuvirus in the family Nanoviridae [13]. It has a multipartite genome consisting of six circular single stranded DNA (cssDNA), each having approximately 1.1 kb length [3]. Each component is encapsidated individually within separate icosahedron virions [27]. Generally each of the six cssDNA components have an open reading frame (ORF) encoding different protein in the virion sense strands [13] such as rolling-circle replication initiation protein (Rep; encoded on DNA R), a protein with unknown function (encoded on DNA U3), a capsid protein (CP; encoded on DNA S), a movement protein (MP; encoded on DNA M), a cell cycle link protein (Clink; encoded on DNA C), a nuclear shuttle protein (NSP; encoded on DNA N). All the components contain a highly conserved, major common region (CR-M) and a stem-loop common region (CR-SL) downstream of CR-M [3]. Few BBTV isolates also carry a satellite DNA component that encodes a protein homologous to Rep encoded in DNA R [9].
Worldwide, BBTV isolates are grouped into two different lineages, Pacific-Indian Oceans (PIO) group and South-East Asian (SEA) group on the basis of phylogenetic relationships amongst DNA R sequences having ~ 1.9–3.0% intra and ~ 10.0% inter-group sequence diversity [12, 28]. The PIO group represents the isolates from Australia, Egypt, Cameroon, Gabon, Democratic Republic of Congo and Angola, Hawaii, India, Myanmar, Pakistan, Sri Lanka and Tonga and the SEA group comprises the isolates from China, Indonesia, Japan, Philippines, Taiwan, Sumatra and Vietnam. The BBTV isolates reported so far from India have been characterized as the members of the PIO group [2, 12, 20, 24]. Only three complete BBTV genome (comprising all six components) sequences have been reported so far from India, one from northern India (BBTV-Lucknow) [24], one from southern India (BBTV-Tamil Nadu) [20] and one from north-eastern (NE) region of India (BBTV-Umiam) [2]. In general, the genetic diversity of BBTV isolates within country is very low [15, 20, 24]. However, the recently reported BBTV isolate from Meghalaya (BBTV-Umiam), India was identified as the most distinct member of the PIO group [2], indicating differential evolution of BBTV in NE India. The recent study on global distribution of BBTV involving 855 newly sequenced full genome components together with all available complete genome sequences indicated that the Indian subcontinent, SEA and the far East are the current BBTV diversity hotspots [21]. It was also suggested that the current distribution of the virus was primarily attained through infrequent movement events over the past 300 years primarily from its diversity hotspots in India and SEA [21].
The NE region of India, comprising eight states, represents a distinct agro-climatic zone of the country. It is a bio-diversity hotspot and possesses diverse germplasm of banana (both wild and cultivated). Although the region shares the boundary with China, Myanmar and Bangladesh, it is being isolated by the hills and mountains at China and Myanmar border restricting vector movement as well as transport of planting materials. However, Tripura, the third-smallest state in the country, is situated in the South-west corner of NE India and bordering Bangladesh to the north, south, and west, and by the Indian states of Assam and Mizoram to the east. In Tripura, banana is the third important fruit crop. However, information on the incidence, distribution and molecular characteristics of BBTV occurring in the state is limited. Therefore, the current study was initiated to determine the distribution and diversity of BBTV population occurring in Tripura. The BBTV isolates from Tripura were characterized on the basis of DNA R and DNA S and compared with those of earlier reported BBTV isolates. Further, we have studied the phylogenetic grouping and population genetics of BBTV from Tripura to understand the evolutionary trend.
Materials and methods
Survey
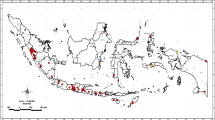
The BBTD was surveyed in several banana growing areas of Tripura, India during 2014–2015 (Fig. 1). Sampling was carried out from commercial orchards, as well as, from roadside banana and residential plantations. The geographical coordinates of each sampled location were recorded using a hand held global positioning recorder. In each district, six to eight locations were surveyed and in each location 5–20 banana mats were observed for “bunchy top” like symptoms. For BBTV diagnosis, district wise 30–70 leaf samples including both symptomatic and symptom-less were collected from the third uppermost leaf from the apex. Samples were stored in plastic bags and transferred to the laboratory after which they were processed immediately. Wherever possible, growers were interviewed in order to gather information on BBTD in specific locations.
