Abstract
The rust fungus Austropuccinia psidii was recently reported from ornamental Corymbia citriodora and plantations of Eucalyptus in Colombia. It is unknown whether the genotypic diversity of the pathogen in Colombia reflects that of other countries in South America or if unique genotypes occur. Multilocus genotypes (MLG) were determined for collections of A. psidii from four host genera, Corymbia, Eucalyptus, Psidium and Syzygium in Colombia and compared to collections from Australia, Brazil, Indonesia, Paraguay and South Africa. The genotypic diversity of 58 samples on 15 genera of Myrtaceae was determined using seven microsatellite markers. Two lineages of A. psidii were detected among Colombian samples. These included a previously unknown genotype on Psidium guajava, different to those sampled from Brazil, as well as the pandemic biotype, which has spread to Pacific countries such as Australia, Hawaii and Indonesia. This is the first time the pandemic biotype of A. psidii has been found in South America where the rust is believed to be native. These findings raise questions with regard to the origin of the pandemic biotype of A. psidii and emphasise the threat that this biotype poses to forestry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Austropuccinia psidii (Sphaerophragmiaceae, Pucciniales) causes rust on approximately 73 genera and 460 species of Myrtaceae (Giblin and Carnegie 2014; Carnegie et al. 2016; Roux et al. 2016). It was formerly known as Puccinia psidii, however its familial and generic position has recently been resolved in the Pucciniales (McTaggart et al. 2016b; Beenken 2017). The disease caused by A. psidii is commonly referred to as eucalyptus, guava, myrtle or ohia rust. Austropuccinia psidii has spread globally, expanding its host range in Central and South America (Coutinho et al. 1998; Pérez et al. 2011; Rodas et al. 2015), North America including Hawaii (Rayachhetry et al. 1997; Uchida et al. 2006), Japan (Kawanishi et al. 2009), China (Zhuang and Wei 2011), Australia (Carnegie et al. 2010), South Africa (Roux et al. 2013), New Caledonia (Giblin 2013) and most recently Indonesia (McTaggart et al. 2016a).
Austropuccinia psidii was first described on Psidium guajava from Brazil (Winter 1884). It caused several outbreaks on all-spice (Pimenta dioica) in Jamaica (MacLachlan 1938) and Florida (Marlatt and Kimbrough 1980) and was found in eucalypt plantations of Brazil in the 1930s (Joffily 1944). Infected eucalypt trees showed a reduction in height and diameter of approximately 25–35% (Silveira and Higashi 2003) and wood-volume losses of 41% (Takahashi 2002). In Australia, A. psidii has brought several native species of Myrtaceae to the brink of extinction (Carnegie et al. 2016). Consequently, this rust is considered a global quarantine threat to commercially propagated and native trees (Tommerup et al. 2003; Glen et al. 2007; Roux et al. 2013; Wingfield et al. 2015; Burgess and Wingfield 2016).
The diversity of A. psidii has been examined using cross inoculation studies and a molecular approach with microsatellite markers. Inoculation studies showed there was intraspecific diversity in A. psidii when cross inoculations revealed variation of pathogenicity between isolates taken from Pimenta dioica and S. jambos (Marlatt and Kimbrough 1980). In other studies, A. psidii taken from P. guajava could not infect S. jambos nor Eucalyptus (Ferreira 1981; Ferreira 1983). Rayachhetry et al. (2001) tested two different isolates of A. psidii from Melaleuca quinquenervia and P. dioica that did not infect S. jambos. And Castro et al. (1983) and Coelho et al. (2001) showed there was variability in cross inoculations made between species of Eucalyptus, Psidium and S. jambos.
Molecular approaches to determine the diversity of A. psidii have mainly used microsatellite markers designed by Zhong et al. (2008). Zhong et al. (2011) showed that the Hawaiian population of A. psidii was a different genotype to samples collected from Florida and Brazil. Graça et al. (2011) found the genotype of A. psidii from California was the same as Hawaii. Ross-Davis et al. (2013) referred to this genotype as the “pandemic biotype”, which has spread through Pacific countries such as Australia, Indonesia and New Caledonia (Graça 2011; Machado et al. 2015; McTaggart et al. 2016a).
