Abstract
Spot blotch (SB) is a major constraint to wheat (Triticum aestivum L.) production in South Asia's warmer plains. It is also known as leaf blight, caused by Bipolaris sorokiniana and causes significant yield loss in Eastern Gangetic Plain Zone of India. The aim of this study was to map SB resistance via composite interval mapping (CIM) in the PBW343/IC252874 population, which comprised of 165 doubled haploid lines (DHLs), across two years in India. The area under the disease progress curve (AUDPC) value for each genotype was calculated using the disease severity. The area under the disease progression curve was developed to determine disease progression. The SB severity was measured at the three different plant growth stages viz. GS63, GS69, and GS77. The phenotypic analysis of these lines revealed a constant variance in disease severity, implying that SB resistance is most likely polygenic. The phenotypic data was used to map Quantitative trait loci (QTL) using SSR markers. The presence of quantitative inheritance with transgressive segregation for SB resistance in the population was also revealed. The QTLs were discovered on 12 chromosomes i.e. 1B, 1D, 2A, 2B, 2D, 3B, 4A, 4D, 5A, 5B, 6A, and 7A. We have also found two consistent QTLs on the chromosomes, 2B and 5B with the average percentage variance explained (PVE) of 17.9% and 19.9% respectively. These current finding reveals new genomic regions linked to spot blotch disease, which could be useful for wheat breeding strategies considering disease resistance with further validation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
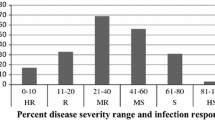
It is necessary to expedite wheat improvement in order to fulfil estimated world food demand by 2050. The latest United Nations projections indicate that world population will reach 10 billion in the year 2056 (Prakash et al. 2019). Spot blotch disease is a significant biotic factor limiting wheat productivity in the warm humid agro-ecosystem of India (Fig. 1), Bangladesh, Nepal, and other such regions of the world (Gurung et al. 2014; Singh et al. 2018). Bipolaris sorokiniana, (Sacc.) Shoemaker [teleomorph: Cochliobolus sativus (Ito and Kuribayashi) Drechsler ex Dastur], a hemi-biotrophic fungus, is the causal agent for the disease. It affects more than 25 million ha of wheat growing area globally. Being a prominent disease in eastern India and adjoining countries of South Asia it threatens the livelihood of many resource poor and marginal farmers of this densely populated region (CIMMYT 2013). Reductions in grain yield due to spot blotch are variable but substantial in warmer areas of South Asia (Saari 1998; Joshi et al. 2007b, a; Duveiller and Sharma 2009). Delayed seeding of wheat, after rice in this part of the world also results in higher losses of grain yield and total kernel weight due to the spot blotch (Duveiller et al. 2005). The disease becomes more severe when the temperature goes beyond 28 °C (Nema and Joshi 1973; Chaurasia et al 2000, Duveiller et al. 2005). This leads to necrosis induced premature leaf senescence thus reduced photosynthetic area (Sharma et al. 2007), and infection in spikes resulting in grain shriveling, black point of the kernels (Sharma et al. 2007; Kumar et al. 2007), affecting both the quantity and quality of the grain negatively. Yield losses are estimated in the range of 1 to 100% (Dubin and Van Ginkel 1991; Siddique et al. 2006; Sharma et al. 2007; Mehta et al. 1998; Saari 1998) depending on the genotypes, sowing time, environmental conditions, soil fertility stresses and soil moisture conditions. Differential responses of wheat genotypes to spot blotch resistance have been reported by Chaurasia et al. 2000. Among the disease management approaches used, the deployment of resistant genotypes is the most effective, economical and sustainable approach for containing the spot blotch in the field (Duveiller and Sharma 2009).
Genetic studies have revealed both the qualitative and quantitative nature of resistance to spot blotch disease. Qualitative resistance is reported to be governed by 2 dominant complementary genes (Srivastava et al., 1971) dominant or partially dominant genes (Adlakha, 1984; Velázquez Cruz et al., 1994; Sharma and Bhatta, 1999; Neupane et al., 2007), and recessive genes (Singh et al. 1998a, 2019; Bhushan et al. 2002; Ragiba et al. 2004). The study by Sharma et al. (1997) has established the quantitative nature of resistance for the disease. Further, Joshi et al. (2007b, a) have reported resistance to spot blotch as polygenic and partial with additive inheritance. Molecular breeding based on marker-assisted selection is a key tool for accelerating the development of the spot blotch-resistant variety. For this, identification of a molecular marker closely linked to the resistance loci is very critical (Duveiller and Sharma 2009). The association mapping approach has been applied to wheat, facilitating the discovery of many quantitative trait loci (Vasantrao et al. 2019). There quite a few studies, which have identified environment-specific QTLs and associated molecular markers in specific biparental mapping populations (Sharma et al. 2007; Kumar et al. 2010, 2015; Singh et al. 2019; Gahtyari et al. 2021; Roy et al. 2021). Genetic variability exists for the pathogen in the area of present investigation, which is also considered a hot spot for the spot blotch disease (Verma et al. 2020). However, none of the study investigated intensively to identify QTL conferring tolerance for the disease in the agro-ecology of this investigation, which is dominated by resource poor and marginal farming community. Hence, the major objective of the current study was to identify QTL associated with the disease tolerance in the agro-ecology of Pusa Bihar using Doubled haploid PBW343/IC252874 population keeping in view the Area Under Disease Progress Curve (AUDPC) for the disease.