Detection and characterization of BBTV isolates from Tripura
Total DNA was extracted using a plant DNA extraction kit (Qiagen, CA). The presence of BBTV was confirmed through polymerase chain reaction (PCR) using primer pairs specific to DNA R [24]. All the samples those tested negative were reanalysed using lower and higher concentrations of DNA to eliminate the possibility of low and high template concentration leading to the negative result.
After initial detection, a total of 31 isolates were selected as the representative population of BBTV from Tripura covering different geographical locations and different varieties found in the state. The BBTV isolates were characterized based on full length DNA R and DNA S. The PCR-mediated fragment amplification was performed to achieve the complete genomic sequence. Two pairs of overlapping primers were used to achieve the full length DNA R and DNA S of BBTV from Tripura. The first pair of primers and PCR conditions for amplification of full-length DNA R and DNA S was similar as described by Vishnoi et al. [24]. The second set of overlapping primer (both forward and reverse) for DNA R (Rep1F 5′ATGGCGCGATATGTGGTATG3′ and Rep1R 5′CGCATATCCTGTATGACATC3′) and DNA S (CPF 5′AAGGTGAAGCCCGGAAGAAT3′ and CPR 5′GTTCTGGTAGTTTATACTTACTCC3′) was designed from the sequence of previously reported BBTV-Lucknow [NCBI Accession # DQ256267 (DNA R) and EF867856 (DNA S)] having the overlapping region of > 100 bp and fragment length of ~ 300 bp. The PCR amplicons of each component were purified from gels using GeneJET gel extraction kit (Fermentas, India) and sequenced bi-directionally (BioLink, New Delhi, India).
Sequence analysis
The DNA sequences of purified PCR products of ~ 1100 and ~ 300 bp for DNA R and DNA S were assembled and evaluated using the BioEdit (Version 7.1.9) software package. The identity and homology of the sequences were first evaluated using the BLASTN program from the NCBI website (www.ncbi.nlm.nih.gov). The genome organization of BBTV DNA R and DNA S of Tripura isolates has been determined considering BBTV-Lucknow [24] as a standard representative. Finally, the complete DNA sequences of each isolate have been deposited in the NCBI Sequence Database designating the complete sequence of DNA R (KR350588-KR350618) and DNA S (KT180271-KT180301).
Phylogenetic analysis
The phylogenetic analysis of DNA R and DNA S was performed to understand the genetic grouping of BBTV isolates from Tripura. The complete nucleotide sequences of DNA R and DNA S of all Tripura isolates were compared with full length BBTV DNA R (127 sequences) and DNA S (96 sequences) available in GenBank representing the global population (Supplementary Table 1). Two full length Abaca bunchy top virus (ABTV) DNA R and DNA S were used as out group member. Sequences were aligned using ClustalW algorithm of MEGA6 (www.megasoftware.net), each beginning at the origin of replication (TATTAC). The Phylogenetic tree for DNAR and DNA S was constructed on the matrices of aligned sequences with 1000 bootstrap replicates following neighbour-joining phylogeny of MEGA6 [23].
Genetic diversity analysis
The genetic distance of the BBTV population from Tripura with those of SEA, PIO and Indian isolates (Supplementary Table 1), as well as, within Tripura districts was calculated to assess the spatial variability. The average genetic distance of DNA R and DNA S was calculated by MEGA6 using the maximum composite likelihood (MCL) method and expressed as the average number of nucleotide substitutions per site in each pair of sequences [22].
