Abstract
Association of 16SrVI group phytoplasmas in symptomatic sweet cherry samples was confirmed in our previous study by amplifying the 16 S rRNA gene in nested PCR assays. However, this method is time-consuming and expensive. Therefore, we developed a loop-mediated isothermal amplification (LAMP) assay based detection protocol from a previously identified 16SrVI group phytoplasma strain in cherry samples. Three sets of primers based on the leucyl tRNA synthetase (leuS) and 16 S rRNA gene sequences were designed and evaluated to establish a rapid and efficient LAMP assay based detection system for 16SrVI-D subgroup associated cherry phytoplasma strains. The sweet cherry phytoplasma DNA was efficiently amplified by employing a set of leuS based LAMP detection primers in a reaction condition of 63o C for 70 min. The phytoplasma positive reactions were visualised by ladder like bands on 2% agarose gel, colour change from violet to blue with hydroxynaphthol blue and presence of fluorescence with ethidium bromide after an hour of LAMP reaction. Furthermore, the designed primers were tested for cross reactivity with other groups of phytoplasma strains belonging to 16SrI, 16SrII and 16SrV but could not amplify any of them. The lowest detection limit for the LAMP detection assay was 10 pg/µL. The developed LAMP method was found to be more robust, reliable and sensitive than the conventional nested PCR assay method. As a result, it has the potential to be used in phytoplasma indexing of sweet cherry germplasm and seedlings for disease free vegetative propagative materials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cherry is one of the most important fruit trees cultivated all over the tropics and native to Europe and Asia regions. Cherry belongs to the genus Prunus and is a rich source of healthy nutrients as well as antioxidants (Hu et al. 2021). In India, cherries are mainly cultivated in Jammu and Kashmir (J&K) and Himachal Pradesh (H.P.) states and some parts of north east India (NHB, 2019). However, the productivity and quality of cherry fruits are severely hampered by several viral and phytoplasmal diseases worldwide (Fiore et al. 2018). The important phytoplasmas such as aster yellows, ‘Candidatus Phytoplasma australasia’, X-disease, elm yellows, ash yellows, apple proliferation, pear decline, stolbur group and European stone fruit yellows have been reported to cause severe losses in cherry production (Fiore et al., 2018; Cieślińska and Smolarek, 2019). Recently, ‘Candidatus Phytoplasma asteris’ and ‘Candidatus Phytoplasma trifolii’ strains have been identified in sweet cherry cultivars in India (Shreenath et al., 2022) by employing 16S rRNA gene specific primer pairs. However, the titre of cherry phytoplasma was relatively quite low in the symptomatic diseased tissues, thus the reliability of routine PCR assay methods was limited as well as being relatively expensive, time-consuming and needing experienced personnel. Another DNA amplification technique known as loop-mediated isothermal amplification (LAMP) has been developed for successful amplification of several phytoplasma strains associated with agriculturally important crops (Notomi et al., 2000; Bekele et al. 2011; Tomlinson et al. 2010; Kogovšek et al. 2015;, Aljafer and Dickinson 2021). The main advantage of LAMP assays over PCR assays, are the involvement of the extended set of 4–6 set of primers which increases specificity, efficiency and the isothermal reaction process at constant temperature, and the use of Bst polymerase, which is less prone to inhibitors found in some DNA extracts (Mori and Notomi, 2009; Njiru, 2012).
LAMP is a more sensitive, rapid and convenient technique for the detection of phytoplasmas than routine PCR amplifications (Mori and Notomi 2009; Parida et al. 2008; Mori et al., 2001). Hence, there is a need to develop a fast, sensitive and efficient LAMP based protocol for detection of the 16SrVI group strain of phytoplasmas infecting cherry trees in India which was achieved in the present study by targeting the leucyl tRNA synthetase gene. We also evaluated the amplification efficiency, sensitivity and specificity of the LAMP assay.