Plant materials and experimental set up
A doubled haploid population, derived from a cross between PBW343 (disease resistant or maternal parent) and IC252874 (disease susceptible or paternal parent) (Table S1) was sown in two experiments over two years viz. 2017–18 and 2018–19. The population contained 165 DH lines. The seeds were sown in alpha lattice design with three replications. Proper agronomic practices and irrigation were provided to avoid yield reduction during the crop cycle. The development or creation of the DH was done in the Washington State University, Pullman, USA (Table S2), and disease assessment was carried out at the research farm of the Rajendra Prasad Central Agricultural University, Pusa, Bihar, India (25° 57′ 08″ N; 85° 40′ 13″ E). An offseason facility at the Research station of Punjab Agricultural University, Keylong, Himachal Pradesh, India, was utilized for the initial seed multiplication.
Irrigation and management practices
Throughout the two years, the required agronomic methods for irrigated and normal fertility (125 kg N; 55 kg P2O5 and 35 kg K2O ha−1) conditions were followed. At the time of seeding, full doses of K2O and P2O5 were administered. The split application of nitrogen was used, with 55 kilogramme N ha−1 applied at sowing, 25 kg N ha−1 applied at first irrigation (20 days after sowing), and 35 kg N ha−1 used at second irrigation (42 days after sowing). To ensure optimum soil moisture, irrigation was applied to the crop at five critical growth stages (at crown root initiation Zadok, GS 21; tiller completion Zadok, GS 29; late jointing Zadok, GS 36; flowering Zadok, GS 61; and milk stage Zadok, GS 75). The weeds were manually picked out.
Disease scoring for spot blotch resistance
Single digit scoring
After the completion of heading in all genotypes, we noted symptoms in the flag leaf (F) and penultimate leaf (F-1) for single digit scoring. In each plot of three replications, single digit scoring was performed using the CIMMYT standard diagram.
(%) Disease severity = (sum of numeric rating/ total numbers of plant observed) × 100.
Double digit scoring
The sensitive reactions (IR = 7–9 on a 0–9 scale) and a 99 score (double-digit score) of SB severity was measured at various stages of crop in the field. The SB severity (Fig. 2) was measured at the GS63, GS69, and GS77 stages of plant development, using Saari and Prescott's double-digit (00–99) severity scale, with the first (D1) and second (D2) digits representing disease progression vertically from the ground and percentage of leaf area infected with SB, respectively.
Using the algorithm provided, the severity of disease was determined for each DH lines (Duveiller et al. 2005).
where,
Estimating area under disease progress curve (AUDPC)
The area under the disease progress curve (AUDPC) value for each genotype was calculated using the disease severity. The area under the disease progression curve was developed to determine disease progression. AUDPC was calculated independently for flag leaf (F), penultimate leaf (F-1), and double digit scoring.
n-1
where,
i.
Yi = Disease severity in the ith date
ti = Date on which the disease was scored
n = number of dates on which disease was recorded
Genotyping
The wheat Coleoptiles were used to extract DNA (http://www.triticarte.com.au/content/DNA-preparation.html). A consensus map was used to do this (Sourdille et al. 2004). In Thermal Cycler22, a 3 min denaturation at 94 °C was followed by 30 s cycles at 50/65 °C, 30 s cycles at 72 °C, and a final 2 min cycle at 72 °C. On 541 SSRs, the parents' SSR markers were evaluated for polymorphism (Fig. 3). The 130 SSR primers (Table S3) were utilized to create wheat chromosome linkage maps. The visual evaluation of polymorphic SSR profiles was done by coding PBW343 alleles as "A" and IC252874 alleles as "B." The letters "H" and "NA" stand for heterozygote and missing bands, respectively. On 3 percent agarose gels, allele bands were seen.
Linkage map construction and QTL mapping
Using the software MapDisto 2.1.7.1, the genotyped data from the DH population was utilised to create twelve linkage maps. For the conversion of recombination frequency to genetic distance, the Kosambi mapping function and interval position type were utilised. QTL Cartographer v2.5 was used to conduct the analyses (Wang et al. 2010). In the composite interval mapping (CIM) approach, the software Windows QTL Cartographer 2.5 was used to perform forward regression using five backdrop markers, a window size of 10.0 cM, and a walking speed of 2 cM. Model 6 was used to set the trait for Composite interval mapping (CIM), which was utilized to determine plausible QTL sites and a 1,000 permutation test threshold of P = 0.05. Two or more linked markers associated with a characteristic with LOD > 3.0 were identified as putative QTLs. Suggestion QTLs were classified as QTLs with two or more connected markers found at a LOD of 2.0 to 3.0. (McIntyre et al. 2010). The LOD value was set at a minimum of 2.0 in order to account for both suggestive and minor QTL.