Population genetic studies
The population genetic parameters were estimated using the DnaSP software version 5.10 [18] including segregating sites (S), haplotype diversity (Hd; the frequency and number of haplotypes in a population), the statistic θ from the number of S [26], the average number of nucleotide differences between the sequences per non-synonymous site (dN) and synonymous site (dS). Depending on dN/dS values, the selection pressure was considered negative or purifying (dN/dS < 1), neutral (dN/dS = 1), and positive or diversifying (dN/dS > 1) for the Rep and CP gene data sets of BBTV population from each district.
Results
Distribution pattern of BBTV in Tripura
A total of 327 samples were collected for detection of BBTV from 48 locations in seven districts of Tripura viz., North Tripura, Dhalai, Khowai, West Tripura, Shipahijala, Gomati, and South Tripura. The detail information about the collected samples is presented in Supplementary Table 2. Banana varieties found in the state are ‘Sabri’ (AAB), ‘Chini Champa’ (AAB), ‘Bangla’ (AB), ‘Dwarf Cavendish’ (AAA), as well as, plantain (AAB). However, ‘Sabri’ was widely grown in five districts (Supplementary Table 2). Typical BBTD symptom (“bunchy top” appearance) were observed in all varieties except ‘Dwarf Cavendish’, while young suckers usually exhibited clear symptoms and provided the best evidence of BBTD occurrence in the field.
The PCR assay confirmed BBTV occurrence in 35 of 48 locations in all the seven districts surveyed in Tripura (Supplementary Table 3). During survey, virus incidence in affected orchards was recorded as 20–50% based on symptomatology. However, the PCR testing of collected samples indicated BBTV infection in 22.02% (72 out of 327) of the samples tested and the infection percentage ranged from 11.76% in South Tripura to 40.63% in North Tripura (Supplementary Table 3). Moreover, PCR assay detected BBTV in some non-symptomatic samples mainly collected from older plants (4–6 year old). BBTV was detected in all the banana samples surveyed and tested from Tripura except in ‘Dwarf Cavendish’ (symptom-less). Maximum BBTV infection (27.03%) was recorded in ‘Chini Champa’ (10 out of 37 collected samples), followed by 24.29% infection in plantain (17 out of 70 collected samples) and 21.43% infection in ‘Bangla’ (9 out of 42 collected samples). While ‘Sabri’ being the popular variety in Tripura showed only 18.94% BBTV infection as PCR assay detected BBTV in 25 samples out of 132 collected samples. Overall, the results indicated widespread distribution of BBTV in Tripura.
Genomic features of BBTV from Tripura
The 31 representative isolates from different geographical locations, as well as, from different varieties were characterized on the basis of full length DNA R and DNA S components (Table 1). The genome organization of BBTV DNA R and DNA S of Tripura isolates has been determined considering BBTV-Lucknow as a standard representative. All the sequences were arranged in a fashion to start with the origin of replication (TATTAC). All the BBTV isolates from Tripura possessed identical features in DNA R and DNA S. The total length of DNA R and DNA S of all BBTV isolates from Tripura was 1111 and 1075 nucleotide, respectively. The BBTV DNA R encoded two open reading frames (ORFs) from 5′- to 3′-end. The first ORF (87-947 nucleotide) encoded the Rep gene of 33.5 kDa (287 amino acids), while the second small ORF (388-516 nucleotide) with unknown function (Un) encoded a small polypeptide of 43 amino acids (5.2 kDa). All the DNA S of BBTV from Tripura possessed a single ORF (198-725 nucleotide) encoding the CP gene of 20.1 kDa (176 amino acids). In each case, the ORFs were flanked by a putative TATA box and a poly A signal sequences.
