Abstract
Ubiquitin-conjugating enzyme E2T (UBE2T) is a member of the E2 family that mediates the ubiquitin-proteasome system and regulates gene expression. It is a major oncogene in several cancers such as lung cancer and breast cancer, while the potential functions of UBE2T in gastric cancer (GC) remains largely unknown. Here, we identified the roles of UBE2T in GC progression and its potential to act as a prognostic marker of GC. Our data demonstrated that UBE2T was significantly upregulated in gastric cancer tissues, and the high expression of UBE2T was significantly correlated with poor differentiation, high T classification, and poor prognosis. In vitro experiments indicated that UBE2T promoted cell proliferation and inhibited cell cycle arrest. In addition, we observed that UBE2T modulated cell mobility by inducing epithelial-mesenchymal transition. Collectively, these findings suggest that UBE2T plays an important role in the tumorigenesis of gastric cancer and could act as a potential independent prognostic factor for cancer therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the fourth most common type of cancer worldwide, and almost two-thirds of these new cases occurred in developing countries [1–3]. Although the multimodal treatment strategies for the disease have significantly improved, the prognosis for advanced GC diseases remains poor, and GC still is a major clinical challenge [4–6]. Therefore, it is imperative to investigate the molecular and genetic mechanisms responsible for GC development.
Ubiquitin-conjugating enzyme E2T (UBE2T), also named HSPC150, is a member of the E2 family. It belongs to the ubiquitin-proteasome degradation system and participates in conjugating ubiquitin to the substrate. This process controls the turnover of regulatory proteins involved in essential cellular processes such as cell cycle progression, differentiation, apoptosis, angiogenesis, and cellular signaling pathways. For instance, UBE2T has been reported to be recruited independently to regulate FANCD2 monoubiquitination and participates in the Fanconi anemia pathway together with FANCD2 [7, 8]. Interestingly, the UBE2T gene is located in 1q32.1 [9], which is amplified in a wide variety of cancers, and overexpression of UBE2T has been reported in various cancers such as lung carcinomas [10, 11], bladder carcinomas [12], and prostate carcinomas [13]. However, whether UBE2T plays any functional roles in GC has not been reported.
In the present studies, we performed immunohistochemistry experiment to evaluate the expressions of UBE2T on GC tissue arrays. We investigated the correlation of UBE2T expression and clinical pathologic factors by χ 2 test and calculated the overall survival rate of UBE2T on GC patients by the Kaplan–Meier analysis. Cox regression model was used to calculate the univariate and multivariate hazard ratios for the variables. We examined the functional roles of UBE2T during GC progression. We found that UBE2T promoted tumor cell growth and metastasis. Thus, UBE2T could be a potential therapeutic target to treat GC patients.
Materials and Methods
Cell Culture
The human GC cell lines, AGS, BGC-823, CRL-5971, CRL-5974, HGC-27, HTB-103, MGC-803, MKN-45, SGC-7901, and non-tumorigenic cell line GES-1 were purchased from the American Type Culture Collection (ATCC). The cells were routinely cultured in RPMI-1640 (Life Technologies, USA) and supplemented with 10 % fetal bovine serum (FBS) (Gibco, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies) in a humidified cell incubator with an atmosphere of 5 % CO2 at 37 °C.
Immunohistochemistry
The GC tissue arrays were obtained from Shanghai Outdo Biotech Company (China). The section slides were baked at 67 °C for 2 h, dewaxed in xylene, and dehydrated in a gradient alcohol. Antigen retrieval was carried out in a citrate buffer (pH 6.0) at 95 °C for 15 min. Endogenous peroxidase activity was deactivated with 3 % hydrogen peroxide at 37 °C for 10 min, and nonspecific binding was blocked with normal goat serum at 37 °C for 10 min. The sections were incubated first with the primary antibodies of UBE2T (1:50, #12992, Cell Signaling Technology, USA) at 4 °C overnight and then subsequently with a secondary biotinylated antibody (SP, China) for 30 min at 37 °C. The PBS instead of an anti-UBE2T antibody was used as negative control.
