Abstract
Imatinib mesylate (IM) is a frontline treatment in the early chronic phase of chronic myeloid leukemia (CML). However, intrinsic and acquired resistance against this drug has been defined and this issue has become a problem and a challenge in CML treatment. According to new findings, the inhibition of Janus kinase 2 (Jak2) in Bcr–Abl+ cells can promote apoptosis in IM-resistant cells. microRNAs (miRNAs) regulate the gene expression by targeting the messenger RNA (mRNA) for degradation. Recently, a growing body of evidence has implicated that dysregulation of miRNAs is associated with cancer initiation and development. In this report, we proposed that miRNA-101 targets Jak2 mRNA and regulates its expression and induces K562 leukemia cell apoptosis. Here, we transduced the K562 cell line with a miR-101-overexpressing vector and evaluated the Jak2 mRNA level. Our results showed that miR-101 overexpression in Bcr–Abl+ cells reduced the Jak2 mRNA level. Moreover, imatinib treatment and miR-101 upregulation led to miR-23a overexpression, which has putative binding site(s) on 3′-untranslated regions (3′-UTRs) of STAT5, CCND1, and Bcl-2 genes. Our results also indicated that miR-101 overexpression inhibited cell proliferation indicated by the MTT assay and promoted apoptosis detected via flow cytometry. Importantly, mRNA expression of NF-kappa B-regulated anti-apoptotic (Bcl-2, Bcl-xl, MCL-1, XIAP, and survivin) and proliferative (c-Myc and CCND1) genes was decreased. These findings suggest that miR-101 acts as a tumor suppressor by downregulating Jak2 expression and sensitizing K562 cells to imatinib. Therefore, restoration of miR-101 may be a therapeutic approach for CML treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder associated with the presence of the Philadelphia chromosome (Ph) that results from a reciprocal translocation between chromosome 9 and 22, t(9;22), that encodes the Bcr–Abl fusion protein [1]. Bcr–Abl activates different signaling pathways including the Ras/Raf/MAPK, PI3K/AKT/mTOR, and JAK/STAT pathways [2–4]; hence, it causes chronic myeloid leukemia cell proliferation, abnormal interactions with the extracellular matrix and stroma [5], and impaired apoptosis [6].
Discovery of tyrosine kinase inhibitors (TKIs) has revolutionized the treatment of CML [7]. The most well known TKI, imatinib, inhibits the Bcr–Abl activity and blocks its downstream signaling pathways. However, a major obstacle to its success is the development of therapeutic resistance as a result of mutations in the Bcr–Abl tyrosine kinase domain (for example, T315I) [8], increasing the copy number of the Bcr–Abl gene [9], and a Bcr–Abl independent mechanism involving Lyn kinase (for example, K562-R cells) [10].
Jak2 tyrosine kinase activated by the Bcr–Abl oncoprotein has a critical role in Bcr–Abl-mediated oncogenicity [11, 12]. There is some evidence that shows that the Bcr–Abl/Jak2 complex stabilizes the Bcr–Abl kinase activity [13]. Therefore, Jak2 inhibition can overcome the resistance to imatinib treatment through the induction of apoptosis in imatinib-resistant cell lines and primary samples of blast crisis (BC) CML patients [14].
MicroRNAs (miRNAs) are a group of endogenously expressed, ~22-nucleotide-long, non-coding small RNAs that are known to negatively regulate the expression of up to 30 % of the human genes by directly binding to the 3′-untranslated region (UTR) of target messenger RNAs (mRNAs) [15]. miRNAs follow two mechanisms for gene silencing: translation repression and/or degradation of the target mRNAs [16]. miRNAs have been found to play important roles in several biological processes including cellular development, apoptosis, differentiation, and tumor growth [17]. Increasing evidence from experimental studies indicates that miRNAs might be involved in cancer by functioning as oncogenes or tumor suppressors [18]. Based on increasing studies, miRNAs may play a role in the chemosensitivity and chemoresistance of human cancer cells [19, 20]; for example, upregulation of miR-424 sensitized K562 cells to imatinib [21]. miR-29b has been shown to augment apoptosis induction by bortezomib in multiple myelomas [22]. miR-101 is a member of a family of miRNAs that are involved in a wide range of cellular activities, like cell proliferation, invasion, and angiogenesis [23]. miR-101-coding genes are located in the genomic fragile regions so that they are prone to being deleted or amplified in several cancers [24]. In particular, several studies have demonstrated that miR-101 can suppress the oncogenic signal transduction in breast [25], ovarian [23], and gastric cancers [26]; hence, miR-101 can play a tumor-suppressive role in tumor cell growth, migration, invasion, and drug resistance. Several studies have indicated that miR-101 downregulation correlates inversely with increased EZH2 and MCL-1 expression levels in many cancer cell types [27]. Lu Wang et al. demonstrated that miR-101 suppressed the tumor cell growth and promoted apoptosis in breast cancer cells by targeting Jak2 [28]. So, it seems that Jak2 inhibition by miR-101 with regard to Jak2’s crucial role in Bcr–Abl-mediated oncogenicity promotes apoptosis and suppresses Jak2-regulated pathways. In the current study, we investigated the therapeutic potential of miR-101 in K562 cells as a chemosensitizer for imatinib treatment. Our results reveal the anti-tumor effects of miR-101 in K562 leukemia cells by targeting Jak2 and NF-κB anti-apoptotic target genes. These findings strongly suggest that upregulation of miR-101 in CML cells has a role in enhancing drug sensitivity to imatinib.