Other studies using microsatellite markers have shown there are host-associated lineages of A. psidii in Brazil on (i) Psidium, (ii) Eucalyptus and S. jambos, and (iii) Eugenia, Myrciaria and Syzygium (Graça et al. 2013). A genotype of A. psidii on P. dioica was different to the pandemic biotype and to other genotypes present in Brazil (Ross-Davis et al. 2013), and a unique genotype of A. psidii was reported in South Africa on native and exotic hosts (Roux et al. 2016). A number of recent reports worldwide indicate that at least two biotypes have spread globally (Graça et al. 2013; Ross-Davis et al. 2013; Roux et al. 2016).
Austropuccinia psidii (as Uredo myrciae) was first reported from Colombia in 1913 on Myrcia cf. acumimata (Mayor 1914; Simpson et al. 2006). Further reports on S. jambos, Psidium sp. and Myrcia sp. were all from the Antioquia Province and within 35 km of each other (Kern et al. 1933; Kern and Toro 1935). There were no new host reports between 1940 and 1995, until the rust was found on Myrcia xylopioides (Buriticá and Pardo-Cardona 1996). Then in 2010, typical symptoms of infection caused by A. psidii were found on ornamental trees of Corymbia citriodora, and a few months later on young E. grandis in plantations (Rodas et al. 2015). Yepes and Buriticá (2012) reported A. psidii on Melaleuca citrina (as Callistemon citrinus), and since 2012 it has regularly been detected on seedlings and clonal gardens of Eucalyptus. In recent years, reports of A. psidii on the commercially planted hybrid E. “urograndis” have increased (C. A. Rodas unpublished).
Austropuccinia psidii has spread in Colombia and poses a potential threat to the plantation forestry sector. Little is known regarding the distribution, host range and genetic diversity of the fungus in Colombia and this hampers breeding programmes to select disease tolerant eucalypts. The aim of this study was to determine (i) which genotypes of A. psidii are present in Colombia on four host genera, including plantation-grown eucalypts, and (ii) whether the pandemic biotype of A. psidii is present.
Materials and methods
Disease distribution in Colombia
The occurrence of Austropuccinia psidii on Myrtaceae in four provinces of Colombia was assessed between 2010 and 2015. Field surveys were conducted every three months throughout plantations of Eucalyptus and private farms. The surveys included 22 sites in the Risaralda, Quindío, Valle del Cauca and Cauca provinces (Fig. 1). These sites were selected based on observations made by foresters who reported the possible presence of rust. Infection was assessed based on uredinial symptoms of A. psidii on young leaves.
Geographic distribution of Austropuccinia psidii in Colombia. Red dots represent the surveys. White dots are from first reports of A. psidii in the early 1920s from the Antioquia province. Additionly, the biggest Pacific Marine (Buenaventura) port close to the surveyed areas, which may serve as an entry point for pests and diseases, is marked on the map as a blue dot
Genotyping of Austropuccinia psidii
Representative single-pustule isolates of A. psidii from the four surveyed Provinces of Colombia and five additional countries were included in the genotype analyses. These included single-pustule isolates from Australia, Brazil, Indonesia, Paraguay and South Africa, from different hosts (Table 1). The Australian isolates were from the same hosts and sampling sites used by Machado et al. (2015), who reported the pandemic biotype as the only biotype present in Australia. Based on this finding, the Australian samples were treated as the pandemic biotype, however, a direct comparison of the samples used in the present study was not made with those from Machado et al. (2015).