Materials and methods
Materials
Total DNA was extracted from the symptomatic leaf midrib or branch portion of cherry samples and in positive phytoplasma controls (brinjal little leaf, Candidatus Phytoplasma trifolii, Accession number KX689234 and Candidatus Phytoplasmas asteris, Accession number KC920747) maintained on periwinkle in glass house using the CTAB protocol (Ahrens and Seemüller 1992). The extracted DNA was subjected to nested PCR assay with universal phytoplasma specific primers, P1/P7 (Deng and Hiruki 1991; Schneider et al., 1995) followed by 3For/3Rev (Manimekalai et al. 2010) and/or R16F2n/R2 primer pairs (Gundersen and Lee 1996). The PCR assays were carried out in a final reaction volume of 25 µl containing of nuclease free water (Sisco Research Laboratories Pvt. Ltd., India), OnePCR™2X PCR Master Mix (GeneDireX, Taiwan), for/rev primer 10 pmol/µl (0.2 µM) and the DNA template (= 100 ng). PCR reactions were performed in a thermal cycler (Mastercycler, Eppendorf, Hamburg, Germany). Reaction mix without DNA was used as negative control.
LAMP primer design and synthesis
To achieve a more specific amplification reaction, six LAMP primers targeting two distinct regions of phytoplasma DNA (16 S rRNA and leucyl tRNA synthetase genes) were used in this study. Furthermore, loop-F and loop-B primers were used to enhance the LAMP reaction (Nagamine et al., 2002; Kogovšek et al., 2017).
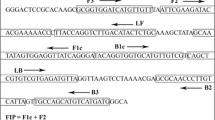
The leucyl tRNA synthetase gene (leuS) (Accession number, KU751799) was chosen to obtain sequences for the design of 16SrVI group specific LAMP primers. Premier Biosoft’s LAMP Designer software (http://www.premierbiosoft.com/ isothermal/ lamp. html) was used to design LAMP primers for 16SrVI group based on leuS. Primer explorer V5 software (https://primerexplorer.jp/e/) was also employed to design two sets of LAMP primers based on the 16 S rRNA gene (Accession number MN861370) for 16SrVI group. The designed LAMP primers were synthesized by AgriGenome, Kerala, India (Table 1).
Establishment of LAMP assay and evaluation of LAMP amplification products
The concentrations of various components in the LAMP reaction such as 100 mM MgSO4 (0.5– 1.5 µl), dNTPs (0.8–1.4 mM) and betaine (0.6–1.4 M) were optimized. Isothermal amplification reactions were carried out in a final volume of 25 µl, which contained 2.5 µl of 10X ThermoPol Buffer (New England Biolabs, UK), 3.5 µl of 10 mM dNTPs (New England Biolabs, UK), 1.5 µl 100 mM MgSO4, 6 µl of 6 M betaine (Sigma-Aldrich, USA), 0.4 to 1.6 µM each of FIP and BIP, 0.1 to 0.4 µM each of F3 and B3 and 0.2 to 0.8 µM each of LF and LB primers [all the three sets of primers based on leuS (MFIP, MBIP, MF3, MB3, MLF and MLB) and 16 S rRNA (HFIP, HBIP, HF3, HB3, HLF and HLB; GFIP, GBIP, GF3, GB3, GLF and GLB) genes were examined), 1 µl of template DNA, 6 µl of nuclease free water and 0.5 µl of 8 units of the Bst DNA polymerase large fragment (New England Biolabs, UK). All reactions were run at 61°- 66 °C for 30–90 min for isothermal amplification. Results were confirmed by gel electrophoresis in 2% agarose gels, or through incorporation of 100–200 µM Hydroxynaphthol blue (HNB) dye into the reaction mixture prior to amplification to visualize the colour change, or through addition of 0.2–0.8 mg of ethidium bromide/ml (Sigma), to 10 µl amplified products after the amplification reaction to observe differences in brightness under a UV transilluminator, along with known positive (Accession number KX689234) and negative controls (phytoplasma-free cherry sample and nuclease free water).
Specificity and sensitivity test of LAMP assay
For determining the specificity of amplification and efficiency of the 16SrVI group sweet cherry phytoplasma LAMP assays, the same primer sets were used to amplify the DNA of known phytoplasma strains of 16SrI (peach strain, Acc. No. OL527667), 16SrII (peach strain, Acc. No. OL454915), 16SrV (plum strain, Acc. No. OL455729) group along with asymptomatic cherry leaves. To test the sensitivity of the LAMP assay, total genomic DNA of cherry phytoplasma diluted to different concentrations viz. 100ng, 50ng, 10ng, 1ng, 100pg, 50pg, 10pg, 500 fg, 100 fg and nuclease free water as non-template control were utilized in LAMP assay at optimized conditions.