Materials and methods
Cell culture
K562 cells were grown in the RPMI-1640 medium supplemented with 2 mM l-glutamine, 10 % fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin in an incubator with 5 % CO2 and 95 % humidity at 37 °C (Memert, Germany). The HEK 293T cells (kindly provided by Dr. Frank Grosveld, Erasmus MC) used for the production of lentiviruses were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % FBS and penicillin–streptomycin.
Cell death analysis by flow cytometry
Cell viability was determined by the propidium iodide (PI)-exclusion test. Because the intact membrane of the live cells excludes charged dyes like PI, short-term incubation of these dyes results in selective labeling of the dead cells, whereas live cells have no or minimal uptake. Cells were incubated with 10 μg/ml PI for 10 min prior to flow cytometric analysis using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Data was analyzed using the CellQuest software (BD Biosciences).
Metabolic activity measured by the MTT assay
The effect of Jak2 downregulation on the proliferation of K562 cells was assessed by the MTT colorimetric method. Briefly, miR-101-transduced cells and negative control (NC)-transduced cells were seeded into a 96-well culture plate at a density of 10 × 103 cells/well. Some of the wells were treated with 250 nM of imatinib and incubated for 72 h. After removing the medium, the cells were incubated with MTT solution (5 mg/ml in PBS) for 4 h and the resulting formazan was solubilized with DMSO (100 μl). The absorbance of each well was measured at 570 nm in an ELISA reader.
Sub-G1 DNA content analysis
Apoptotic cells were detected using PI staining of miRNA-transduced cells followed by flow cytometry to detect the so-called sub-G1 peak. Briefly, 7 days after removal of the virus-containing medium, 0.4 × 106 cells were detected by flow cytometry. Transduced cells were then harvested and washed once in PBS and fixed with 70 % ethanol. Then, the cells were treated with 0.5 μg/ml RNase in PBS and incubated at 37 °C for 30 min before staining with 50 μg/ml PI for 30 min. The cells were analyzed using a FACScan flow cytometer (Becton Dickinson).
Phosphatidylserine externalization (Annexin V assay)
The Annexin V binding assay was performed to detect apoptosis in Jak2 downregulated cells. The K562 transduced cells were treated with 250 nM of imatinib for 72 h and were then washed with PBS after the incubation time. A total of 0.4 × 106 cells were washed with PBS and resuspended in a total volume of 100 μl of the incubation buffer. Annexin V (5 μl per sample) (BD Biosciences, USA) was added, and the cell suspension was incubated for 20 min in the dark. Fluorescence was then measured using flow cytometry. The data was evaluated using the CellQuest software (Becton Dickinson) and expressed as percentage of the cells positive for Annexin V (early apoptotic phase).
Lentiviral production and infection
On the first day, 5 × 106 HEK-293T cells were seeded in a 10-cm dish. In the next morning, the HEK293T cells were transfected with GFP hsa-miR-101-1 Lentivector and pLenti-III-mir-GFP-Blank (ABM Inc., Canada) together with two packaging vectors (psPAX2 and pMD2.G) using X-tremeGENE™ HP DNA Transfection Reagent (Roche Diagnostics). Lentivirus-containing supernatants were harvested at 48 and 72 h after transfection and filtered through a low-protein binding 0.45-μM sterile filter (Merck Millipore Ltd). To infect the cells, a virus-containing medium supplemented with 8 μg/ml SureENTRY™ Transduction Reagent (Qiagen) was added to 2 × 105 K562 cells in 24-well plates. The viral supernatant was replaced with normal culture medium after 24 h, and infection efficiency was monitored by GFP expression at 48 h after infection.