Analysis of sequence data confirmed the existence of CR-M and CR-SL in the intergenic region of DNA R (948-86 nucleotide) and DNA S (726-197 nucleotide) in all BBTV isolates from Tripura. The 64 nucleotide long CR-M of DNA R was located 41 nucleotide downstream of Rep gene spanning 985-1049 nucleotide, while the 90 nucleotide long CR-M of DNA S was located at 771-860 nucleotide. The second conserved region (CR-SL) was found to be 69 nucleotide in both DNA R (1072-29) and DNA S (1036-29). In silico analyses revealed the existence of stem-loop sequences in CR-SL with highly conserved nonanucleotide, TATTATTAC, within each isolate. The presence of three iterated sequences such as F1, F2 (GGGAC) and R (GTCCC), acting as Rep sequence-specific binding motifs was also identified within the CR-SL region. Overall, the genomic components of BBTV DNA R and DNA S of Tripura isolates were found to be similar to that of BBTV-Lucknow.
Phylogenetic analysis
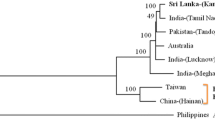
The phylogenetic analysis of DNA R and DNA S was performed to understand the molecular grouping of BBTV isolates from Tripura. The complete nucleotide sequences of DNA R and DNA S were compared with full length reference sequences available in GenBank including two full length ABTV DNA R and DNA S as outgroup member. The grouping of the virus isolates used in the current study has been presented in Figs. 2 (for DNA R) and 3 (for DNA S). All BBTV isolates clustered separately to that of the ABTV isolates. Two sub-clusters, corresponding to PIO group and SEA group were found within the BBTV isolates both for DNA R and DNA S. The BBTV isolates from Tripura clustered along with the PIO isolates (Figs. 2, 3). They showed genetic distance of 10.19% (DNA R) and 12.22% (DNA S) with SEA group. Within the PIO cluster Tripura isolates formed two sub-clusters for DNA R (Fig. 2). The first sub-cluster was composed of 27 isolates including all isolates from Dhalai, West Tripura, Shipahijala, Gomati, South Tripura, one isolate from North Tripura (Trp-N4) and one isolate from Khowai (Trp-KH4). On the other hand, the second sub-cluster was composed of four isolates, three from North Tripura (Trp-N3, Trp-N5, Trp-N6) and one from Khowai (Trp-KH1). The phylogenetic tree based on DNA S showed four sub-clusters of Tripura isolates within PIO group (Fig. 3), where first sub-cluster showed close association with isolates from Myanmar, India, Australia and Africa. The second and third cub-clusters grouped with Pakistan isolates. Interestingly, the four isolates (Trp-N3, Trp-N5, Trp-N6 and Trp-KH1) forming the second sub-cluster in DNA R also showed similar grouping in case of DNA S (fourth sub-cluster) including a new isolate from Gomati (Trp-GO8). These findings suggest a possible monophyletic origin of majority of BBTV isolates present in Tripura. However, the grouping of four isolates (Trp-N3, Trp-N5, Trp-N6 and Trp-KH1) in close association with distinct PIO isolate from Meghalaya (BBTV-Umiam) at the distant end of PIO cluster indicated possibility of variants of BBTV in North Tripura and Khowai (Figs. 2, 3).