Stability of LAMP detection system
Using the established LAMP detection system, the two symptomatic sweet cherry samples each from three cultivars (BN, BNG and CITH-Cherry − 9) were evaluated. Total DNA was extracted from the sweet cherry samples and LAMP amplification was performed after PCR assay verification. The LAMP reaction results were observed after the reaction to ensure the stability of the system.
Results
Molecular detection and identification of phytoplasma by routine PCR assays
About ~ 1.3 kb amplicons were amplified from the three symptomatic sweet cherry cvs BN, BNG and CITH-Cherry − 9 in nested PCR assays with the primers 3For/3Rev along with positive sample (data not shown). These PCR products were cloned, sequenced and submitted to GenBank (Acc. No. OM019094 - OM019099) (Table 2). Two sets of LAMP primers based on the 16 S rRNA gene (Accession number MN861370) were designed for LAMP amplification assays along with a leucyl tRNA synthetase gene sequence from the 16SrVI-D brinjal little leaf phytoplasma held at the University of Nottingham phytoplasma collection (Accession Number: KU751799).
Optimization and establishment of stability in LAMP detection system
Among the three sets of primers designed, sweet cherry phytoplasma DNA was efficiently amplified from the primers designed based on the leuS gene (Accession Number: KU751799). For optimization of the LAMP assay, different concentrations of MgSO4 (2–10 mM) and betaine (0.6–1.4 M) were tested in the final reaction and better results were obtained at 8 mM of MgSO4 and 1.2 M of betaine (Fig. 1). The optimised concentration of different components in 25 µl reaction buffer was: 2.5 µl of 10X Thermopol buffer, 8 U of Bst polymerase, 1.5 µl MgSO4 (100 mM), 3.5 µl dNTPs (10 mM), betaine 6 µl (5 M, Sigma), 20 µM each of FIP, BIP, LF and LB primers, 5 µM of each of F3 and B3 primers. Temperature gradient showed that reaction mix incubated at 63 °C for 70 min was found optimum for the assay developed in this study. Use of 120 µM hydroxy naphthol blue (HNB) dye was optimal for the visual detection of LAMP reactions giving a colour change from violet to sky blue in positive reactions; however, no colour change was visible in negative reactions (Fig. 1 A). At 0.6 mg of EtBr/ml, brightness of the positive reaction tube was clearer when compared to the negative reaction (no fluorescence) visualised on a UV transilluminator (Fig. 1B). Furthermore, the results showed laddering pattern in phytoplasma positive samples in gel electrophoresis using 2% agarose gels (Fig. 1 C), whilst no laddering patterns were seen from the asymptomatic cherry samples and nuclease free water (negative control) (Table 2). Also, no amplification was obtained in any of the cherry DNA samples with the LAMP primers designed based on 16 S rRNA gene (Accession number MN861370) in the present study (data not shown).
A Visual detection of LAMP reaction using HNB dye: The colour changes from violet (negative reaction) to sky blue (positive reaction). Tubes from left to right; DNA template of cherry phytoplasma (1–6); P-Positive (Cathranthus roseus) sample; N1- healthy cherry sample N2- nuclease free water. B Visual detection of LAMP reaction using EtBr: The light colour seen in negative reaction and bright colour (fluorescence) seen in positive reaction. Tubes from left to right (photo shown in different angle); C Agarose gel electrophoresis of LAMP products. Lanes from left to right; Lane M (1 kb molecular marker); Lane M (1 kb molecular marker)
Specificity and sensitivity test of LAMP assay
For testing specificity of standardized LAMP protocol assay, an experiment was performed with template DNAs of sweet cherry phytoplasma belonging to 16SrVI, along with the earlier identified peach phytoplasma strains of 16SrI (Acc. No. OL527667), 16SrII (Acc. No. OL454915) and plum phytoplasma 16SrV (Acc. No. OL455729) groups as well as DNA isolated from healthy sweet cherry samples. A positive amplification was achieved in the LAMP assay only in sweet cherry phytoplasma infected with 16SrVI samples and no amplifications were observed in any other groups of phytoplasmas (16SrI, 16SrII and 16SrV) associated with peach and plum. These results were further confirmed by negative results in HNB and EtBr dye based detection assays and gel electrophoresis (Fig. 2). To assess the sensitivity of the LAMP assay, a dilution series of sweet cherry phytoplasma DNAs was tested by the optimized LAMP assay. The sensitivity of the LAMP detection assay up to 10 pg of cherry phytoplasma DNA concentration was observed and further established through laddering pattern in agarose gel electrophoresis (Fig. 3 C). Similar results were also confirmed by using HNB and EtBr dyes (Fig. 3 A, B).