Statistical analysis
The disease severity scores across different years were used to obtain best linear unbiased estimates (BLUEs) and predictions (BLUPs) by fitting linear mixed effects models in R v4.0.3 using where, Yik is the trait of interest, m is the mean effect, Yeari is the effect of the ith year, Linek is the effect of the kth line, and + ik is the error associated with the kth line. The genotypes were treated as fixed effects in the BLUEs model, while all of the effects in the BLUPs model were treated as random effects. When genotypes are treated as random effects, the impact of screening time and other environmental factors on SB severity is reduced (Tomar et al. 2021). To explore the distribution across the DHLs, the disease severity scores generated by fitting the BLUPs and BLUEs models were displayed using ggplot2 v3.3.3 and ggpubr v0.4.0 in R v4.0.3.
Result
Phenotypic evaluation of DHLs
The analysis of variance for AUDPC values revealed a significant variation for genotypes and genotype-by-year interaction (Table 1). Large variation in disease severity was observed across the different growth stages with disease pressure increasing from GS63, GS69 and GS77 (Table 2). Across the environments, overall disease pressures were the lowest in E1 and highest in E2. Within the same year, both populations responded similarly as observed from their comparable disease severity/AUDPC score ranges. To enhance the accuracy and map stable QTLs across the environments, linear mixed-effects models were used to obtain fitted values of disease severity, accounting for G X E effect. These values are termed as BLUPs (genotypes as random effects) and BLUEs (genotypes as fixed effects) from here onward. The BLUPs showed lower variance than the BLUEs which meant BLUPs were able to reduce the environmental variance across the years to a larger extent.
Mean spot blotch severity (%) of the disease susceptible parent (IC252874) at GS 77 stage (Zadoks scale, Zadoks et al. 1974) ranged from 80% (2017– 2018) to 87% (2018–2019) and resistant parent (PBW343) at GS 77 stage (Zadoks scale, Zadoks et al. 1974) ranged from 13% (2017– 2018) to 17% (2018–2019). The disease severity of DHLs ranged from 52% (2017–2018) to 59% (2018–2019). The continuous distribution of spot blotch AUDPC and the test of normality using Shapiro– Wilk test (W = 0.973, P = 0.327) revealed that the DHLs data fit a normal distribution. The parental lines exhibited contrasting phenotypes for spot blotch mean AUDPC for all the two years. The spot blotch AUDPC of the DHLs showing large phenotypic variation in the population.
The resistant parent PBW343 was found to be immune to SB showing highly resistant disease severity score of 00, on the double-digit scale, across all the growth stages studied whereas susceptible parent IC252874 showed high susceptibility across the growth stages and AUDPCs when compared with range of disease scores of respective data sets. At GS63, disease score BLUE of the susceptible parent IC252874 were 40.0 while for the DHLs, it ranged from 0.29 to 41.00. The disease score, BLUP of IC252874 was 24.20 with DHLs showing a range from 08.23 to 26.71. At GS69, disease score BLUEs of the susceptible parent, IC252874 was 71.43 while for the DHLs, it ranged from 09.61 to 71.32. The disease score BLUP of IC252874 was 56.21 with DHLs showing a range from 23.72 to 56.48. At GS77, disease score BLUE of susceptible parent IC252874 were 87.51 while for the DHLs, it ranged from 34.00 to 79.36. The disease score BLUPs IC25287 was 81.34, respectively, with DHLs showing a range from 40.13 to 76.14. The AUDPC values showed a similar trend, where the disease score BLUEs of the resistant parent PBW343 and susceptible parent IC252874 were 200.13 and 1400.39, respectively, while for the DHLs, it ranged from 323.45 to 1217.32. The disease score BLUPs of PBW343 and IC252874 were 221.34 and 1323.27 respectively, with DHLs showing a range from 245.61 to 1235.59. The genotypes, DH10, DH125, DH80 and DH134 were found highly resistant across all the growth stages. Overall, less than 5% of lines were categorized under highly resistant category while 25 and 30% of genotypes showed moderate to high susceptibility, respectively. The rest of the lines fell under resistant to moderately resistant category (Fig. 4). The data from different years were used separately for QTL mapping.