Seven microsatellite loci were scored for samples of A. psidii. The seven microsatellite markers (EF523501, EF523502, EF523503, EF523504, EF523508, EF523511 and EF523513) were developed by Zhong et al. (2008) and modified by Graça et al. (2013). Genomic DNA was extracted from a single uredinium per host using the Ultraclean® Microbial DNA Isolation Kit (MoBio Laboratories, Carlsbad, California, USA). The markers were labelled with either NED™, FAM™, PET® or VIC™ fluorescent dye on the forward primer. PCR mixtures included 1× PCR Fast Taq Buffer with MgCl2 (Sigma-Aldrich, St. Louis, Missouri, USA), 200 μM dNTPs, 0.1 μM primers, 1 unit Fast Taq DNA polymerase (Sigma-Aldrich) and DNA template in 12.5 μL reaction volumes. PCR products were amplified with the following conditions: one cycle at 95 °C for 5 min, followed by three cycles at 95 °C for 30s, 52–56 °C (depending on the primer pair) for 30s, 72 °C for 80s, 35 cycles at 94 °C for 15 s, 52–56 °C (depending on the primer pair) for 15 s and 45 s at 72 °C. Fragment analyses were performed using an ABI Applied Biosystems PRISM 3500xl (Life Technologies) at the Sequencing Facility of the Faculty of Natural and Agricultural Sciences, University of Pretoria. Samples were run with Liz500 (−250) size standard and scored with Genemapper® Software 4.1 (Applied Biosystems, Thermo Fischer Scientific, Carlsbad, USA).
Data analyses
Clonal determination
GENECLONE 2.0 was used to calculate the minimum, average and maximum multilocus genotypes (MLGs) present in the data set from each sampling area (Arnaund-Haond and Belkhir 2007; Arnaud-Haond et al. 2007). The frequency distribution of the calculated genetic distance (Nei 1978) was used to separate each MLG. Low genetic distance was treated as an indication that samples belonged to the same “clonal lineage” or the same genetic individual (Arnaund-Haond and Belkhir 2007; Arnaud-Haond et al. 2007).
To determine whether identical MLGs occured by chance or from distinct reproductive events given the allelic frequency within the population, Psex was calculated using the “round robin” approach (Parks and Werth 1993). The software MLGsim 2.0 (www.rug.nl/research/gelifes/tres/software) (Stenberg et al. 2003) was used to estimate the statistical significance values for the Psex probability using Monte Carlo simulation of each Psex value. Simulations (10000) were run to estimate a P-value for each MLG. P-values higher than 0.001 were considered significant.
Population genetic indices
The indices to determine diversity and heterozygosity were calculated from the whole data set. The Simpson Index of clonal diversity and evenness was obtained using GENECLONE 2.0 (Arnaund-Haond and Belkhir 2007). Clonal richness was estimated as R = (G – 1) / (N – 1) where G is the number of MLGs, and N the number of individuals (Dorken and Eckert 2001). The genotypic richness corresponded to the number of observed MLGs. The aggregation index estimated if the different MLGs belonged to the same lineage. The edge effect index tested whether clonal lineages were over represented based on the area sampled. All indices were calculated using GENECLONE 2.0 (Arnaud-Haond et al. 2007).
Population structure
Isolation by distance (IBD) was used to determine if geographic distance and genetic distance present in the samples collected from the six different countries were correlated. A Mantel test was used to correlate the matrices generated from GenAlex V. 6.5, assuming 10,000 permutations (Peakall and Smouse 2006, 2012).
The isolates of A. psidii collected in six different countries of the Southern Hemisphere were clustered using two methods. Firstly, Principal Coordinates Analysis (PCoA), implemented in GenAlEx V. 6.5 (Peakall and Smouse 2006, 2012), was used to construct a covariance matrix based on genetic and geographic distance. The sample data consisted of isolates that represented different MLGs found per location. Adobe Illustrator was used to modify the graphics for presentation. The second method, a median-joining (MJ) network, constructed the relationships between MLGs of all isolates of A. psidii. This analysis was made in NETWORK V. 5.0 (www.fluxus-engineering.com), implementing the default option (Bandelt et al. 1999). To identify the best shortest tree, the output file was subjected to post processing with Farri’s Maximum Parsimony calculation that calculates all possible shortest trees (Farris 1970).
Results
Disease distribution in Colombia
Results of the survey showed that A. psidii occurred across a broad range of agroecological conditions in provinces of Colombia (Fig. 1). These included high altitudes (1015–2116 m.a.s.l.) and across an approximate average temperature of 17–20 °C in the four surveyed Provinces. Austropuccinia psidii was confirmed from all 22 of the sites surveyed and detected in one commercial plantation nursery in Valle del Cauca Province. It affected trees in young established plantations between six-months and three-years-old in 17 of the surveyed sites. Affected hosts included C. citriodora, E. grandis, the hybrid E. urograndis, P. guajava and S. jambos (Fig. 1).