Testing of specificity of LAMP assay: A Visual detection of LAMP reaction using HNB dye: The colour changes from violet (negative reaction) to sky blue (positive reaction). B Visual detection of LAMP reaction using EtBr: The light colour seen in negative reaction and bright colour (fluorescence) seen in positive reaction. C Agarose gel electrophoresis of LAMP products. Lane M (1 kb molecular marker) P-Positive (C. roseus), N- nuclease free water Lane C1, C2: DNA template of cherry phytoplasma; Lane 1: Peach sample (16SrI); Lane 2: Peach sample (16SrII); Lane 3: plum (16SrV)
Testing of sensitivity of LAMP assay: A Visual detection of LAMP reaction using HNB dye: The colour changes from violet (negative reaction) to sky blue (positive reaction). B Visual detection of LAMP reaction using EtBr: The light colour seen in negative reaction and bright colour (fluorescence) seen in positive reaction. C Agarose gel electrophoresis of LAMP products. Lane M (1 kb molecular marker), Lane N- nuclease free water, Lane 1–11: DNA template of cherry, 1(100ng); 2 (50ng); 3 (10ng); 4 (1ng); 5 (100pg); 6 (50pg); 7 (25pg); 8 (10pg); 9 (1pg); 10 (500 fg); 11 (100 fg)
Comparison of PCR and LAMP assay sensitivity (detection limit)
Nested PCR assays of DNAs of sweet cherry phytoplasma with 3For/3Rev primers pairs showed amplification up to 100 pg of DNA for about 4–5 h of duration as compared to the LAMP detection assay which was efficient in amplification up to 10 pg of DNA concentration within 70 min observed by DNA laddering pattern in agarose gel electrophoresis (Fig. 4).
Comparision of sensitivity of PCR and LAMP assays: A Sensitivity of PCR assay using 3For/3Rev primers B Agarose gel electrophoresis of LAMP products. Lane M (1 kb molecular marker), Lane N- nuclease free water, Lane 1–11: DNA template of cherry, 1(100ng); 2 (50ng); 3 (10ng); 4 (1ng); 5 (100pg); 6 (50pg); 7 (25pg); 8 (10pg); 9 (1pg); 10 (500 fg); 11 (100 fg) N (nuclease free water)
Discussion
Several isothermal nucleic acid based amplification technologies have been described in recent years, with the potential to be turned into in-field plant pathogen detection systems (Lau and Botella 2017; Donoso and Valenzuela 2018). Tomlinson et al. (2010) reported the first use of LAMP for phytoplasma identification, when six-primer set assays based on the 16-23 S rRNA gene were reported for the detection of 16SrI group (‘Ca. P. asteris’) and the 16SrXXII group (Ca. P. palmicola) in coconut. In these assays, identification of product was shown using gel electrophoresis as well as the addition of HNB dye into the reaction mixture, which resulted in valid detection of phytoplasma in positive reactions with high sensitivity and reliability.
Based on the leuS gene, this study could develop and validate LAMP assays for detecting phytoplasma strains enclosed in 16SrVI groups. Due to the relatively well-conserved nature of ribosomal sequences (16-23 S rRNA genes), it is difficult to find regions that are specific for each of the phytoplasma ribosomal groups. The leuS gene has a unique sequence for most of the phytoplasma groups and this has been reflected in the ability to develop LAMP protocol assays for phytoplasmas belonging to a variety of specific ribosomal groups (Abeysinghe et al. 2016). Our results also support these findings and we have detected 16SrVI group in cherry witches’ broom phytoplasma by employing leuS gene specific designed LAMP based primers.