Circular cladogram representing phylogenetic relationship based on nucleotide sequences of DNA R of 31 BBTV isolates from Tripura along with previously reported PIO and SEA isolates of BBTV rooted with ABTV. The evolutionary history was inferred using the neighbour-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (shown only when > 50%). The evolutionary distances were computed using the ‘Maximum Composite Likelihood’ method and are in the units of the number of base substitutions per site. Each sequence is labelled with the GenBank accession number followed by origin and isolate name. The blue coloured branches indicate the ABTV isolates, the orange coloured branches indicate PIO isolates and the green coloured branches indicate SEA isolates of BBTV
Circular cladogram representing phylogenetic relationship based on nucleotide sequences of DNA S of 31 BBTV isolates from Tripura along with previously reported PIO and SEA isolates of BBTV rooted with ABTV. The evolutionary history was inferred using the neighbour-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (shown only when > 50%). The evolutionary distances were computed using the ‘Maximum Composite Likelihood’ method and are in the units of the number of base substitutions per site. Each sequence is labelled with the GenBank accession number followed by origin and isolate name. The blue coloured branches indicate the ABTV isolates, the orange coloured branches indicate PIO isolates and the green coloured branches indicate SEA isolates of BBTV
Genetic diversity
The BBTV isolates from Tripura shared comparatively higher nucleotide variations for DNA R (4.48%) and DNA S (4.70%) with BBTV-Umiam compared to the BBTV isolates reported from rest of India (DNA R: 2.31% and DNA S: 4.23%) (Supplementary Table 4). Thus, Tripura isolates were more identical with the isolates reported from the rest of India rather than the distinct isolate reported from NE India (Meghalaya). However, the isolates from North Tripura (DNA R: 1.21–4.86%; DNA S: 1.98–9.24%) and Khowai (DNA R: 01.21–04.17%; DNA S: 02.08–07.06%) shared comparatively higher nucleotide variation with isolates from rest of India. Moreover, the North Tripura and Khowai isolates recorded higher nucleotide variability in DNA R (3.29 and 3.42%, respectively) and DNA S (5.50 and 5.35%, respectively) even within district (Supplementary Table 5). Further comparison showed higher nucleotide variability in Rep gene (0.64–1.21%) than the intergenic region (0.48–0.60%) in case of majority of the identical BBTV isolates from Dhalai, West Tripura, Shipahijala, Gomati and South Tripura (Supplementary Table 5). The Rep gene of those identical isolates showed higher amino acid variability (0.66–1.75%) than the nucleotide variability (0.64–1.21%) indicative of non-synonymous substitution in Rep gene of majority of the BBTV isolates from Tripura. Interestingly, the variant isolates identified from North Tripura and Khowai showed higher nucleotide variability in the intergenic region (on an average 6.79 and 4.12%, respectively) than the Rep gene (on an average 2.31 and 3.22%, respectively) (Supplementary Table 5). Moreover, the Rep gene of those variant isolates from North Tripura and Khowai showed higher nucleotide variability (on an average 2.31 and 3.22%, respectively) than the amino acid variability (on an average 1.39 and 3.10%, respectively) indicating the synonymous substitution in Rep gene of the variant isolates identified from North Tripura and Khowai.
Population genetics and selection pressure
District wise population genetic parameters in each coding region are provided in Supplementary Table 6. Haplotype diversity values were high for both coding regions (Rep and CP) ranging from 0.833 to 1.000. Similarly, both Rep (0.0–3.09%) and CP (0.0–3.18%) showed similar nucleotide variation (Supplementary Table 5). However, it was evident that the BBTV population from North Tripura and Khowai had higher diversity than the other areas. Similarly, the statistic θ (S) value was greater for the variant BBTV population from North Tripura (Rep: 0.034 and CP: 0.055) compared to those from other areas (0.007–0.023). The population genetic parameters couldn’t be analysed for Khowai district as only two isolates were available.
The pattern of selection constraint on the coding region of DNA R and DNA S was estimated by analysing the dN/dS ratio (Supplementary Table 6). In case of Rep gene, majority of the BBTV population from Tripura showed neutral to positive or diversifying selection pressure, while a negative or purifying selection appeared to be acting on the CP gene except the population from West Tripura (Supplementary Table 6). However, the variant BBTV population from North Tripura showed negative selection for both Rep (0.091) and CP (0.124). This indicated variable degree of selection pressure depending on the area and coding regions possibly because of the diversity of their functions.