Linkage map
The parents ‘PBW343’ and ‘IC252874’ were screened for SSR polymorphism. Once the polymorphism was identified between the parents, 165 double haploid lines were screened with the polymorphic SSR primer pairs. The anchored markers helped to form 12 linkage groups representing wheat chromosomes in the DH mapping population. Out of 541 SSR markers, 130 (24%) marker loci segregated in the expected 1:1 ratio (P\0.05) (Table S4) with few exceptions, some marker loci did not fit the 1:1 ratio. The linkage maps consisting of 10 markers on chromosome 1B, 5 markers on1D, 12 markers on 2A, 7 markers on 2B and 13 markers on chromosome 2D, 14 markers on chromosome 3B, 15 markers on chromosome 4A, 8 markers on chromosome 4D, 12 markers on chromosome 5A, 8 markers on chromosome 5B, 13 markers each on chromosome 6A and 7A were developed. The rate of polymorphism was highest in the A genome (40%) as compared to genomes B (30%) and D (20%).
QTL detection
A total no. of eighteen QTLs was detected for spot blotch AUDPC over the two years (Table 3). We found the QTLs for spot blotch resistance on chromosomes 1B, 1D, 2A, 2B, 2D, 3B, 4A, 4D, 5A, 5B, 6A, and 7A over the course of two years (Fig. 5). The LOD values ranged from 2.61 to 15.39 (Fig. 6) and the corresponding R2 ranged from 7.43 to 21.12 in the individual years. Individual QTLs explained between 11.31 and 39.15% of phenotypic variance in the composite interval mapping. Using composite interval mapping, two most consistent QTLs mapped on the chromosome 2B and 5B flanked by the marker, Wmc109-gwm312 and Cfd71-wmc173 respectively and the alleles for reduced disease severity were derived from the resistant parent PBW343. On the other hand, QTLs present in at least single year were located on the chromosome 1D, 3B, 2D, 4A and 4D. The QTL on 2A explained the largest part of phenotypic variance in the second year (22.13%). In the first year, maximum phenotypic variation (18.75%) was controlled by the QTL located on chromosome 5B.
The PBW343/IC252874 linkage map for spot blotch resistance. QTL was discovered in both the years 2017–18-19. The Kosambi mapping function was used to calculate marker positions, which are listed in cM position from the top of each linkage group. C1-1B, C2-1D, C3-2A, C4-2D, C5-3B, C6-4A, C7-4D, C8-5A, C9-6A, C10-7A, C11-2B, C12-5B
Discussion
Spot blotch is one of the major constraints to the global wheat production, especially in areas with hot and humid climate (Tomar et al. 2021). To counter the constraints from foliar diseases like SB, there is a need for constantly identifying and introgressing new sources of resistance. The DHLs used in the present study showed wide range of variation for different traits and has already been reported to possess various QTLs for heat tolerance (Pankaj 2021; Pankaj et al. 2022a, b). In the present study, during phenotypic evaluation of disease severity for SB, four DH viz. DH10, DH125, DH80 and DH134 were identified to be highly resistant against SB. Because no wheat cultivar presently grown in North-Eestern plains of India possesses resistance to SB, these lines become an important resource for transfer of SB resistance. The phenotypic evaluation of the segregating population showed a wide range of SB severity scores from highly resistant to highly susceptible, which indicated that more than one locus for resistance was segregating in the population. The continuous distribution of spot blotch AUDPC and the test of normality using revealed that the DHLs data fit a normal distribution (Fig. 7). The distribution of 165 DHLs for spot blotch AUDPC suggested that spot blotch resistance is polygenic and not controlled by a single gene in the PBW343 and IC252874 cross. Earlier studies on the inheritance of resistance to spot blotch (Joshi et al. 2004) also suggested a polygenic control. The resistant parent, PBW343 have two genes for disease resistance and resistance was found to be dominant over susceptibility. It revealed that, for spot blotch resistance, duplicate and complementary gene effects are contributing significantly along with additive gene effects. Singh et al. (2019) unravelled the genetics and map the resistance to Tan spot and Septoria nodorum blotch in the PBW343/Kenya Nyangumi derived recombinant inbred line (RIL) population. Therefore, PBW343 could be the ideal donor or resistant parent in deciphering disease resistance. To achieve the highest possible disease pressure, sowing was carried out during the second week of December which allows the post-anthesis stage to coincide with warm temperature conducive to the disease. Similarly, Kumar et al. (2009) sown seeds in the third or fourth week of December led the post-anthesis stages to coincide with relatively higher temperature that favoured disease development. It has been reported that spot blotch disease becomes more severe when the mean temperature exceeds 26 °C (Chaurasia et al. 2000). The phenotypic evaluation of the segregating population showed a wide range of SB severity scores from highly resistant to highly susceptible, which indicated that more than one locus for resistance was segregating in the population. AUDPC was calculated using the disease severity (%) data, recorded at three growth stages (GS63, GS69, GS77). Kaur et al. (2021) also conducted disease scoring at three different growth stages (GS) on Zadoks’ scale (Zadoks et al. 1974), which are GS55 (flowering stage or FS), GS75 (medium milk/dough stage or DS), and GS87 (hard dough stage or HDS).