Scores of UBE2T immunostaining were conducted by two pathologists according to the staining strength and stained cell proportion on each section as described previously [14]. Briefly, the intensity of staining was scored as 0 (<15 %), 1 (15–50 %), 2 (50–75 %), and 3 (>75 %), and the percentage was scored as 0 (<15 %), 1 (15–50 %), 2 (50–75 %), and 3 (>75 %). Then, the final score was calculated by multiplying the two scores and classified them into two groups which were low expression and high expression.
Quantitative Real-time PCR
The GC cells were incubated in 60-mm dishes, and the total RNA was extracted from the GC cells using TRIzol (Takara, Japan) according to the manufacturer’s instructions. Amplification of cDNA was performed using the Prime Script RT reagent Kit (Takara, Japan). Real-time PCR was conducted using the FastStart Universal SYBR Green Master Mix kit (Roche, USA). Thermal cycling was executed by initial incubation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, and annealing at 60 °C for 34 s. Amplification and detection were implemented using the ABI Prism 7700 system (Applied Biosystems, USA) according to the manufacturer’s instructions. The primer sequences were showed in Supplementary Table S1.
Western Blot
The proteins from GC cells were extracted in a RIPA lysis buffer. Equal amounts of proteins were separated by 10 % polyacrylamide SDS gels (SDS-PAGE) and blotted onto polyvinylidene fluoride (PVDF) membranes (Amersham, UK). After blocked in 5 % BSA for 1 h at room temperature, membranes were incubated with primary antibodies overnight. The membranes were incubated for 1 h at room temperature in secondary HRP-conjugated antibodies (Perbio Science, Belgium). Immunoreactive proteins were visualized using the ECL immunodetection system (Immobilon, USA). The following antibodies were used for Western blot: anti-UBE2T antibody (10105-2-AP, Proteintech Group, USA), anti-E-cadherin (610404, BD Bioscience, US), anti-N-cadherin (610920, BD Bioscience, US), anti-P-cadherin (610227, BD Bioscience, US), anti-vimentin (Carpentaria, Australia), and anti-β-actin (Biotechnology, US).
Plasmid Construction and Cell transfection
The full-length UBE2T ORF was amplified from CL1-0 cDNA by PCR. The primers used were as follows: forward: 5′-TACGAATTC ATCATGCAGAGAGCTTCACGTCT-3′; reverse: 5′-ACACTCGAG CTAAACATCAGGATGAAA-3′. The PCR product was gel purified, digested, and then inserted into the EcoRI and XhoI restriction site of the pcDNA3.1 (+) vector (Invitrogen, USA). The sequences of siRNA used were as follows: UBE2T-siRNA: GAAGAGAGAGCTGCACATGTT; Control-siRNA: GAGUGGGUCUGGGUCUUCCCGUAGATT. The GC cells were transfected with UBE2T-siRNA or pcDNA3.1-UBE2T using the Lipofectamine 2000 Transfection Reagent (Invitrogen, USA), according to the manufacturer’s instructions. Stable cell lines were established by selection with gentamicin (Life Technologies, USA).
Cell Proliferation Assay
Cell growth was estimated by using CCK-8 (Cell Counting Kit-8) (Dojindo, Japan) assay. The cells were seeded in 96-well plates at a density of 3 × 103 cells per well and were allowed to adhere. Ten microlitre of CCK-8 solution was added into each well of the plate, and the plate was subsequently incubated for 3 h at 37 °C. The cell proliferation rate was documented every 24 h using a Coulter Counter (Beckman, USA). Each sample was run in triplicate.
Colony Formation Assay
The GC cells were plated at a density of 500 cells per well in triplicate and grown for 2 weeks. Subsequently, after fixed with 20 % methanol and stained with Giemsa (Sigma, USA), the colonies were counted under a microscope.