Genotyping of Austropuccinia psidii
A total of 58 isolates of A. psidii were genotyped, of which 29 were from Colombia. The seven microsatellite markers were polymorphic and three to six alleles were detected per locus among all samples (Table 2). A total of seven MLGs were detected for the 58 isolates of A. psidii. Two distinct MLGs were obtained from four host genera in Colombia. The genotype of A. psidii on C. citriodora, species of Eucalyptus and S. jambos from Colombia was identical to that of isolates from Australia and Indonesia at the seven tested microsatellite loci. These samples from Colombia represent the pandemic biotype. The second MLG in Colombia was of a unique genotype found only on P. guajava. This MLG is different to the MLG obtained on P. guajava in Brazil. Isolates from Brazil included four MLGs, one of which was shared with Paraguay. The seven isolates of A. psidii from South Africa had a genotype not found in any of the other samples tested, as described in Roux et al. (2016).
The recovered Psex values (P < 0.001) indicated that identical MLGs of A. psidii used in the present study did not occur from independent sexual reproductive events. This is an indication that the studied samples were from different clonal populations because clonal richness values were low at R = 0.105.
Population genetic indexes
The genotypic richness of A. psidii was seven, which corresponded to the number of studied MLGs. Samples from Brazil had the highest genetic diversity with four MLGs present on two host species. The clonal heterogeneity represented by the Simpson Index in this study was 0.664, with a medium probability that two individuals selected at random from the sample would belong to the same genotype. The maximum probability that this could occur was 0.691.
The aggregation and edge effect indices rejected the probability that neighboring samples did not differ on average from the global and Multi Locus Lineages (P < 0.001). This provides an indication that there is no correlation between the distance of the samples and the genotype or genetic distance obtained. IBD results were weak and non-significant (r2 = 0.0057, P = 0.156) and showed that the IBD model does not apply to these results. The geographic distance among the isolates suggested a Long Distance Dispersal (LDD) model for the spread of A. psidii across the Southern Hemisphere.
Population structure
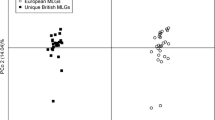
The PCoA analyses revealed the MLGs differed among countries without differentiation by host. The two axes explained 77.6% of the observed variation based on a covariance matrix. The first axis explained 46.1% of variation and the second axis explained the remaining 31.5%. Seven distinct groups were observed (Fig. 2). The first group included the isolates from Colombia (C. citriodora, E. grandis and S. jambos), Australia and Indonesia, known as the pandemic biotype. The second group included the isolates from P. guajava in Colombia. The third, fourth and fifth groups included isolates from Brazil on S. jambos with three MLGs, one of which was shared with Paraguay. The sixth group included the isolate on P. guajava from Brazil. The seventh group included the MLG from South Africa. Furthermore, the MJ network provided similar results to the PCoA, where the pandemic biotype was the most dominant MLG and included isolates from Australia, Colombia and Indonesia (Fig. 3).