RNA preparation and quantitative real-time PCR
For reverse transcription PCR (RT-PCR), the total RNA with miRNA was purified using the miRNeasy Mini Kit (Qiagen) following the manufacturer’s instruction. One microgram of the isolated RNA was used for the preparation of cDNA using the miScript II RT Kit (Qiagen). The level of miRNAs was quantified using the miScript SYBR Green PCR Kit and miScript Primer Assay (Qiagen) in the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, USA). The data was normalized to U6 expression (Qiagen). The qRT-PCR of mRNA templates (1 μg) was performed using the RealQ Plus 2× Master Mix Green High ROX™ (Ampliqon, PCR enzymes & Bioreagents, Denmark) in the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, USA). Values are expressed as relative expression of mRNA normalized to the housekeeping GAPDH mRNA. Relative quantification (RQ) values were calculated as 2−ΔΔCt by the comparative Ct method. The experiment was repeated three times and representative data sets are shown. Primer sequences for determining mRNA level are available upon request.
Statistical analysis
All data were analyzed using the IBM SPSS software (version 20). All numerical data were generated from three independent experiments and are presented as means ± SEM. Differences between groups were examined using the Student’s t test. A two-tailed difference with P < 0.05 was considered statistically significant.
Results
miR-101 has inhibitory effect on K562 cell viability index
In order to investigate the effect of miR-101 on the induction of cell death, two groups of cells including miR-101-transduced and NC-transduced cells were analyzed using PI staining followed by flow cytometry. As shown in Fig. 1a, miR-101 overexpression induced cell death as indicated by the increased percentage of dead cells. To evaluate cell proliferation, the MTT assay was performed on miR-101-transduced and negative control cells 7 days after the removal of the lentivirus-containing medium. As shown in Fig. 1b, the population of miR-101-overexpressed cells was significantly decreased compared to the NC-transduced cells.
Effects of miR-101 overexpression on cell proliferation and cell death in K562 cells. a miR-101 induces cell death in K562. Cell viability was detected by PI assay in K562-transduced and NC-transduced groups and analyzed by flow cytometery. One representative experiment of at least three performed is presented. b Cell proliferation assay of K562-tansduced cells. Transduced cells were grown in complete medium, and cell growth was measured by MTT assay. The metabolic activity of miR-101-transduced cells was decreased. The results are expressed as mean ± SD of at least three independent experiments (*P < 0.05 relative to NC-transduced cells). Con = control
miR-101 induces a cytotoxic effect through apoptosis in K562 cells
To address whether miR-101-induced cell death in K562 cells was due to apoptosis, we evaluated the modulation of phosphatidylserine externalization as an indicator of early apoptosis in K562 leukemia cells by flow cytometry. Our results showed that the percentage of Annexin V-positive transduced cells was significantly increased compared to NC-transduced cells (Fig. 2). Altogether, these findings suggest that miR-101 overexpression induces apoptosis in chronic myeloid leukemia CML K562 cells.
Apoptotic effect of miR-101 overexpression on K562 cells. K562 cells were transduced with miR-101 and NC vectors. Cells were analyzed for Annexin V by flow cytometry. Results showed increased apoptosis in K562 cells overexpressing miR-101 compared with those in the vector control cells. One representative experiment of at least three performed is presented
miR-101 influences cell cycle progression of K562 leukemia cells
As miR-101 overexpression influences K562 leukemia cell proliferation, we evaluated cell cycle changes in leukemia cells after miR-101 upregulation. The results of flow cytometry showed that miR-101 overexpression caused an arrest in the G0/G1 phase and blocked cells before the S phase (Fig. 3). As shown in Fig. 3, overexpression of miR-101 led to G0/G1 arrest and the proportion of sub-G1 apoptotic cells was also significantly increased. These findings suggest that miR-101 arrests K562 leukemia cells at the G0/G1 phase, thereby inhibiting cell cycle progression.
Effects of miR-101 on cell cycle distribution in K562 cells. K562 cells were transduced with miR-Con or miR-101. Cells were collected, stained with PI, and processed for analysis by flow cytometry to measure DNA content. Flow cytometry data was analyzed to determine cell cycle distribution (G0/G1, S, and G2/M) in the surviving cell populations. One representative experiment of at least three performed is presented (*P < 0.05 relative to NC-transduced cells)
Transduction of miR-101 augments the effect of imatinib in inducing apoptosis of K562 cells
Imatinib treatment reduced cell viability and increased the apoptotic rate of K562 cells compared to untreated cells. miR-101 overexpression enhances the apoptosis-inducing effect of imatinib on K562 cells. To assess the effect of miR-101 and imatinib on apoptosis in K562 cells, we evaluated the modulation of phosphatidylserine externalization after transduction and imatinib treatment. We found that imatinib treatment of miR-101-transduced cells increased apoptosis rather than either alone (Figs. 2 and 4a). Moreover, cell viability and cell proliferation were more affected after treatment of miR-101-trancduced cells by imatinib compared to untreated miR-101-transduced cells and NC-transduced cells with or without imatinib treatment (Figs. 1a, b and 4b, c). Collectively, these results indicate that transduction of miR-101 enhances the imatinib-induced cytotoxicity.