Cell Cycle Assay
The GC cells were harvested 48 h after transfection, washed with cold PBS (phosphate buffer saline), and fixed in 70 % ethanol. After incubation at 4 °C overnight, cells were resuspended in 300 μL binding buffer containing 5 μL of PI (propidine iodide) for 30 min. Populations in G0-G1, S, and G2-M phases were measured by flow cytometry (BD Biosciences Clontech) and analyzed by Multicycle-DNA Cell Cycle Analyzed Software.
Migration Assay
The migrated ability of cells was measured in Transwell with 8.0-mm Pore Polycarbonate Membrane Insert (Corning, NY) according to the manufacturer’s instructions. The bottom chambers were filled with 800 μL RPMI-1640 with 10 % fetal bovine serum, and the top chamber was filled with 200 μL GC cells serum-free-starved overnight and resuspended in RPMI-1640 medium with 1 % FBS. After incubated for 24 h, the non-invading cells remained on the upper surface of membrane insert. The migrated cells on the lower surface of the membrane insert were fixed with methanol and stained with Giemsa (Sigma, USA). The numbers of migrated cells were counted in a microscope.
Statistical Analysis
Each experiment was repeated in triplicate to ensure quantitative accuracy. Statistical comparisons were conducted by use of the SPSS 19.0 software (SPSS, USA). The Kaplan–Meier analysis was used to calculate the overall survival rate; χ2 test was used to investigate the correlation of UBE2T expression and clinical pathologic factors, and the Student’s t-test was used to compare the differences of various groups. Cox regression model was used to calculate the univariate and multivariate hazard ratios for the variables. All the data were presented as mean ± SD. A P value less than 0.05 was considered to be statistically significant.
Results
Elevated Expression of UBE2T is Correlated with Histological Stages, T Classification, and Poor Prognosis in Patients with GC
To investigate roles played by UBE2T in GC, we first examined the expression level of UBE2T in GC tissue microarrays by immunochemical staining. As shown in Fig. 1a, UBE2T expression was observed in the cytoplasm of GC cells. Of the 180 total GC cases, 106 (58.9 %) were UBE2T-positive. Moreover, UBE2T expression was most highly expressed in GC tissues, but hardly detectable in the corresponding adjacent noncancerous tissues. Next, we investigated the clinical pathologic roles of UBE2T expression in GC. Table 1 showed the correlation between UBE2T expression and clinical pathologic characteristics. Interestingly, the analysis revealed that the expression of UBE2T was strongly elevated in the cases with poor differentiation (histological stages III) than in high or medium differentiation cases (histological stages I–II) (P = 0.0001). Additionally, the expression of UBE2T was upregulated significantly in high T classification (T3-T4) than in low T classification (T1-T2) (P = 0.0386). Whereas, no significant correlation was observed with gender, age, N classification, and clinical stages. Furthermore, we found that the overall survival rate for the UBE2T-high group was significantly worse than that for the UBE2T-low group by the Kaplan–Meier analysis (Fig. 1b). More importantly, we applied Cox regression model to calculate the univariate and multivariate hazard ratios for the variables and found that UBE2T was an independent prognostic factor of gastric cancer (Table 2). Taken together, these results clearly show that elevated UBE2T expression is correlated with histological stages and T classification and implies poor prognosis in patients with GC.