Principal Coordinates Analysis of 58 isolates of Austropuccinia psidii from different hosts in the Southern Hemisphere. The two axes explain 77.55% of the observed variation based in a covariance matrix with data standardization. Purple diamonds correspond to the South African (SA) samples, red squares to the Australian isolates (AUS), blue triangles to Indonesia (IND) and light blue circles to Colombian isolates (COL). Brazil (BRA) is represented by green diamonds and orange squares are Paraguayan isolates (PAR). Each country contains the number of isolates (N). Black circles indicate the pandemic biotype and the genotypes of A. psidii on Psidium
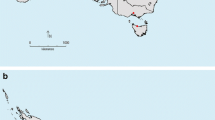
Graphic representation of the different populations of A. psidii and their location. a Median-joining network of 58 isolates of A. psidii, the size of the circles is proportional to the frequency of different Multilocus genotypes (MLG), the colour represents each of the sampling sites. b Map of the Southern Hemisphere showing the A. psidii collection sites. *refers to the isolates of P. guajava exclusively from Colombia. **represents the isolate of P. guajava from Brazil
Discussion
Studies on global populations of A. psidii have recorded a pandemic biotype present in areas of the Pacific, including Australia, California, China, Hawaii, Indonesia, Japan and New Caledonia (Ross-Davis et al. 2013; Machado et al. 2015). Results of the present study provide the first report of the pandemic biotype of A. psidii from South America. This is particularly interesting because South America has been suggested as the most likely centre of diversity of the pathogen (Joffily 1944; Coutinho et al. 1998; Glen et al. 2007; Zhong et al. 2011). The origin of the pandemic biotype is unknown, but Graça et al. (2013) determined that it most likely did not spread from the populations on Eucalyptus and S. jambos, or guava, in Brazil. The hosts of the pandemic biotype in Colombia are C. citriodora, E. grandis and S. jambos. The pandemic biotype has caused minor symptoms on species of Eucalyptus in Australia but very serious damage to various other species of Myrtaceae (Carnegie 2014). Eucalyptus trees were infected at 17 of the surveyed sites in the present study, but disease incidence was not assessed. Consequently, it is not possible to make conclusions regarding the threat of the disease to Eucalyptus plantation forestry in Colombia. Our results do, however, show that Eucalyptus may be at risk from the pandemic biotype of A. psidii if high levels of disease incidence occur in the field. This could have implications for management of the disease in Colombia and elsewhere in the world where Eucalyptus and the pandemic biotype of A. psidii co-occur.
Two genotypes of A. psidii in Colombia were recovered from 29 isolates. These included the pandemic biotype and a unique genotype on P. guajava. The genotype of A. psidii on guava has not been found in other studies and is the second known genotype on P. guajava in the native range of this host. The difference between the genotypes of A. psidii on guava in Brazil and Colombia could be explained by the natural, allopatric barrier of the Amazon jungle, which provides a 4700 km buffer from admixture between populations.
Austropuccinia psidii has been known from Colombia for over a century (Mayor 1914; Kern et al. 1933; Kern and Toro 1935). Despite intensive surveys of Eucalyptus and S. jambos since 1993, it was not found to cause consipicuous infections on these trees until 2010, when it was observed in plantations by Rodas et al. (2015). There are two possible hypotheses for the presence of the pandemic biotype of A. psidii in Colombia. One possibility is that this genotype is native to Colombia and is the result of a host shift from native to non-native species of Myrtaceae. Alternatively the pandemic genotype is a recent introduction to Colombia.
If the pandemic biotype of A. psidii is native to Colombia, it could have evolved with a native plant. This would be a plausible hypothesis because Central and South America are the likely centers of diversity for A. psidii and the probable origin of all genotypes of the pathogen. There are several host-associated genotypes of A. psidii reported by Graça et al. (2013) on Eugenia and Syzygium, and a previously unknown genotype on P. guajava is reported in the present study. It is probable that different genotypes of A. psidii are host-specific and that they evolved on native South American taxa, which are largely species in the Myrteae. Naïve genera of Myrteae such as Rhodamnia and Rhodomyrtus in Australia and other countries are highly susceptible hosts and most at risk (Pegg et al. 2014; Carnegie et al. 2016). It is plausible that many other host-associated genotypes of A. psidii will be detected in the future and these could be the source of the pandemic biotype. The hypothesis is supported because there was no correlation between location, genotype or genetic distance in the present study. It could thus be an indication of local adaptation and reproductive isolation of A. psidii on its hosts (Giraud et al. 2006). Barrès et al. (2008) suggested that when IBD is not significant, plant pathogens might have adapted locally to their host plants. This applies if recombination and selection occur on the same host. If this is the case, genetic diversity of A. psidii will be shaped by the location of different species of host plants rather than by geographic distances among the genotypes. However, the fact that the pandemic biotype was not observed to cause severe symptoms on the exotic and highly susceptible S. jambos in Colombia until 2010 (Rodas et al. 2015) does not support the hypothesis that this biotype is from Colombia.
The hypothesis that the pandemic biotype of A. psidii represents a recent introduction into Colombia must be considered. In this case, the distance between one of the most important trade ports and the site where A. psidii was first recorded on C. citriodora is approximately 94 km. This would be a possible entry point from the Pacific. While quarantine measures are enforced, failures of biosecurity programs globally have resulted in the entry of many pests and diseases of planted forests (Wingfield et al. 2015), and this would also be possible for the pandemic biotype in Colombia.