In this study, the time required to amplify phytoplasma DNA for the LAMP assay was typically around 70 min. However, it could provide better and clear visible amplification consistently from 60 to 70 min. The amplification time was slightly differ from previous reports which were amplified within 20–45 min for leuS gene and 16-23 S rRNA gene assays (Aljafer and Dickinson, 2021; Kogovšek et al. 2015).
The LAMP assays appeared to have good specificity for the phytoplasmas in different groups for which they were designed and developed. The detection sensitivity of LAMP assays showed that it can amplify phytoplasma DNA at dilutions as low as 10 pg/µl; however, nested PCR has efficiency to amplify only up to 100 pg/ µl sensitivity. Another positive point is that LAMP assays provide results in 60–70 min, whereas nested PCR takes 4–5 h. The low titer of phytoplasmas in woody host plants experienced problematic and prevents the efficient detection of phytoplasma from diseased hosts (Nejat and Vadamalai 2010). Our findings suggest that the LAMP assay is a superior alternative to traditional molecular detection methods based on targeted the phytoplasma leuS gene. The use of four primers that bind to six different regions of the target ensures high specificity. The use of additional loop primers enhances the amplification efficiency as suggested by Nagamine et al. (2002). This ensures that the phytoplasma is detected even if it is present in very low titers, and not detected consistently by using 16 S rRNA gene universal primer pairs, as is the case with the phytoplasmas associated with sweet cherry in the present study. The closed tube detection system employing HNB in the reaction mixture eliminates the possibility of cross contamination, which can be a negative aspect of LAMP assays due to high target amplification (Goto et al. 2009). By optimizing HNB in the reaction (120 µM), we were able to clearly distinguished the positive results from the negative one which is slightly different from the previous studies (150 µM) (Nair et al., 2016).
Finally, in the present study, an optimised and validated LAMP assay was established for the detection of phytoplasma strain associated with sweet cherry which is an economic important fruit for international trade in India and other sweet cherry grown countries. The method is particularly useful for on-site surveys and point-of-care screening because the entire analysis process takes about an hour and does not necessitate the facility of PCR machine. The developed LAMP assay can also be applied for disease indexing and management of sweet cherry phytoplasma diseases for establishing disease free necessary for clonal propagation or grafting.
References
Abeysinghe S, Kanatiwela-de Silva C, Abeysinghe PD, Udagama P,Warawichanee K, Aljafer N, Kawicha P, Dickinson M (2016) Refinement of the taxonomic structure of 16SrXI and 16SrXIV phytoplasmas of gramineous plants using multilocus sequence typing. PlDis 100(10):2001–2010.
Aljafer N, Dickinson M (2021) Evaluating LAMP assays for detection of phytoplasmas classified in different ribosomal groups. J Pl Pathol, 103(4):1315–1321.
Bekele B, Hodgetts J, Tomlinson J, Boonham N, Nikolic´ P, Swarbrick P, Dickinson M (2011) Use of a real-time LAMP isothermal assay for detecting 16SrII and XII phytoplasmas in fruit and weeds of the Ethiopian Rift Valley. Pl Pathol 60:345–355.
Cieślińska M, Smolarek T (2019) Multilocus sequence analysis of phytoplasmas detected in cherry trees in Poland. Zemdirbyste-Agriculture 106(1).
Deng S, Hiruki C (1991) Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J of Microbiol Meth 14: 53–61.
Donoso A, Valenzuela S (2018) In-field molecular diagnosis of plant pathogens: recent trends and future perspectives. Pl Pathol 67(7): 1451–1461.
Fiore N, Bertaccini A, Bianco PA, Cieślińska M, Ferretti L, Hoat TX, Quaglino F (2018). Fruit crop phytoplasmas. Rao GP, Bertaccini A, Fiore N, Liefting LW(eds) In Phytoplasmas: Plant pathogenic bacteria-I, Springer, Singapore. pp 153–190.