UBE2T expression in GC tumor tissues and adjacent tissues and the correlation of UBE2T expression and overall survival in GC. a Immunohistochemical staining showed UBE2T expression was upregulated in GC cancer tissues compared with adjacent tissues. The arrows indicated UBE2T-positive cancer cells, which were located in the cell cytoplasm. Magnification: upper panel, ×400; under panel, ×200. b The 5-year overall survival curves of 180 patients with GC, stratified by UBE2T expression by the Kaplan–Meier analysis. There was a significant difference in the survival curves between low expression of UBE2T and high expression of UBE2T (P = 0.01). Blue lines depict low levels of UBE2T expression, and green lines depict high levels of UBE2T expression in GC samples. Scale bar, 50 μm
Upregulated Expression of UBE2T in GC cells
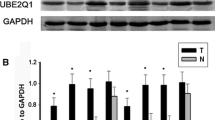
To further confirm our hypothesis, we examined UBE2T expression in different human GC cell lines by real-time PCR. We collected ten various types of gastric cells, including nine GC cell lines (AGS, BGC-823, CRL-5971, CRL-5974, HGC-27, HTB-103, MGC-803, MKN-45, SGC-7901) and the non-tumorigenic cell line GES-1. As we expect, UBE2T expression was extremely low in the normal gastric mucosa cell line GES-1, while all GC cell lines displayed high UBE2T expression (Fig. 2a). To better explore the roles of UBE2T in GC, retroviral vectors were used to establish GC cell lines AGS and CRL-5974 stably overexpressing UBE2T and GC cell lines BGC-803 and HTB-103 were transiently transfected with UBE2T-siRNA. Then the efficiencies were monitored using Western blot (Fig. 2b, c) and real-time PCR (Fig. 2d). The results showed that GC cell lines AGS and CRL-5974 stably overexpressing UBE2T expressed a significantly higher level of UBE2T compared with control groups, respectively (P < 0.05, Fig. 2b, c, d). Similar results were shown in GC cell lines BGC-803 and HTB-103 transiently transfected with UBE2T-siRNA (P < 0.05, Fig. 2b, c, d).
UBE2T expression in GC cell lines compared with one non-tumorigenic cell line. a The overexpression of UBE2T in GC cell lines was examined by real-time PCR using specific primers compared with one non-tumorigenic cell line. b UBE2T overexpression and knockdown efficiencies were determined by real-time PCR. c UBE2T overexpression and knockdown efficiencies were determined by Western blot. d UBE2T protein levels relative to β-actin were quantified. *P < 0.05, **P < 0.01, and ***P < 0.001
UBE2T Promotes GC Cell Proliferation
The above clinical findings suggest UBE2T may participate in GC progression. Hence, we speculated that UBE2T could participate in the proliferation of GC cells. CCK-8 assay was performed to investigate the potential effect of UBE2T on the vitality and the motility in GC cells. Significantly, increased proliferation rates were observed in AGS (P < 0.001, Fig. 3a) and CRL-5974 (P < 0.001, Fig. 3b) cells stably overexpressing UBE2T as compared to cells stably overexpressing vector. On the contrary, significantly decreased proliferation rates were showed in GC cell lines BGC-803 (P < 0.001, Fig. 3c) and HTB-103 (P < 0.001, Fig. 3d) transiently transfected with UBE2T-siRNA as compared to control cells. Meanwhile, colony formation assay showed that overexpression of UBE2T in AGS and CRL-5974 cells dramatically increased the numbers of colonies (P < 0.001, Fig. 3e). Oppositely, silencing UBE2T expression by UBE2T-siRNA in BGC-803 and HTB-103 cells significantly suppressed the colonies of the both cell lines (P < 0.001, Fig. 3f). Taken together, these observations suggest that UBE2T plays a growth-promoting activity in GC.