The presence of the pandemic biotype of A. psidii in Colombia on plantation-grown species of Corymbia and Eucalyptus could have repercussions for commercial forestry. The genotype of A. psidii on Eucalyptus in Brazil has been an important constraint to forestry in the past, but it has not been reported outside of that country. The discovery of a second genotype of A. psidii from these trees on the South American continent emphasizes the importance of applying more effective quarantine for Eucalyptus and related plants in Myrtaceae. This should include assessing the risk of a pathogen based on its genotype (McTaggart et al. 2016c) and developing a global collaborative strategy (Wingfield et al. 2015) to limit the spread of this rust.
References
Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA (2007) Standardizing methods to address clonality in population studies. Mol Ecol 16:5115–5139
Arnaund-Haond S, Belkhir K (2007) GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7:15–17
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barrès B, Halkett F, Dutech C, Andrieux A, Pinon J, Frey P (2008) Genetic structure of the poplar rust fungus Melampsora larici-populina: evidence for isolation by distance in Europe and recent founder effects overseas. Infect Genet Evol 5:577–587. doi:10.1016/j.meegid.2008.04.005
Beenken L (2017) Austropuccinia: a new genus name for the myrtle rust Puccinia psidii placed within the redefined family Sphaerophragmiaceae (Pucciniales). Phytotaxa 297:53–61
Burgess TI, Wingfield MJ (2016) Pathogens on the move: a 100-year global experiment with planted eucalypts. Bioscience. doi:10.1093/biosci/biw146
Buriticá CP, Pardo-Cardona VM (1996) Flora urediniana Colombiana. Revista Acad Colomb Ci Exac 20:845–236
Carnegie AJ (2014) First report of Puccinia psidii (Myrtle rust) in Eucalyptus plantations in Australia. Plant Dis 99:161. doi:10.1094/PDIS-09-14-0901-PDN
Carnegie AJ, Lidbetter JR, Walker J, Horwood MA, Tesoriero L, Glen M, Priest MJ (2010) Uredo rangelii, a taxon in the guava rust complex, newly recorded on Myrtaceae in Australia. Australas Plant Pathol 39:463–466. doi:10.1071/ap10102
Carnegie AJ, Kathuria A, Pegg GS, Entwistle P, Nagel M, Giblin FR (2016) Impact of the invasive rust Puccinia psidii (myrtle rust) on native Myrtaceae in natural ecosystems in Australia. Biol Invasions 18:127–144
Castro HA, Krugner TL, Ideriha CHF, Capello MSC, Marchi AB (1983) Inoculação cruzada de Eucalyptus, goiaba (Psidium guajava) e jambeiro (Syzygium jambos) com Puccinia psidii. Fitopatol Bras 8:491–497
Coelho L, Alfenas AC, Ferreira FA (2001) Variabilidade fisiológica de Puccinia psidii – Ferrugem do Eucalipto. Summa Phytopathol 27:295–300
Coutinho TA, Wingfield MJ, Alfenas AC, Crous PW (1998) Eucalyptus rust: a disease with the potential for serious international implications. Plant Dis 82:819–825
Dorken ME, Eckert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol 89:339–350
Farris JS (1970) Methods for computing Wagner trees. Syst Zool 19:83–72
Ferreira FA (1981) Ferrugem do eucalipto – ocorrências, temperatura para germinação de uredosporos, produção de teliosporos, hospedeiro alternativo e resistência. Fitopatol Bras 6:603–604
Ferreira FA (1983) Ferrugem do eucalipto. Rev Árvore 7:91–109
Giblin F (2013) Myrtle rust report: New Caledonia. University of the Sunshine Coast. Maroochydore, Queensland
Giblin F, Carnegie AJ (2014) Puccinia psidii (Myrtle rust) – global host list. Version current at 24 sept. 2014. https://www.Anbg.Gov.au/anpc/resources/Myrtle_Rust.Html. Accessed 06/12/2016