In this study, DHLs were classified into four groups based on days to heading and disease severity was recorded at specific growth stages when days to headings were synchronized by differential sowing of the DHLs. Hence, the problem of variation in earliness was overcome. Similarly, RILs were classified into three groups based on days to heading and disease severity was recorded at specific growth stages when days to headings were synchronized by differential sowing of the RILs (Kumar et al. 2015).
Following two years of disease recording at different growth stages, an accurate evaluation of the population for resistance to spot blotch was obtained under field conditions. The results suggest that accuracy and reproducibility of experimental conditions and of the scoring method used for spot blotch evaluation is reliable. Most of the recent studies on spot blotch (Joshi et al. 2007b, a) are also based on AUDPC. We deployed approximately 10–20 microsatellite markers covering wheat chromosomes. The ratio of polymorphic markers of nearly 30% was consistent with results of Prasad et al. (1999) and Roy et al. (1999). The order and orientation of the mapped microsatellite markers in our study was in agreement with those in the map of the ITMI population (Ganal and Ro¨der 2007). There were five loci that did not segregate in 1:1 mendelian ratio showed segregation distortion. These loci were randomly distributed throughout the genome. However, the linkage map was not affected by the distortion and we did not include these loci in the linkage map. Since only 35% of the markers were polymorphic, gaps in certain region were not covered. Across two years, we identified eighteen QTLs for spot blotch resistance on chromosomes 1B, 1D, 2A, 2B, 2D, 3B, 4A, 4D, 5A, 5B, 6A and 7A. This study supports the finding of Kaur et al. (2021), discovered five QTLs, Q.Sb.pau-2A, Q.Sb.pau-2B, Q.Sb.pau-3B, Q.Sb.pau-5B, and Q.Sb.pau-6A, linked to SB resistance were mapped across chromosomes 2A, 2B, 3B, 5B, and 6A. Similar results were obtained by Tomar et al. (2021) where Physical map of candidate QTLs for spot blotch were mapped on 1A, 1B,1D, 2A, 2D, 4A, 5B, and 6D chromosomes. Kaur et al. (2021) reported the QTL, Q.Sb.pau-5B, linked to SNP S5B_703858864, was validated on this BC2F1 population. Therefore, marker on the nearby position could prove be a potential diagnostic marker for SB resistance.
The published data of Sourdille et al. (2004), about deletion mapping of more than 700 microsatellite markers on specific chromosome segments enabled us to check the physical location of markers linked to the detected QTLs. The order and orientation of markers on the maps developed in our mapping population were also in agreement with the IWGSC physical maps (http://www.wheatgenome.org). Therefore, it could be possible to assign the QTL on the physical map. Most of the alleles for reduced disease severity were derived from the resistant parent PBW343 for the QTLs except Qsb_rpcau_2D, Qsb_rpcau_2B, Qsb_rpcau_2A, Qsb_rpcau_4D detected in the PBW343/IC252874 mapping population. Two most consistent QTLs mapped on the chromosome 2B and 5B flanked by the marker, Wmc109-gwm312 (18.4 cM) and Cfd71-wmc173 (9.4Cm) respectively. The average phenotypic variances for both the QTL were 17.9% and 19.9%. QTLs on 2B and 2D are similar to the ones reported in the Mexican conditions as deduced either by their physical positions or by the contribution of the SB resistance allele. Similarly, Kumar et al. (2009) reported two consistent QTLs mapped on the short arm of chromosome 2B and the long arm of chromosome of 5B detected in all 3 years. Tomar et al. (2021) also reported two significant chromosomal regions/QTLs on 2B and 5B that were consistent between the locations viz. Pusa, India, and Jamalpur, Bangladesh. We found a total no. of two QTLs on chromosome 1B. However, the flanking markers were different in both the years. Gurung et al. (2014) also reported a QTL on 1BL for SB in an association mapping panel. Comparing the QTL positions of both studies by a BLAST of the marker sequences to the IWGSC RefV1.0 genome sequence of Chinese Spring, indicated the QTL to be the different. The results showed that the QTL in Gurung et al. (2014) was in the proximal region of chromosome 1BL, whereas the QTL of this study was in the far distal region of chromosome 1BL. In a mapping population derived from the cross Avocet × Saar, Lillemo et al. (2013) demonstrated the major effect of pleiotropic multi-fungal resistance gene Lr46 located on chromosome 1BL on SB resistance, the former gene being named as Sb1. In our case, chromosome, 3B and 5A were also associated with SB. Similarly, Zhu et al. (2014) identified QTL for SB resistance on chromosomes 1B, 3BS and 5AL. A QTL, Qsb_rpcau_6A was found on chromosome 6A in a single year only. The QTL is flanked by gwm570-wmc553 and accounted for the phenotypic variance of 12.52%. In a review report of Gupta et al. 2018, the marker, gwm570 was associated with SB resistance in a F2 population. Several simple satellite repeat (SSR) markers gwm67, gwm570, and gwm469, were linked to SB resistance in early marker-trait association studies. (Sharma et al. 2007).