Goto M, Honda E, Ogura A, Nomoto A, Hanaki KI (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniq 46:167–172
Hu T, Subbiah V, Wu H, Amrit BK, Rauf A, Alhumaydhi FA, Suleria HAR (2021) Determination and characterization of phenolic compounds from Australia-grown sweet cherries (Prunus avium L.) and their potential antioxidant properties. ACS Omega 6 (50): 34687–34699
Kogovšek P, Hodgetts J, Hall J, Prezelj N, Nikolić P, Mehle N, Lenarčič R, Rotter A, Dickinson M, Boonham N, Dermastia M (2015) LAMP assay and rapid sample preparation method for on-site detection of favescence dorée phytoplasma in grapevine. Pl Pathol 64(2):286–296.
Kogovšek P, Mehle N, Pugelj A, Jakomin T, Schroers HJ, Ravnikar M, Dermastia M (2017) Rapid loop-mediated isothermal amplification assays for grapevine yellows phytoplasmas on crude leafvein homogenate has the same performance as qPCR. Eur J Pl Pathol 148:75–84.
Lau HY, Botella JR (2017) Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front Pl Sci 8: 2016.
Manimekalai R, Soumya VP, Sathish Kumar R, Selvarajan R, Reddy K, Thomas GV, Baranwal VK (2010) Molecular detection of 16SrXI group phytoplasma associated with root (wilt) disease of coconut (Cocos nucifera) in India. Pl Dis 94(5): 636
Mori Y, Nagamine K, Tomita N, Notomi T (2001) Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun 289: 150–154.
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16: 223–229.
Nair S, Manimekalai R, Ganga Raj P, Hegde V (2016) Loop mediated isothermal amplification (LAMP) assay for detection of coconut root wilt disease and arecanut yellow leaf disease phytoplasma. World J Microbiol Biotechnol 32(7):1–7.
Nejat N, Vadamalai G (2010) Phytoplasma detection in coconut palm and other tropical crops. Pl Pathol J (Faisalabad) 9(3):112–121.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K,Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):e63–e63.
Njiru ZK (2012) Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS neglected tropical diseases, 6(6):e1572.
Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K (2008) Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 18:407–421.
Schneider B, Seemüller E, Smart CD, Kirkpatrick BC (1995) Phylogenetic classification of plant pathogenic mycoplasma like organisms or phytoplasmas. In Razin S & Tully JG (eds). Molecular and diagnostic procedures in mycoplasmology. Academic Press pp 369–380
Tomlinson JA, Boonham N, Dickinson M (2010) Development and evaluation of a one-hour DNA extraction and loop-mediated isothermal amplification assay for rapid detection of phytoplasmas. Pl Pathol 59: 465–471.
Acknowledgements
The authors are grateful for the financial help provided by the Director, ICAR-Indian Agricultural Research Institute, New Delhi, India. We would also like to thank the Head, Division of Plant Pathology, Indian Agricultural Research Institute for providing laboratory facilities and Dr A I Bhat ICAR-Indian Institute of Spice Research, Kerala for his help in designing LAMP specific primers and corrections in the manuscript.
Table 1 LAMP primers designed for 16SrVI group phytoplasma based on 16SrRNA and leuS genes.
Author information
Authors and Affiliations
Contributions
The authors conceived the idea, analysed data and drafted manuscript for publication. The first author, Shreenath Y S did the survey, sample analysis, sequence analysis, sequence submission, design of primer and also contributed significantly in the preparation of draft of the manuscript. Sunil Kumar Sunani and M Dickinson have contributed for the design of primers and finalization of the manuscript and G P Rao contributed significantly in preparation of draft of the manuscript, finalization of the manuscript and formatting.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human studies and participants
The current study did not include any human or animal volunteers or animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shreenath, Y.S., Sunani, S.K., Rao, G.P. et al. Development of loop-mediated isothermal amplification (LAMP) assay for the detection of clover proliferation phytoplasma-associated with sweet cherry witches’ broom disease. Australasian Plant Pathol. 51, 467–474 (2022). https://doi.org/10.1007/s13313-022-00871-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-022-00871-y