Giraud T, Jonot O, Shykoff JA (2006) Common sex-linked deleterious alleles in a plant parasitic fungus alter infection success but show no pleitropic advantage. J Evol Biol 19:970–980
Glen M, Alfenas AC, Zauza EAV, Wingfield MJ, Mohammed C (2007) Puccinia psidii: a threat to the Australian environment and economy – a review. Australas Plant Pathol 36:1–16
Graça RN (2011) Genetic diversity of Puccinia psidii populations. Universidade de Viçosa, PhD Dissertation
Graça RN, Ross-Davis A, Klopfenstein N, Sook M, Peever T, Cannon P, Uchida J, Alfenas AC (2011) Tracking down worldwide Puccinia psidii dispersal. BMC Proc 5:14
Graça RN, Ross-Davis AL, Klopfenstein NB, Kim M, Peever TL, Cannon PG, Aun CP, Mizubuti EG, Alfenas AC (2013) Rust disease of eucalypts, caused by Puccinia psidii, did not originate via host jump from guava in Brazil. Mol Ecol 22:6033–6047
Joffily J (1944) Ferrugem do eucalipto. Bragantia 4:475–487
Kawanishi T, Uemastu S, Kakishima M, Kagiwada S, Hamamoto H, Horie H, Namba S (2009) First report of rust disease on ohia and the causal fungus, Puccinia psidii, in Japan. J Gen Plant Pathol 75:428–431
Kern FD, Toro RA (1935) Notes on some fungi from Colombia. Mycologia 27:615–617
Kern FD, Thurston HW Jr, Whetzel HH (1933) Annotated index of the rusts of Colombia. Mycologia 25:448–503
Machado PS, Alfenas AC, Alfenas RF, Mohammed CL, Glen M (2015) Microsatellite analysis indicates that Puccinia psidii in Australia is mutating but not recombining. Australas Plant Pathol 44:455. doi:10.1007/s13313-015-0364-5.9
MacLachlan JD (1938) A rust of the pimento tree in Jamaica, British West Indies. Phytopathology 28:157–170
Mayor E (1914) Contribution à l’étude des Urédinées de Colombie. Mem Soc Sci Nat Neuchâtel 5:442–599
Marlatt RB, Kimbrough JW (1980) Puccinia psidii on Pimento dioica in South Florida. Plant Dis 68:510–512
McTaggart AR, Roux J, Granados GM, Gafur A, Tarrigan M, Santhakumar P, Wingfield MJ (2016a) Rust (Puccinia psidii) recorded in Indonesia poses a threat to forests and forestry in southeast Asia. Australas Plant Pathol 45:83–89
McTaggart AR, Shivas RG, van der Nest MA, Roux J, Wingfield BD, Wingfield MJ (2016b) Host jumps shaped the diversity of extant rust fungi (Pucciniales). New Phytol 209:1149–1158
McTaggart AR, van der Nest MA, Steenkamp ET, Roux J, Slippers B, Shuey LS, Wingfield MJ, Drenth A (2016c) Fungal genomics challenges the dogma of name-based biosecurity. PLoS Pathog 12:e1005475. doi:10.1371/journal.ppat.1005475
Nei M (1978) Estimation of heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Parks JC, Werth CR (1993) A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae). Am J Bot 80:537–544
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analyses in excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pegg GS, Giblin FR, McTaggart AR, Guymer GP, Taylor H, Ireland KB, Shivas RG, Perry S (2014) Puccinia psidii in Queensland, Australia: disease symptoms, distribution and impact. Plant Pathol 63:1005–1021
Pérez AC, Wingfield MJ, Altier NA, Simento S, Blanchette RA (2011) Puccinia psidii infecting cultivated Eucalyptus and native myrtaceae in Uruguay. Mycol Prog 10:273–282
Rayachhetry MB, Elliott ML, Van TK (1997) Natural epiphytotic of the rust Puccinia psidii on Melaleuca quinquenervia in Florida. Plant Dis 81:831
Rayachhetry MB, Van TK, Center TD, Elliot ML (2001) Host range of Puccinia psidii, a potential biological control agent of Melaleuca quinquenervia in Florida. Biol Control 22:38–45
Rodas CA, Roux J, Granados GM, Bolaños MD, McTaggart AR, Wingfield MJ (2015) First report of Puccinia psidii on Corymbia citriodora and Eucalyptus in Colombia. For Pathol 45:534–536