Kaur et al. (2021) discovered a QTL, QSb.pau-6A and it was mapped 53 Mb from QTL SNP_3021829 (Bainsla et al. 2020) mapped in the same genomic region. A gene for Ubiquitin family protein was found flanking the QTL. Ubiquitin-related proteins implant plant resistance by degrading flagellin-sensing 2 (FLS2) receptor, which binds the microbe associated molecular pattern (MAMP), flagellin. Ubiquitin, which is a part of the ubiquitin–proteasome system (UPS), controls various pathways including response to biotic and abiotic stresses (Sadanandom et al. 2012), and acts as one of the major systems in plant immunity (Üstün et al. 2016).The QTL mapping achieved in this study should therefore serve as a starting point for developing a more detailed map and initiating a marker assisted selection approach. Successful MAS and cloning of the key resistance QTL will critically depend on the introduction of novel flanking markers and high-resolution mapping populations in the future. Thus, it is necessary to investigate additional marker systems, such as genotyping by sequencing, in order to develop markers for marker-assisted transfer of additional QTLs found in this work.
Data availability
Not Applicable.
References
Adlakha KL (1984) Resistance of wheat to leaf spot caused by Bipolaris sorokiniana. Plant Dis 68:320. https://doi.org/10.1094/pd-69-320
Bainsla NK, Phuke RM, He X, Gupta V, Bishnoi SK, Sharma RK (2020) Genome-wide association study for spot blotch resistance in Afghan wheat germplasm. Plant Pathol 69:1161–1171
Bhushan B, Singh K, Kaur S, Nanda GS (2002) Inheritance and allelic relationship of leaf blight resistance genes in three bread wheat varieties in the adult-plant stage. J Genet Breed 56:6–76
Chaurasia S, Chand R, Joshi AK (2000) Relative dominance of Alternaria triticina Pras. et Prab. and Bipolaris sorokiniana (Sacc.) Shoemaker in different growth stages of wheat (T. aestivum L.). J Plant Dis Prot 107:176–181
CIMMYT (2013) Research battles wheat spot blotch disease. Available on: https://www.cimmyt.org/research-battles-wheat-spot-blotch-disease/. Retrieved on 17th June 2017
Dubin HJ, van Ginkel M (1991) The status of wheat diseases and disease research in warmer areas. In: Saunders DA (ed) Wheat for the Nontraditional Warmer Areas. CIMMYT, Mexico, D.F, pp 125–145
Duveiller E, Sharma RC (2009) Genetic improvement and crop management strategies to minimize yield losses in warm non-traditional wheat growing areas due to spot blotch pathogen Cochliobolus sativus. J Phytopathol 157:521–534
Duveiller E, Kandel YR, Sharma RC, Shrestha SM (2005) Epidemiology of foliar blights (spot blotch and tan spot) of wheat in the plains bordering the Himalayas. Phytopathology 95:248–256
Gahtyari NC, Roy C, He X, Roy KK, Reza MMA, Hakim MA et al (2021) Identification of qtls for spot blotch resistance in two bi-parental mapping populations of wheat. Plants 10:973. https://doi.org/10.3390/plants10050973
Ganal MW, Ro¨der MS (2007) Microsatellite and SNP markers in wheat breeding. In: Varshney RK, Tuberosa R (eds) Genomics assisted crop improvement, Genomics applications in crops. Springer Netherlands 2:1–24
Gupta PK, Chand R, Vasistha NK, Pandey SP, Kumar U, Mishra VK, Joshi AK (2018) Spot Blotch Disease of Wheat: the Current Status of Research on Genetics and Breeding 17:508–531
Gurung S, Mamidi S, Bonman JM, Xiong M, Brown-Guedira G, Adhikari TB (2014) Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS ONE 9:e108179. https://doi.org/10.1371/journal.pone.0108179
Joshi AK, Kumar S, Chand R, Ortiz-Ferrara G (2004) Inheritance of resistance to spot blotch caused by Bipolaris sorokiniana in spring wheat. Plant Breed 123:213–219
Joshi AK, Kumari M, Reddy VPS, Kumar S, Rane J, Chand R et al (2007a) Stay green trait: variation, inheritance and its association with spot blotch resistance in spring wheat (Triticum aestivum L.). Euphytica 153:59–71. https://doi.org/10.1007/s10681-006-9235-z
Joshi AK, Kumari M, Singh VP, Reddy CM, Kumar S, Rane J, Chand R (2007b) Stay green trait: variation, inheritance and its association with spot blotch resistance in spring wheat (Triticum aestivum L.). Euphytica 153:59–71
Kaur J, Kaur J, Dhillon GS, Kaur H, Singh J, Bala R, Srivastava P, Kaur S, Sharma A, Chhuneja P (2021) Characterization and Mapping of Spot Blotch in Triticum durum–Aegilops speltoides Introgression Lines Using SNP Markers. Front Plant Sci 12:650400