The effect of UBE2T on cellular proliferation in GC cell lines. CCK-8 assay demonstrated the obviously increased proliferative rates in AGS (a) and CRL-5974 cells (b) overexpression of UBE2T and significantly decreased proliferation rates in BGC-823 (c) and HTB-103 cells transfected with UBE2T-siRNA (d). e Colony formation assay showed an increase in the number of colonies by overexpression of UBE2T in AGS and CRL-5974 cells. f Colony formation assay showed a decrease in the number of colonies by knockdown of UBE2T in BGC-823 and HTB-103 cells. *P < 0.05, **P < 0.01, and ***P < 0.001
UBE2T Downregulation Induces GC Cell Cycle Arrest
Subsequently, we investigated the impact of UBE2T on cell cycle distribution in GC cell lines by flow cytometry. The results showed that AGS and CRL-5974 cells stably overexpressing UBE2T significantly increased cell percentage at G0/G1 as well as reduced cell percentage at S phase (P < 0.01, Fig. 4a, b). In contrary, GC cell lines BGC-803 and HTB-103 transiently transfected with UBE2T-siRNA had a smaller part of the population at G0-G1 phase and an enhanced proportion at S phase (P < 0.01, Fig. 4c, d). We infer from that si-UBE2T can induce G1/S phase arrest in GC cells, which in turn contributes to the stimulating growth properties of GC cells.
The effect of UBE2T on cellular cycle in GC cell lines. Representative histograms from flow cytometry analysis depicted cell cycle profiles of AGS cells (a) and CRL-5974 cells (b) by overexpression of UBE2T, as well as BGC-823 cells (c) and HTB-103 cells (d) by knockdown of UBE2T. *P < 0.05, **P < 0.01, and ***P < 0.001
UBE2T Induces Epithelial-Mesenchymal Transition in GC cells
Changes were observed in cell morphology after UBE2T overexpression in GC cells. This hinted that UBE2T possibly induced EMT of GC cells. To confirm our hypothesis, we investigated the expression patterns of genes involved in EMT by real-time PCR. AGS and CRL-5974 cells stably overexpressing UBE2T significantly inhibited the expressions of E-cadherin, yet significantly enhanced the level of N-cadherin, P-cadherin, and vimentin, respectively (Fig. 5a). Consistently, BGC-803 and HTB-103 cells transiently transfected with UBE2T-siRNA remarkably increased E-cadherin mRNA levels, yet decreased the expressions of N-cadherin, P-cadherin, and vimentin, respectively (Fig. 5a). Additionally, the same results were found through Western blot to examine expressions of EMT markers after UBE2T overexpression or knockdown in GC cells (Fig. 5b, c). Moreover, both the AGS and CRL-5974 cells overexpressing UBE2T triggered a transition from a cobblestone-like to an elongated fibroblast-like morphology compared with control cells (Fig. 5d) under the microscope. Furthermore, the transwell assay showed that the AGS and CRL-5974 cells overexpressing UBE2T promoted more cells migrating to the lower surface of the membrane insert (Fig. 5e, f). Collectively, these results indicated that UBE2T induces EMT of GC cells.
The effect of UBE2T on EMT in GC cell lines. a The expression of E-cadherin, N-cadherin, P-cadherin, and vimentin were examined by the real-time PCR after overexpression of UBE2T in AGS cells and CRL-5974 cells or knockdown of UBE2T in BGC-823 cells and HTB-103 cells. b The expression of E-cadherin, N-cadherin, P-cadherin, and vimentin were examined by the Western blot after overexpression of UBE2T in AGS cells or knockdown of UBE2T in BGC-823 cells. c E-cadherin, N-cadherin, P-cadherin, and vimentin protein levels relative to β-actin were quantified. d The morphologies of AGS and CRL-5974 cells overexpressing UBE2T and vector. e, f The AGS and CRL-5974 cells overexpressing UBE2T possessed more migrating abilities by transwell assay. Scale bar, 50 μm. *P < 0.05, **P < 0.01, and ***P < 0.001
Discussion
GC is still the fourth most common malignancy, and the overall prognosis for advanced disease remains poor, although declining in incidence in recent decades [15, 16]. Carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 19–9, and CA72-4 are widely used for GC, however they are relatively low sensitive and specific in the diagnosis and prognosis of GC [17, 18]. Hence, it is urgently required to identify more specific and sensitive markers for early detection, monitoring recurrences, and prediction of survival.