Ross-Davis AL, Graça RN, Alfenas AC, Peever TL, Hanna JW, Uchida JY, Hauff RD, Kadooka CY, Kim MS, Cannon PG, Namba S, Minato N, Simeto S, Pérez CA, Rayamajhi MB, Móran M, Lodge DJ, Arguedas M, Medel-Ortiz R, López-Ramirez A, Tennant P, Glen M, Klopfenstein NB (2013) Tracking the distribution of Puccinia psidii genotypes that cause rust disease on diverse Myrtaceous trees and shrubs. In: http://www.Fs.Fed.Us/rm/pubs_journals/2014/rmrs_2014_ross_davis_a001.Pdf. Accessed 10/03/2016
Roux J, Greyling I, Coutinho TA, Verleur M, Wingfield MJ (2013) The myrtle rust pathogen, Puccinia psidii, discovered in Africa. IMA Fungus 4:155–159
Roux J, Granados GM, Shuey L, Barnes I, Wingfield MJ, McTaggart AR (2016) A unique genotype of the rust pathogen, Puccinia psidii, on Myrtaceae in South Africa. Australas Plant Pathol. doi:10.1007/s13313-016-0447-y
Silveira RLVA, Higashi EN (2003) Aspectos nutricionais envolvidos na ocorrência de doeças com ênfase para o eucalipto. Instituto de pesquisas e estudios florestais. Circular Técnica IPEF 200:1–13 ISSN 0100-3453
Simpson JA, Thomas K, Grgurinovic CA (2006) Uredinales species pathogenic on species of Myrtaceae. Australas Plant Pathol 35:549–562
Stenberg P, Lundmark M, Saura A (2003) MLGSIM: a program for detecting clones using a simulation approach. Mol Ecol Notes 3:329–331
Takahashi SS (2002) Ferrugem do eucalipto: Índice de infecção, análise e estimativas de danos relacionadas à intensidade da doença no campo. MSc Thesis. Universidade Estadual Paulista. Julio de Mesquita Filho, Botucatu, Brazil
Tommerup IC, Alfenas AC, Old KM (2003) Guava rust in Brazil – a threat to Eucalyptus and other Myrtaceae. N Z J For Sci 33:420–428
Uchida J, Zhong S, Killgore E (2006) First report of a rust disease on Ohia caused by Puccinia psidii in Hawaii. Plant Dis 90:524
Wingfield MJ, Brockerhoff EG, Wingfield BD, Slippers B (2015) Planted forest health: the need for a global strategy. Science 349:832–836
Winter G (1884) Repertorium. Rabenhorstii fungi europaei et extraeuropaei exsiccati cura Dr. G. Winter, Centuria XXXI et XXXII. Hedwigia 23:164–172
Yepes MS, Buriticá P (2012) Nuevos registros de Royas (Pucciniales) en plantas de interés agronómico y ornamental en Colombia. RFNAM 65:6691–6696
Zhong S, Yang B, Alfenas AC (2008) Development of microsatellite markers for the guava rust fungus, Puccinia psidii. Mol Ecol Resour 8:348–350
Zhong S, Yang B, Puri KD (2011) Characterization of Puccinia psidii isolates in Hawaii using microsatellite DNA markers. J Gen Plant Pathol 77:178–181
Zhuang JY, Wei SX (2011) Additional materials for the rust flora of Hainan Province, China. Mycosystema 30:853–860
Acknowledgements
We are grateful to Ms. Ximena Silva (University of Helsinki, Finland) and Dr. Geoff Pegg (Department of Agriculture, Fisheries and Forestry, Horticulture and Forestry Science, Agri-Science Queensland, Australia) for providing samples from Paraguay and Australia, respectively. We also acknowledge Smurfit Kappa Colombia (SKC), the National Research Foundation (NRF) of South Africa (Grant UID 78566), and members of the Tree Protection Co-operative Program (TPCP) based at the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa for logistical and financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Granados, G.M., McTaggart, A.R., Barnes, I. et al. The pandemic biotype of Austropuccinia psidii discovered in South America. Australasian Plant Pathol. 46, 267–275 (2017). https://doi.org/10.1007/s13313-017-0488-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-017-0488-x