Discussions
In this study, we have investigated the distribution pattern and genetic structure of BBTV in NE Indian states of Tripura. Overall results indicated wide spread distribution of BBTD in all parts of Tripura with an average incidence of 22.02%. A diverse range of banana and plantain cultivars (AAA, AAB, AB) known by different local names were observed in various regions. Typical BBTD symptom was identified in all (AAB, AB) except ‘Dwarf Cavendish’ (AAA) and PCR detection confirmed BBTV infection in all cultivars except ‘Dwarf Cavendish’. Generally, cultivars with the B genome (AAB and ABB) were found to be less susceptible than those with the A genome (AA and AAA) [11]. However, the negative reaction in ‘Dwarf Cavendish’ was due to lack of infection, as none of the cultivars of banana, plantain and hybrids were found resistant to BBTV [11].
The comparison of 31 sequence data of DNA R and DNA S components of BBTV from Tripura revealed identical genomic features as of earlier reported BBTV isolate from India (BBTV-Lucknow). The existence of hairpin structure with highly conserved nonanucleotide, TATTATTAC, within the loop of each BBTV isolate from Tripura confirmed the localization of origin of BBTV virion-strand replication in CR-SL similar to that of ssDNA viruses like geminiviruses [7]. Three iterated sequences acting as Rep sequence-specific binding motifs, was also found to be present in CR-SL as reported earlier [8].
The BBTV isolates from Tripura was supposed to be a PIO group member due to its geographical origin (Tripura, India). The clear cut separation of BBTV isolates into two groups has also been reported by several workers [2, 12, 20, 24, 28]. In general, the genetic diversity of BBTV isolates within the country is very low [15]. However, in India, relatively greater diversity for BBTV was observed in the NE region [2] as the isolate from Meghalaya (BBTV-Umiam) was identified as the most distinct member of the PIO group. In the current study, BBTV isolates from Tripura grouped within PIO cluster regardless of genomic component under consideration. However, Tripura isolates of BBTV formed two sub-clusters for DNA R and four sub-clusters for DNA S within the PIO group. The sub-clustering pattern suggested that majority of BBTV population in Tripura perhaps came from a common ancestor that was introduced subsequently to this region from rest part of India and neighboring countries through infected planting materials. Further dissemination of BBTV through infected propagules and spread by the banana aphid vector could be responsible for the genetic homogeneity of the Tripura isolates. However, four isolates, three from North Tripura (Trp-N3, Trp-N5, Trp-N6) and one from Khowai (Trp-KH1) always formed a sub-cluster in close association with distinct BBTV-Umiam for both DNA R and DNA S indicating presence of variants of BBTV in North Tripura and Khowai.
Tripura isolates showed on an average 10.19% (DNA R) and 12.22% (DNA S) nucleotide variation with SEA group, while they showed only 2.79% (DNA R) and 3.49% (DNA S) nucleotide sequence variation with PIO group. Earlier studies on PIO and SEA isolates of BBTV also reported ~ 10% inter-group and ~ 1.9–3.0% intra group sequence variation [12]. BBTV-Umiam showed relatively higher nucleotide diversity with PIO group for both the genomic components (4.60–15.00%) and ORFs (2.50–6.20%) resulting the grouping of each genomic component of BBTV-Umiam at the distant end of PIO cluster [2]. Thus, Tripura isolates were more identical with the BBTV isolates reported from rest of India rather than that from NE India, while BBTV isolates from North Tripura (DNA R: 3.30% and DNA S: 6.35%) and Khowai (DNA R: 2.53% and DNA S: 4.60%) shared comparatively higher nucleotide variation even with isolates from rest of India. Both phylogeny and genetic variability supported the variable nature of four isolates (Trp-N3, Trp-N5, Trp-N6 and Trp-KH1) from Tripura. Moreover, the isolates from North Tripura (DNA R: 4.36–4.96% and DNA S: 4.81–7.34%) and Khowai (DNA R: 4.46–4.55% and DNA S: 4.62–5.01%) showed similar range of nucleotide variation with distinct BBTV isolate as of other isolates characterized from rest part of Tripura (DNA R: 2.99–5.54% and DNA S: 3.15–5.21%). Therefore, the genetic variability of the variant isolates identified from North Tripura and Khowai was somewhat unique than that of BBTV-Umiam.