Ubiquitin-proteasome system is involved in the GC progression by mediating important oncogenes, tumor suppressor genes, and signaling pathways. For instance, the key regulator of wnt/β-catenin signaling, β-catenin is critically regulated by the ubiquitin E2 protein which binds to APC protein and promotes ubiquitination. In our study, UBE2T, a novel E2 gene, has been reported to be serum stimulated by a DNA microarray and its promoter bound to E2F by ChIP-on-chip experiments [7]. Researchers have found that UBE2T is the characteristic E2 in the Fanconi anemia pathway and regulates the Fanconi anemia pathway negatively [7, 8, 19–21]. More importantly, UBE2T has been proved to be the major E2 ubiquitin-conjugating enzyme required for the monoubiquitination of FANCD2 and FANCI [8]. In addition, hypoxia sensitizes cells to chemotherapy through disruption of the Fanconi anemia pathway and regulation of UBE2T [22]. Alternatively, UBE2T has essential functions other than the Fanconi anemia pathway [23]. Recently, UBE2T has been demonstrated to be upregulated in different tumors, such as lung cancer, breast cancer, and prostate cancer [10, 13, 24]. However, the relationship between UBE2T and GC has not been investigated.
In the present study, we firstly examined that UBE2T was upregulated in GC tissues by immunohistochemistry and found that elevated expression of UBE2T was correlated with poor differentiation (76/106) and high T classification (96/155) (Table 1). These results suggested a clear relationship between elevated UBE2T expression and GC. We identified that patients with overexpressed UBE2T had worse outcomes. Besides, using Cox regression model, we indicated that clinical stage, N classification, and UBE2T could be the independent prognostic factor of GC in our study (Table 2).
Meanwhile, the correlation between upregulated UBE2T and poor prognosis in patients with GC hinted that UBE2T could be an independent prognostic factor in GC. Consistently, UBE2T was also upregulated in GC cells. It has been reported that UBE2T regulated the proliferation, migration, and EMT of tumor cells [10, 13]. Therefore, we explored the functional roles of UBE2T in GC cells. The results conveyed that UBE2T promoted the proliferation and inhibited the cell cycle arrest of GC cells. More than that, the changes in cell morphology after UBE2T overexpression pointed out that UBE2T might induce EMT. It was reported that UBE2T elevated vimentin expression in prostate cancer cells, and vimentin may be a potential downstream protein of UBE2T in UBE2T-induced EMT and metastasis [13]. Recently, EMT has been reported to participate in GC cell invasions and metastasis, by which GC cells lose epithelial phenotype and cell-cell adhesion and gain invasive mesenchymal properties [25–27]. Actually, EMT has been demonstrated to be closely associated with cancer progression, and most cancers have partial features of EMT [28]. In tumor cells, EMT-inducing transcription factors (EMT-TFs) may primarily redefine assigning stem cell characteristics to dedifferentiated tumor cells or cancer stem cells [29]. If the cancer cell has acquired the necessary genetic aberrations and receives the appropriate signals, the dissemination of tumor cells from the solid tumor after breakdown of the basement membrane can be achieved [29]. EMT features can lead to tumor recurrence and a poor prognosis by promoting resistance during tumor therapy [30, 31]. To verify our assumption, we conducted a series of experiments. Firstly, UBE2T inhibited the expression of E-cadherin, which is a well-established condition for malignant cancer progression, yet induced the expressions of EMT-related genes, such as N-cadherin, P-cadherin, and vimentin in both mRNA and protein level by real-time PCR and Western blot. Secondly, UBE2T triggered a transition from a cobblestone-like to an elongated fibroblast-like morphology. Finally, UBE2T facilitated the metastasis, which might be induced by EMT. In short, UBE2T might induce EMT and metastasis in GC cells.
In conclusion, our study shows that UBE2T overexpression is related to high T classification and advanced pathologic stage of GC and may be an independent prognostic factor in GC. We also demonstrate that UBE2T overexpression promotes tumor growth and induces EMT of GC cells, which may be the mechanism caused by UBE2T upregulation.