Effects of imatinib on apoptosis and cell proliferation in K562-transduced cells. a Transduced cells were treated with 250 nM imatinib for 72 h. Cells were analyzed for Annexin V by flow cytometry. Results showed that miR-101 overexpression enhances imatinib-induced apoptosis. One representative experiment of three performed is shown. b Imatinib induces cell death in miR-101 and NC-transduced cells. Transduced cells were treated with 250 nM of imatinib and cultured for 72 h. Then, cell death was analyzed for PI by flow cytometery. One representative experiment of at least three performed is presented. c Cell proliferation assay of imatinib treated K562-transduced cells. Cells were cultured in complete medium with 250 nM of imatinib in 96-well plates for 72 h, and cell growth was measured by MTT assay. The results are expressed as mean ± SD of at least three independent experiments (*P < 0.05 relative to NC-transduced cells)
Jak2 and MCL-1 may be targets for miR-101
In order to identify the potential targets of miR-101, we performed gene expression analysis using the miR-101-transduced K562 cells relative to NC-transduced cells. First, we identified 11 genes to be downregulated by the miR-101 overexpression. Then, using two online prediction software programs (TargetScan.org and miRNA.org) that have high precision and sensitivity [29, 30], we found that only two of them could be predicted targets for miR-101. To show that miR-101 negatively regulated these two genes, we transduced K562 cells by the miR-101 expression vector (Fig. 5a, b) Interestingly, we noticed that Jak2 and MCL-1 that contained putative binding site(s) on their 3′-UTRs (Fig. 5c) were considerably downregulated in miR-101-transduced K562 cells (Fig. 5d). In agreement with this finding, a negative correlation was seen between endogenous miR-101 level and Jak2 and MCL-1 mRNA expression in K562 cells transduced by miR-101. Our data also showed that imatinib treatment of miR-101-transduced K562 cells and NC-transduced cells resulted in increased Jak2 and MCL-1 mRNA levels as compared to untreated cells, which was associated with the effect of imatinib on miR-101 downregulation (Fig. 5e, f). These observations indicate that imatinib treatment leads to miR-101 downregulation that negatively regulates Jak2 and MCL-1 expression.
Upregulated miR-101 significantly affected Jak2 and MCL-1 expression. a Verification of miR-101 transduction. The qRT-PCR showed that expression level of miR-101 is nine times than the normal time. The difference was statistically significant. b The fluorescence microscopy image showed miR-101 expression in K562 cells. c Two predicted miR-101 target sites in the 3′-UTR of Jak2 and one predicted miR-101 target site in the 3′-UTR of MCL-1. The seed sequences of the two miRNAs are bolded, and the matched or complementary nucleotides between the miRNAs and the Jak2 and MCL-1 3′-UTR are indicated. d Jak2 and MCL-1 downregulation (normalized to GAPDH) in microRNA-transduced cells analyzed by real-time PCR. e, f Imatinib treatment of miR-101 and NC-transduced K562 cells leads to Jak2 and MCL-1 upregulation in qRT-PCR compared to untreated cells, which is associated with miR-101 expression. One representative experiment of at least three performed is presented (*P < 0.05 relative to NC-transduced cells)
Transduction of CML cell line (K562) with miR-101 reduced c-Myc and hTERT expression
It has been shown that the transcription factor c-Myc is required for Bcr–Abl transformation [31]. Furthermore, Xie et al. showed that Jak2 was involved in c-Myc overexpression [12]. A growing body of data indicates that various transcription factors regulate the human telomerase reverse transcriptase (hTERT) expression by acting on the hTERT promoter, and it seems that c-Myc is a strong regulator of the hTERT gene expression [32, 33]. Due to Jak2 downregulation in miR-101-transduced cells and given the regulatory role of Jak2 in c-Myc and hTERT mRNA expression, we evaluated c-Myc and hTERT mRNA levels 7 days after removal of the lentivirus-containing medium using the qRT-PCR. As presented in Fig. 6a, c-Myc and hTERT mRNA levels decreased in miR-101-transduced cells compared to NC-transduced cells. Imatinib treatment reduced the hTERT mRNA level rather than miR-101 overexpression (Fig. 6a, c). We also showed that imatinib treatment enhanced the effect of miR-101 overexpression in c-Myc and hTERT mRNA downregulation (Fig. 6b, c).