The dN/dS ratio is an estimator of the evolutionary constraints imposed on a coding region at the intra-specific level and a dN/dS ratio of > 1 is considered as evidence for positive selection pressure [10]. It was observed that different selection pressure appears to be acting on coding regions of BBTV population from Tripura as of earlier reports [4, 14]. This indicated independent evolution of Rep and CP. Moreover, the CP gene of BBTV population from Tripura appeared to be under purifying selection except the isolates from West Tripura. Since BBTV is transmitted by aphid in a persistent manner, it is acceptable that such constraint is imposed on the CP gene in order to avoid the accumulation of deleterious mutations that might interfere with the complex virus-vector interactions [14]. Hypothetically, the Rep gene of BBTV should be under purifying selection considering DNA R as the minimal replicative unit [9]. However, the dN/dS ratio of Rep gene of majority of isolates from Tripura suggested diversifying selection as of previous report from sub-Saharan Africa [14]. Only, the Rep gene of variant BBTV isolates from North Tripura was identified to be under purifying selection similar to the SEA isolates reported from Sumatra Island, Indonesia [4].
Generally, the BBTV isolates in the PIO group are distributed across the natural geographical range of Musa balbisiana, whereas the SEA group isolates occur across M. balbisiana and M. acuminata range. However, the banana germplasm of NE India comprises mostly the hybrids of M. balbisiana from Indian subcontinent and M. acuminata from south-east Asia [17]. The identification of the most divergent BBTV isolate from NE Indian state of Meghalaya indicated differential evolution of BBTV in NE India [2]. The geographic structuring of global BBTV populations confirmed India as one of the current global hotspots of BBTV diversity [21]. The current study provided strong molecular support for presence of unique variants of BBTV in North Tripura (Trp-N3, Trp-N5, Trp-N6) and Khowai (Trp-KH1), which in turn strengthened the possibility of differential evolution of BBTV in NE India. However, the monophyletic origin of majority of BBTV isolates from Tripura indicated introduction and expansion of PIO isolates from other parts of India, as well as, from neighbouring countries (Bangladesh, Myanmar). According to the information collected from the local farmers during survey, most of the cultivars of banana existing in Tripura are also cultivated in bordering Bangladesh. Although, till date no authentic report has been found on molecular characteristics of BBTV from Bangladesh, but the neighbouring country Myanmar showed presence of PIO isolates of BBTV. DNA S based phylogeny showed close association of some BBTV isolates from Tripura with Myanmar isolates. Therefore, a through survey and detail analysis involving representative BBTV isolates from entire NE India will help in proper understanding on evolution of BBTV in this distinct agro-climatic zone.
References
Anhalt MD, Almeida RPP. Effect of temperature, vector life stage, and plant access period on transmission of Banana bunchy top virus to banana. Phytopathology. 2008;98:743–8.
Banerjee A, Roy S, Behere GT, Roy SS, Dutta SK, Ngachan SV. Identification and characterization of a distinct Banana bunchy top virus isolate of Pacific-Indian Oceans group from North-East India. Virus Res. 2014;183:41–9.
Burns TM, Harding RM, Dale JL. The genome organization of Banana bunchy top virus: analysis of six ssDNA components. J Gen Virol. 1995;76:1471–82.
Chiaki Y, Nasir N, Herwina H, Sonoda A, Fukumoto T, Nakamura M, Iwai H. Genetic structure and diversity of the Banana bunchy top virus population on Sumatra Island, Indonesia. Eur J Plant Pathol. 2015;143:113–22.
Dale JL. Banana bunchy top: an economically important tropical plant virus disease. Adv Virus Res. 1987;33:301–26.
Drew RA, Moisander JA, Smith MK. The transmission of Banana bunchy top virus in micropropagated bananas. Plant Cell Tissue Organ Cult. 1989;16:187–93.
Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Biochem Mol Biol. 2000;35:105–40.