Kumar D, Chand R, Prasad LC, Joshi AK (2007) A new technique for monoconidial culture of the most aggressive isolate in a given population of Bipolaris sorokiniana, cause of foliar spot blotch in wheat and barley. World J Microbiol Biotechnol 23:1647–1651. https://doi.org/10.1007/s11274-007-9410-y
Kumar U, Joshi AK, Kumar S, Chand R, Röder MS (2009) Mapping of resistance to spot blotch disease caused by Bipolaris sorokiniana in spring wheat. Theor Appl Genet 118:783–792
Kumar U, Joshi AK, Kumar S, Chand R, Röder MS (2010) Quantitative trait loci for resistance to spot blotch caused by Bipolaris sorokiniana in wheat (T. aestivum L.) lines ‘Ning 8201’ and ‘Chirya 3.’ Mol Breed 26:477–491
Kumar S, Röder MS, Tripathi SB, Kumar S, Chand R, Joshi AK, Kumar U (2015) Mendelization and fine mapping of a bread wheat spot blotch disease resistance QTL. Mol Breed 35:218
Lillemo M, Joshi AK, Prasad R, Chand R, Singh RP (2013) QTL for spot blotch resistance in bread wheat line SAAR co-locate to the biotrophic disease resistance loci Lr34 and Lr46. Theor Appl Genet 126:711–719
McIntyre CL, Mathews L, Ratty A, Chapman SC, Drenth J, Ghaderi M, Reynolds M, Shorter R (2010) Molecular detection of genomic regions associated with grain yield and yield- related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor Appl Genet 120:527–541
Mehta YR (1998) Constraints on the integrated management on spot blotch of wheat. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Helminthosporium Blight of Wheat: Spot Blotch and Tan Spot. CIMMYT, El Batán, DF, Mexico, pp 18–27
Nema KG, Joshi LM (1973) The spot blotch disease of wheat caused by Helminthosporium sativum. In: Jaiswal PL, Wadhwani AM (eds) Annotated Compendium on Wheat Diseases in India. Offset Press, India, pp 223–224
Neupane RB, Sharma RC, Duveiller E, Ortiz-Ferrara G, Ojha BR, Rosyara UR et al (2007) Major gene controls of field resistance to spot blotch in wheat genotypes “Milan/Shanghai #7” and “Chirya. 3.” Plant Dis 91:692–697. https://doi.org/10.1094/PDIS-91-6-0692
Pankaj YK, Kumar R, Pal L, Nagarajan R, Gill KS, Kumar V, Panigrahi S (2021) Performance and yield stability of doubled haploid population of wheat (Triticum aestivum L.) under high temperature regime. Cereal res comm. https://doi.org/10.1007/s42976-022-00247-4
Pankaj YK, Kumar R, Pal L, Nagarajan R, Gill KS, Kumar V, Panigrahi S (2022a) Mapping QTLs for morpho-physiological traits related to grain yield under late sown conditions in wheat (Triticum aestivum L.). Cereal res comm. https://doi.org/10.1007/s42976-021-00234-1
Pankaj YK, Kumar R, Nagarajan R, Gill KS (2022b) Mapping QTLs for physiological traits associated with grain yield under timely and delayed sown conditions in wheat (Triticum aestivum L.). Preprint. https://doi.org/10.21203/rs.3.rs-1291146/v1
Prakash N, Pankaj YK, Kumar R, Kumar P, Jat RK (2019) Genetic diversity for submergence tolerance in rice (Oryza sativa L.) utilizing SSR Markers. Appl Biol Res 21:151–157
Prasad M, Varshney RK, Kumar A, Balyan HS, Sharma PC, Edwards KJ, Singh H, Dhaliwal HS, Roy JK, Gupta PK (1999) A microsatellite marker associated with a QTL for grain protein content on chromosome arm 2DL of bread wheat. Theor Appl Genet 99:341–345
Ragiba M, Prabhu KV, Singh RB (2004) Recessive genes controlling resistance to Helminthosporium leaf blight in synthetic hexaploid wheat. Plant Breed 123:389–391. https://doi.org/10.1111/j.1439-0523.2004.00939.x
Roy JK, Prasad M, Varshney RK, Balyan HS, Blake TK, Dhaliwal HS, Singh H, Edward KJ, Gupta PK (1999) Identification of a microsatellite on chromosome 6B and STS on 7D of bread wheat showing an association with preharvest sprouting tolerance. Theor Appl Genet 99:336–340
Roy C, Gahtyari NC, He X, Mishra VK, Chand R, Joshi AK et al (2021) Dissecting quantitative trait loci for spot blotch resistance in South Asia using two wheat recombinant inbred line populations. Front Plant Sci 12:641324. https://doi.org/10.3389/fpls.2021.641324
Saari EE (1998) Leaf blight disease and associated soil-borne fungal pathogens of wheat in South and South East Asia. Helminthosporium Blights of wheat: spot blotch and tan spot 37–51
Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S (2012) The ubiquitinproteasome system: central modifier of plant signalling. New Phytol 196:13–28
Sharma RC, Bhatta MR (1999) Inheritance of field resistance to spot blotch in three wheat crosses. J Inst Agric Anim Sci 19:111–118