miR-101 expression executes transcriptional suppression of c-Myc and hTERT. a Real-time PCR showed c-Myc and hTERT (normalized to GAPDH) downregulation in K562 cells after miR-101 overexpression. b, c Down modulation of c-Myc and hTERT following imatinib treatment. miR-101 and NC-transduced cells were treated with 250 nM imatinib for 72 h, after which RNA was harvested, and expression of the indicated genes was measured using quantitative RT-PCR and normalized to the expression of GAPDH. The figure shown is a representative of at least three independent experiments (*P < 0.05 relative to NC-transduced cells)
miR-101 overexpression downregulates STAT5, Bcl-2, and CCND1 mRNA levels via upregulation of miR-23a
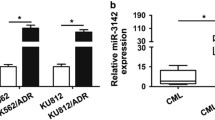
As shown in Fig. 7a, miR-101 overexpression and imatinib treatment led to upregulation of the miR-23a expression. A bioinformatic analysis (http://www.targetscan.org/) found putative target sites of miR-23a in the 3ʹ-UTR of signal transducer and activator of transcription (STAT) 5, cell cycle regulator gene CCND1, and anti-apoptotic gene Bcl-2 (Fig. 7b). STAT5 is constitutively active in Bcr–Abl+ cells [34] that support a leukemogenic role for this transcription factor in CML. It has been reported that CCND1 is a key regulator of the G1/S transition [35] and Bcl-2 is known as an anti-apoptotic gene [36]. Our results showed that STAT5, CCND1, and Bcl-2 mRNA levels decreased in miR-101-transduced cells (Fig. 7c). We also showed that imatinib enhanced the miR-101 overexpression effect on upregulation of miR-23a and downregulation of these three mRNAs. These observations indicate that STAT5, CCND1, and Bcl-2 genes may serve as targets for miR-23a, which was overexpressed upon miR-101 upregulation or imatinib treatment in K562 cells.
Upregulated miR-101 and imatinib treatment leads to miR-23a overexpression. a miR-23a upregulation (normalized to RNU6) in K562 and microRNA-transduced cells analyzed by real-time PCR. miR-101 and NC-transduced cells were treated with 250 nM imatinib for 72 h, and then the overexpression of miR-23a was measured using quantitative RT-PCR and normalized to the expression of RNU6. b STAT5, CCND1, and Bcl-2 are putative targets of miR-23a. Sequence alignments of miR-23a with their corresponding potential target sites in the 3′-UTR of STAT5, CCND1, and Bcl-2. The seed sequence of the miRNA is bolded. c STAT5, CCND1, and Bcl-2 relative mRNA expression (normalized to GAPDH) in transduced cells analyzed by real-time PCR. Imatinib treatment of miR-101 and NC-transduced K562 cells leads to STAT5, CCND1, and Bcl-2 downregulation in qRT-PCR compared to untreated cells, which is associated with miR-101 expression. The figure is a representative of three independent qPCRs (*P < 0.05 relative to NC-transduced cells)
miR-101-induced downregulation of the STAT5 expression in the K562 cell line promotes apoptosis
It has been reported that increased STAT5 expression results in resistance to TKI treatment [37]; the molecule is now considered a potential therapeutic target in CML CD34+ cells. We observed a significantly decreased expression of STAT5 accompanied by decreased STAT5 target genes Bcl-xL, MCL-1, and Bcl-2 (Figs. 8, 5d, and 7c). Furthermore, STAT5 was downregulated upon imatinib treatment in both miR-101-transduced and NC-transduced cells. Interestingly, the miR-101 overexpression augmented the imatinib effect on STAT5 downregulation. The data confirmed that miR-101 overexpression in combination with imatinib treatment resulted in the downregulation of STAT5 and its anti-apoptotic target genes Bcl-xL, MCL-1, and Bcl-2, inducing apoptosis (Figs. 8, 5e, f, and 7c).