Herrera-Valencia VA, Dugdale B, Harding RM, Dale JL. An iterated sequence in the genome of Banana bunchy top virus is essential for efficient replication. J Gen Virol. 2006;87:3409–12.
Horser CL, Harding R, Dale J. Banana bunchy top nanovirus DNA-1 encodes the ‘master’ replication initiation protein. J Gen Virol. 2001;82:459–64.
Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–7.
Jose PC. Reaction of different varieties of banana against bunchy top disease. Agric Res J Kerala. 1981;1981(19):108–10.
Karan M, Harding RM, Dale JL. Evidence for two groups of Banana bunchy top virus isolates. J Gen Virol. 1994;75:3541–6.
King AM, Adams MJ, Lefkowitz EJ, Carstens EB, Ball LA. Virus taxonomy: IXth report of the international committee on taxonomy of viruses. London: Elsevier Academic Press; 2011.
Kumar PL, Hanna R, Alabi OJ, Soko MM, Oben TT, Vangu GHP, Naidu RA. Banana bunchy top virus in sub-Saharan Africa: investigations on virus distribution and diversity. Virus Res. 2011;159(2):171–82.
Kumar PL, Selvarajan R, Iskra-Caruana ML, Chabannes M, Hanna R. Biology, etiology, and control of virus diseases of banana and plantain. Adv Virus Res. 2015;91:229–69.
Magee CJP. Transmission studies on the banana bunchy-top virus. J Aust Inst Agric Sci. 1940;6:109–10.
Molina AB, Kudagamage C. The international network for the improvement of banana and plantain (INIBAP): PGR activities in South Asia. In: South Asia network on plant genetic resources (SANPGR) meeting held on December 9–11 at Plant Genetic Resources Center (PGRC), Peradeniya, Sri Lanka; 2002. pp. 1–7.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–7.
Rybicki EP. A top ten list for economically important plant viruses. Arch Virol. 2015;160:17–20.
Selvarajan R, Mary Sheeba M, Balasubramanian V, Rajmohan R, Lakshmi Dhevi N, Sasireka T. Molecular characterization of geographically different banana bunchy top virus (BBTV) isolates in India. Indian J Virol. 2010;21(2):110–6.
Stainton D, Martin DP, Muhire BM, Lolohea S, Halafihi M, Lepoint P, Blomme G, Crew KS, Sharman M, Kraberger S, Dayaram A, Walters M, Collings DA, Mabvakure B, Lemey P, Harkins GW, Thomas JE, Varsani A. The global distribution of Banana bunchy top virus reveals little evidence for frequent recent, human-mediated long distance dispersal events. Virus Evol. 2015;1(1):1–16.
Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;2004(101):11030–5.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Vishnoi R, Raj SK, Prasad V. Molecular characterization of an Indian isolate of Banana bunchy top virus based on six genomic DNA components. Virus Genes. 2009;38(2):334–44.
Wardlaw CW. Mosaic, Infectious chlorosis and other virus diseases. Banana diseases, including plantains and abaca. London: Longmans; 1961.
Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–76.
Wu RY, Su HJ. Purification and characterization of Banana bunchy top virus. J Phytopathol. 1990;125:153–60.
Yu NT, Zhang YL, Feng TC, Wang JH, Kulye M, Yang WJ, Liu ZX. Cloning and sequence analysis of two Banana bunchy top virus genomes in Hainan. Virus Genes. 2012;44:488–94.
Acknowledgements
This work is partially carried out under M.Sc. thesis programme by Tanmoy Das. The authors are thankful to Dr. Satish Chandra, Dr. G. T. Behere and Dr. T. Rajesh for their critical suggestions during the work. The work was supported by IXX09855 project of Indian Council of Agricultural Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Das, T., Banerjee, A. Distribution, molecular characterization and diversity of banana bunchy top virus in Tripura, India. VirusDis. 29, 157–166 (2018). https://doi.org/10.1007/s13337-018-0451-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-018-0451-7