Sharma RC, Dubin HJ, Bhatta MR, Devkota RN (1997) Selection for spot blotch resistance in four spring wheat populations. Crop Sci 37:432–435. https://doi.org/10.2135/cropsci1997.0011183X003700020021x
Sharma RC, Duveiller E, Jacquemin JM (2007) Microsatellite markers associated with spot blotch resistance in spring wheat. J Phytopathol 155:316–319. https://doi.org/10.1111/j.1439-0434.2007.01238.x
Siddique AB, Hossain MH, Duveiller E, Sharma RC (2006) Progress in wheat resistance to spot blotch in Bangladesh. J Phytopathol 154:16–22
Singh PK, He X, Sansaloni CP, Juliana P, Dreisigacker S, Duveiller E et al (2018) Resistance to spot blotch in two mapping populations of common wheat is controlled by multiple QTL of minor effects. Int J Mol Sci 19:4054. https://doi.org/10.3390/ijms19124054
Singh D, Singh RV, Singh AK, Singh BN (1998a) Identification and inheritance of resistance to foliar blight of wheat. In: Duveiller E, Dubin HJ, Reeves J, McNab A (eds) Helminthosporium blights wheat Spot blotch tan spot. CIMMYT, Mexico, pp 259–262
Singh PK, Singh S, Deng Z, He X, Kehel Z, Singh RP (2019) Characterization of QTLs for Seedling Resistance to Tan Spot and Septoria Nodorum Blotch in the PBW343/Kenya Nyangumi Wheat Recombinant Inbred Lines Population. Int J Mol Sci 20:5432
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite based deletion bin system for the establishment of genetic physical map relationships in wheat (Triticum aestivum L.). Funct Integr Gen 4:12–25
Srivastava OP, Luthra TK, Narula PN (1971) Inheritance of seedling resistance to leaf blight of wheat. Indian Journal of Genetics and Plant Breeding 31(2):209–211
Tomar V, Singh D, Dhillon GS, Singh RP, Poland J, Joshi AK, Singh PK, Bhati PK, Kumar S, Rahman M, Tiwari BS (2021) New QTLs for spot blotch disease resistance in wheat (Triticum aestivum L.) using Genome-Wide Association Mapping. Front Genet 11:613217
Üstün S, Sheikh A, Gimenez-Ibanez S, Jones A, Ntoukakis V, Börnke F (2016) The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol 172:1941–1958
Velázquez Cruz C, Galán M, Dconsejer J, Rajaram S, Castillo González F, Gilchrist Saavedra L et al (1994) Genética de la Resistencia a Bipolaris sorokiniana en Trigos Harineros. M.Sc. Thesis. Colegio de Posgraduados, Montecillo, México 84
Vasantrao JM, Pankaj YK, Kumar R (2019) Characterization of wheat (Triticum aestivum L.) genotypes unraveled by molecular markers considering heat stress. Indian J Biotechnol 18:204–213
Verma SK, Chaurasia SK, Pankaj YK, Kumar R (2020) Study on the genetic variability and pathogenicity assessment among isolates of spot blotch causing fungi (Bipolaris sorokiniana) in wheat (Triticum aestivum L.). Plant Physiol Rep 25:255–267
Wang S, Basten CJ and Zeng ZB (2010) Windows QTL Cartographer 2.5. Department of statistics, North Carolina State University, Raleigh, NC
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Zhu Z, Bonnett D, Ellis M, Singh P, Heslot N, Dreisigacker S, Gao C, Mujeeb-Kazi A (2014) Mapping resistance to spot blotch in a CIMMYT synthetic-derived bread wheat. Mol Breed 34:1215–1228
Acknowledgements
The Acknowledgement to the Department of Agricultural Biotechnology and Molecular Biology, Rajendra Prasad Central Agricultural University, Pusa, Bihar and Department of Biotechnology, Government of India.
Funding
The work was supported by United States Agency for International Development-Biotechnology Industry Research Assistance Council (Reference number: BIRAC/TG/USAID/08/2014).
Author information
Authors and Affiliations
Contributions
RK and YKP involved in conceptualization of research; RK and YKP involved in designing of the experiments; KSG involved in contribution of experimental materials; YKP involved in execution of field/laboratory experiments and data collection; YKP, RK, and RN involved in analysis of data and interpretation; YKP and RK involved in preparation of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate (Human Ethics, Animal Ethics or Plant Ethics)
Not Applicable.
Consent for publication
All authors are in agreement of the publication.
Competing interests
The authors declare no competing interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pankaj, Y.K., Kumar, R., Gill, K.S. et al. Discovering QTLs related to spot blotch disease in spring wheat (Triticum aestivum L.) genome. Australasian Plant Pathol. 51, 441–452 (2022). https://doi.org/10.1007/s13313-022-00873-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-022-00873-w