Upregulated miR-101 exerts transcriptional suppression of STAT5 target gene. Relative Bcl-xL mRNA expression (normalized to GAPDH) after transduction with miR-101 by qPCR. miR-101 and NC-transduced cells were treated with 250 nM imatinib for 72 h. The relative expression of the indicated gene was measured using quantitative RT-PCR and normalized to the expression of GAPDH. The figure shown is a representative of at least three independent experiments (*P < 0.05 relative to NC-transduced cells)
miR-101 induced downregulation of NF-κB-regulated anti-apoptotic genes
Chronic myeloid leukemia is characterized by the Bcr–Abl fusion protein that can activate the NF-κB pathway [38]. Interestingly, it has been reported that Jak2 is involved in NF-κB activation by Bcr–Abl [39]. It is of interest to note that NF-κB can promote the cellular growth process through the regulation of specific target genes like CCND1 and c-Myc [40]. Furthermore, NF-κB directly regulates a range of anti-apoptotic genes including Bcl-2, Bcl-xL, MCL-1, XIAP, and survivin [41]. In our study, qRT-PCR showed that CCND1, c-Myc, Bcl-2, MCL-1, Bcl-xL, XIAP, and survivin mRNA levels were significantly downregulated in miR-101-transduced cells compared to NC-transduced cells (Figs. 7c, 6a, 5d, 8, and 9). These findings revealed that miR-101 overexpression led to the downregulation of anti-apoptotic NF-κB target genes in K562 cells, thereby promoting apoptosis. We also showed that imatinib treatment enhanced the effect of miR-101 overexpression on NF-κB-regulated anti-apoptotic genes except for MCL-1 (Figs. 7c, 6b, c, 5e, f, 8, and 9).
miR-101 overexpression decreases the NF-κB-regulated anti-apoptotic genes. Survivin and XIAP downregulation (normalized to GAPDH) in miR-101-transduced cells analyzed by real-time PCR. miR-101 and NC-transduced cells were treated with 250 nM imatinib for 72 h. The relative expression of survivin and XIAP was measured using qPCR and normalized to the expression of GAPDH. The figure is a representative of three independent qPCRs (*P < 0.05 relative to NC-transduced cells)
Discussion
Based on previous studies, Jak2 is a critical signaling molecule in Bcr–Abl+ cells [14, 42]. Xie et al. showed that Jak2 activation by Bcr–Abl induced c-Myc expression [12]. So, the c-Myc mRNA level is inhibited either by the selective Bcr–Abl tyrosine kinase inhibitor (imatinib) or by the Jak2 kinase inhibitor (AG490). Hence, the Jak2/c-Myc pathway may play an important role in CML progression that can be inhibited by Jak2 inhibition.
Interestingly, Bcr–Abl activates the NF-κB pathway, which leads to NF-κB-regulated gene expression [38]. It has been reported that Jak2 is involved in NF-κB activation by Bcr–Abl [39]. Given the Jak2 role in Bcr–Abl+ cells and its critical role in tumorgenicity, it is not surprising that Jak2 inhibition controls oncogenic mechanisms. Samanta et al. showed that Jak2 inhibition resulted in the apoptosis induction in leukemia cells expressing imatinib-resistant forms of Bcr–Abl [14]. Therefore, Jak2 inhibitors have a potential for the treatment of imatinib-resistant forms of CML.
miRNAs are involved in multiple cellular processes including proliferation, differentiation, and apoptosis [43]. Increasing amounts of data have shown that miRNAs can act as oncogenes or tumor suppressors and participate in cancer development and progression [44].
According to the results of online prediction software, it was found that miR-101 might pair the Jak2 3′-UTR. Given the ectopic expression of miR-101 in the numerous cancer cell lines and tissues [45], miR-101 acts as a cancer-related miRNA. All target genes regulated by miR-101 have been found to be involved in tumorigenesis; therefore, miR-101 can be considered a tumor suppressor in cancer therapy [45, 46]. Our present study is consistent with the results of a study by Wang et al. [28] reporting that Jak2 is a direct target of miR-101. In this study, we showed Jak2 upregulation and miR-101 downregulation as a result of K562 leukemia cell imatinib treatment. Our results also indicated that miR-101 overexpression in K562 cells resulted in Jak2 downregulation, induced apoptosis, and enhanced imatinib-induced apoptosis. However, whether miR-101 upregulation has any effect on the Jak2 function and how the miR-101 mRNA level is regulated during imatinib treatment remain to be investigated.
It has been reported that hTERT has a pivotal role in cellular immortalization. In most somatic cells, hTERT has a very low expression and activity, while it is highly expressed and has a high activity level in approximately 85 % of the tumors. Uziel et al. demonstrated that imatinib repressed the telomerase activity in both Bcr–Abl-positive and Bcr–Abl-negative cells [47] and telomerase inhibition could inversely augment imatinib-induced apoptosis in K562 cells [48].