References
Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singap Med J. 2014;55:621–8.
Zilberstein B et al. Gastric cancer trends in epidemiology. Arq Gastroenterol. 2012;49:177–8.
Karimi P et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2014;23:700–13.
Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol WJG. 2015;21:9838–52.
Zhang WH et al. Outcomes of surgical treatment for gastric cancer patients: 11-year experience of a Chinese high-volume hospital. Med Oncol. 2014;31:150.
Zhai R et al (2014) miR-152 suppresses gastric cancer cell proliferation and motility by targeting CD151. Tumour Biol J Int Soc Oncodev Biol Med
Machida YJ et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96.
Alpi A et al. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–30.
Corson TW et al. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–53.
Hao J et al. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumour Biol J Int Soc Oncodev Biol Med. 2008;29:195–203.
Kikuchi T et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192–205.
Takata R et al. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin Cancer Res Off J Am Assoc Cancer Res. 2005;11:2625–36.
Wen M et al. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6:25226–39.
Zhai J et al. miR-129 suppresses tumor cell growth and invasion by targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;464:161–7.
Lin Y et al. Gene mutations in gastric cancer: a review of recent next-generation sequencing studies. Tumour Biol J Int Soc Oncodev Biol Med. 2015;36:7385–94.
Tsunoda S et al (2014) Laparoscopic gastrectomy for remnant gastric cancer: a comprehensive review and case series. Gastric Cancer Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc
Takahashi Y et al. The usefulness of CEA and/or CA19-9 in monitoring for recurrence in gastric cancer patients: a prospective clinical study. Gastric Cancer Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc. 2003;6:142–5.
Kochi M et al. Evaluation of serum CEA and CA19-9 levels as prognostic factors in patients with gastric cancer. Gastric Cancer Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc. 2000;3:177–86.
Hira A et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015;96:1001–7.
Rickman KA et al. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 2015;12:35–41.
Virts EL et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum Mol Genet. 2015;24:5093–108.
Ramaekers CH et al. Hypoxia disrupts the Fanconi anemia pathway and sensitizes cells to chemotherapy through regulation of UBE2T. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2011;101:190–7.
Kelsall IR et al. The Fanconi anaemia components UBE2T and FANCM are functionally linked to nucleotide excision repair. PLoS One. 2012;7, e36970.
Ueki T et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69:8752–60.
Huang N et al. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6:15222–34.
Yoo YA et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–70.
Yang Y et al. C23 protein meditates bone morphogenetic protein-2-mediated EMT via up-regulation of Erk1/2 and Akt in gastric cancer. Med Oncol. 2015;32:76.
Bullock MD et al. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell Under Auspices Eur Cell Biol Organ. 2012;104:3–12.
De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110.
Iwanaga R et al. Expression of Six1 in luminal breast cancers predicts poor prognosis and promotes increases in tumor initiating cells by activation of extracellular signal-regulated kinase and transforming growth factor-beta signaling pathways. Breast Cancer Res BCR. 2012;14:R100.
Che J et al. Decreased expression of claudin-3 is associated with a poor prognosis and EMT in completely resected squamous cell lung carcinoma. Tumour Biol J Int Soc Oncodev Biol Med. 2015;36:6559–68.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was supported by the Ethics Committee of Binhu Traditional Chinese Medicine Hospital. All patients agreed to participate in the study and gave written informed consent. The tissue microarray (TMA) containing 180 cases of gastric cancer specimens used in this study was purchased from Shanghai Outdo Biotech Inc, and the clinicopathological features were complete.
Conflicts of Interest
None
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table S1
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Yu, H., Xiang, P., Pan, Q. et al. Ubiquitin-Conjugating Enzyme E2T is an Independent Prognostic Factor and Promotes Gastric Cancer Progression. Tumor Biol. 37, 11723–11732 (2016). https://doi.org/10.1007/s13277-016-5020-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5020-3