C-Myc, a potent regulator of telomerase, positively regulates hTERT expression, which has been demonstrated in different cancers [49]. A positive correlation between c-Myc and hTERT expression has been shown in leukemia cells by silencing c-Myc mRNA leading to the inhibition of the telomerase activity [50]. It has been reported that Jak2 activation by Bcr–Abl induces the expression of transcription factor c-Myc, which is required for Bcr–Abl transformation [12, 31].
Given the role of Jak2 in enhancing c-Myc expression [12], it seems that Jak2 downregulation may result in c-Myc and hTERT mRNA repression. Our findings indicated that upregulation of miR-101 resulted in c-Myc and hTERT mRNA repression due to Jak2 downregulation in k562 cells compared to control cells. Furthermore, our data showed that miR-101 overexpression augmented the effect of imatinib on decreasing c-Myc and hTERT mRNA levels. With regard to these results, miR-101 overexpression can regulate the cell growth by c-Myc and hTERT mRNA expression through Jak2 downregulation.
Across the various signaling pathways activated by Bcr–Abl in CML cells, NF-κB may be the substantial pathway. Interestingly, it has been reported that Jak2 is involved in NF-κB activation by Bcr–Abl [39]. NF-κB promotes cell growth through upregulation of key regulators of cell proliferation such as c-Myc and CCND1 [51, 52]. In addition, NF-κB suppresses apoptosis via regulating anti-apoptotic genes such as Bcl-2, Bcl-xL, MCL-1, XIAP, and survivin [38, 41] which are frequently overexpressed in human cancers. Given the fundamental role of NF-κB in the regulation of apoptosis and cell survival [53], it is not surprising that the inhibition of NF-κB leads to apoptosis. Our results indicated that miR-101 could suppress anti-apoptotic and proliferative genes regulated by NF-κB. In addition, miR-101 enhances the effect of imatinib on NF-κB-regulated genes. Furthermore, we showed that miR-101 overexpression and imatinib treatment resulted in miR-23a upregulation. Interestingly, online prediction tools show that miR-23a is a predicted miRNA for transcription regulation of STAT5, CCND1, and Bcl-2 genes. Therefore, the most straightforward interpretation of our results is that miR-23a may target key signals involved in the survival of CML cells.
In conclusion, our findings showed that miR-101 sensitized K562 leukemia cells to imatinib treatment through Jak2 repression and miR-23a upregulation. Therefore, restoration of miR-101 expression alone, or in combination with imatinib, may be a potential therapeutic strategy for CML treatment.
References
Kantarjian HM, Talpaz M, Giles F, O’Brien S, Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med. 2006;145(12):913–23.
Sawyers CL, McLaughlin J, Witte ON. Genetic requirement for Ras in the transformation of fibroblasts and hematopoietic cells by the Bcr-Abl oncogene. J Exp Med. 1995;181(1):307–13.
Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16(20):6151–61.
Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159(10):4720–8.
Helgason GV, Karvela M, Holyoake TL. Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood. 2011;118(8):2035–43.
Goldman JM, Melo JV. BCR-ABL in chronic myelogenous leukemia—how does it work? Acta Haematol. 2008;119(4):212–7.
Savage DG, Antman KH. Imatinib mesylate—a new oral targeted therapy. N Engl J Med. 2002;346(9):683–93.
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001; 293(5531):876–880.
Saffroy R, Lemoine A, Brezillon P, Frenoy N, Delmas B, Goldschmidt E, et al. Real-time quantitation of bcr-abl transcripts in haematological malignancies. Eur J Haematol. 2000;65(4):258–66.
Donato NJ, JY W, Stapley J, Lin H, Arlinghaus R, Aggarwal BB, et al. Imatinib mesylate resistance through BCR-ABL independence in chronic myelogenous leukemia. Cancer Res. 2004;64(2):672–7.
Xie S, Wang Y, Liu J, Sun T, Wilson MB, Smithgall TE, et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 2001;20(43):6188–95.
Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene. 2002;21(47):7137–46.
Chen M, Gallipoli P, DeGeer D, Sloma I, Forrest DL, Chan M, et al. Targeting primitive chronic myeloid leukemia cells by effective inhibition of a new AHI-1-BCR-ABL-JAK2 complex. J Natl Cancer Inst. 2013;105(6):405–23.
Samanta AK, Chakraborty SN, Wang Y, Kantarjian H, Sun X, Hood J, et al. Jak2 inhibition deactivates Lyn kinase through the SET-PP2A-SHP1 pathway, causing apoptosis in drug-resistant cells from chronic myelogenous leukemia patients. Oncogene. 2009;28(14):1669–81.
Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–50.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14(1):1–6.
Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7(1):1–9.
Salerno E, Scaglione BJ, Coffman FD, Brown BD, Baccarini A, Fernandes H, et al. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol Cancer Ther. 2009;8(9):2684–92.
Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, et al. Restoration of miR-424 suppresses BCR-ABL activity and sensitizes CML cells to imatinib treatment. Cancer Lett. 2015;360(2):245–56.
Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012:3e436.
Semaan A, Qazi AM, Seward S, Chamala S, Bryant CS, Kumar S, et al. MicroRNA-101 inhibits growth of epithelial ovarian cancer by relieving chromatin-mediated transcriptional repression of p21(waf(1)/cip(1)). Pharm Res. 2011;28(12):3079–90.
Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, et al. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105(2):296–303.
Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72(12):3091–104.
He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, et al. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. Febs J. 2012;279(22):4201–12.
Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60(3):590–8.
Wang L, Li L, Guo R, Li X, Lu Y, Guan X, et al. miR-101 promotes breast cancer cell apoptosis by targeting Janus kinase 2. Cell Physiol Biochem. 2014;34(2):413–22.
Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25(23):3049–55.
Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11(2):93–109.
Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70(6):901–10.
Wojtyla A, Gladych M, Rubis B. Human telomerase activity regulation. Mol Biol Rep. 2011;38(5):3339–49.
Cerni C. Telomeres, telomerase, and myc. An update. Mutat Res. 2000;462(1):31–47.
Carlesso N, Frank DA, Griffin JD. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183(3):811–20.
CL S, Deng TR, Shang Z, Xiao Y. JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. Int J Hematol. 2015;102(1):76–85.
Tsujimoto Y, Croce CM. Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc Natl Acad Sci U S A. 1986;83(14):5214–8.
Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011;117(12):3409–20.
Reuther JY, Reuther GW, Cortez D, Pendergast AM, Baldwin Jr AS. A requirement for NF-kappaB activation in Bcr-Abl-mediated transformation. Genes Dev. 1998;12(7):968–81.
Digicaylioglu M, Lipton SA. Erythropoietin-mediated neuroprotection involves cross-talk between Jak2 and NF-kappaB signalling cascades. Nature. 2001;412(6847):641–7.
Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–30.
Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40.
Samanta AK, Lin H, Sun T, Kantarjian H, Arlinghaus RB. Janus kinase 2: a critical target in chronic myelogenous leukemia. Cancer Res. 2006;66(13):6468–72.
Patel N, Tahara SM, Malik P, Kalra VK. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem J. 2011;434(3):473–82.
Pouladi N, Kouhsari SM, Feizi MH, Gavgani RR, Azarfam P. Overlapping region of p53/wrap53 transcripts: mutational analysis and sequence similarity with microRNA-4732-5p. Asian Pac J Cancer Prev. 2013;14(6):3503–7.
Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–9.
Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82(18):9065–74.
Uziel O, Fenig E, Nordenberg J, Beery E, Reshef H, Sandbank J, et al. Imatinib mesylate (Gleevec) downregulates telomerase activity and inhibits proliferation in telomerase-expressing cell lines. Br J Cancer. 2005;92(10):1881–91.
Tauchi T, Nakajima A, Sashida G, Shimamoto T, Ohyashiki JH, Abe K, et al. Inhibition of human telomerase enhances the effect of the tyrosine kinase inhibitor, imatinib, in BCR-ABL-positive leukemia cells. Clin Cancer Res. 2002;8(11):3341–7.
Latil A, Vidaud D, Valeri A, Fournier G, Vidaud M, Lidereau R, et al. htert expression correlates with MYC over-expression in human prostate cancer. Int J Cancer. 2000;89(2):172–6.
Fujimoto K, Takahashi M. Telomerase activity in human leukemic cell lines is inhibited by antisense pentadecadeoxynucleotides targeted against c-myc mRNA. Biochem Biophys Res Commun. 1997;241(3):775–81.
Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–6.
Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107(3):241–6.
Burstein E, Duckett CS. Dying for NF-kappaB? Control of cell death by transcriptional regulation of the apoptotic machinery. Curr Opin Cell Biol. 2003;15(6):732–7.
Acknowledgments
This study was supported by the grant 24412 from Iran University of Medical Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Farhadi, E., Zaker, F., Safa, M. et al. miR-101 sensitizes K562 cell line to imatinib through Jak2 downregulation and inhibition of NF-κB target genes. Tumor Biol. 37, 14117–14128 (2016). https://doi.org/10.1007/s13277-016-5205-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5205-9