Abstract
While p53 mutation is found in the majority of triple-negative breast cancer (TNBC) and despite recent developments in p53-targeting agents, their therapeutic application is still limited by the absence of standard biomarkers and ambiguousness of its essential biological role in cancer. Whole sections from 305 TNBC cases were stained for p53 to determine the correlation with lymph node metastasis and clinical outcomes in the whole cohort as well as in stratified patient groups according to AJCC stage and the use of adjuvant chemotherapy. Reduced immunohistochemical expression of p53 was an independent risk factor for lymph node metastasis. p53 overexpression was predictive of better clinical outcome in all patients (P = 0.012, disease-free survival and P = 0.008, overall survival) and the stratified cohorts of those who had early breast cancer and received adjuvant chemotherapy. Suppression of endogenous mutant p53 by siRNA and induction of wild-type p53 repressed TNBC cell invasion in vitro. In TNBC, increased immunohistochemical expression of p53 may reflect the accumulation of wild-type p53 rather than the mutant form. Strong p53 protein expression may serve as a favorable prognostic indicator and provide evidence for the use of specific agents targeting p53.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triple-negative breast cancer (TNBC) is characterized by the absence of hormone receptor expression and lack of human epidermal growth factor receptor 2 (HER2) overexpression. The absence of a recognized therapeutic target and its biologically aggressive characteristics lead to poor prognosis with high rates of metastasis and short relapse-free survival in TNBC, compared with luminal or HER2 type breast carcinomas [1].
The TP53 gene, known as the “guardian of the genome,” is a tumor suppressor gene that regulates cell cycle progression, DNA repair, cellular senescence, and apoptosis [2]. However, when TP53 is either mutated or lost its tumor suppressive function, the protein p53 shows pro-oncogenic properties in up to 50 % of cancers [3]. Interestingly, a number of recent studies have indicated that p53, particularly its mutant form, promotes tumor cell migration, invasion, and metastasis through various biological pathways in several cancers [4–8].
In breast cancer, TP53 mutation is the most common single genetic alteration observed in as much as 30 % of all breast carcinomas [9]. Moreover, in TNBC, p53 mutations occur more frequently compared to the other intrinsic subtypes of breast cancer (60–70 %) [10], making p53 an attractive candidate for therapeutic target in TNBC [11]. Previous studies have shown that TP53 mutations show distinctive prognostic significance in breast cancer [12, 13]. However, the prognostic significance of p53 protein expression remains controversial [14–17] since it is unclear whether its expression is reflective of wild-type p53 or mutant p53 [18]. In addition, only a few studies have described the predictive ability of p53 protein expression and its association with metastasis in human TNBC cohorts [19, 20] utilizing immunohistochemistry while its role in cell migration and invasion is well known from several preclinical analyses using human TNBC cell lines or animal models [21–25].
Since its introduction three decades ago, immunohistochemistry has provided enormous benefits in evaluating prognostic and predictive markers in neoplasms, examples of which include agents specifically targeting estrogen receptor (ER) and HER2 in breast cancer [26, 27]. p53 is considered a challenging yet attractive target in drug development, and a few agents have shown promising results in recent clinical trials [11].
In the present study, we examined the clinicopathological significance of p53 protein expression in a large cohort of TNBC patients. We also evaluated the predictive function of p53 protein expression level for lymph node metastasis (LNM) as well as clinical outcome according to AJCC stage and adjuvant therapeutic modalities. In addition, the biologic effects of p53 gene manipulation on cancer invasion and metastasis were evaluated in multiple human TNBC cell lines. We evaluated the biological characteristics of p53 phenotypes and described potential utilization of measuring p53 protein expression levels in clinical contexts for predicting metastasis and prognostic outcome, which may provide a new insight in applying p53-targeted therapeutic options in TNBC.
Materials and methods
Patient selection and study design
A total of 305 TNBC patients who received surgical resection between 2003 and 2006 were retrospectively selected from the pathologic archive in Seoul National University Hospital. All of the enrolled TNBC cases had been diagnosed previously by an immunohistochemistry panel of ER (1:100, 1D5; Novocastra Laboratories, Newcastle, UK) and progesterone receptor (PR, 1:200, PgR636; Dako, Glostrup, Denmark). ER and PR expression was considered as positive when ≥1 % of staining was observed in tumor cells. HER2 immunohistochemistry (4B5; Ventana, Medical System, Tucson, AZ, USA) and HER2 fluorescence in situ hybridization using PathVysion assay (Abbott Molecular, Downers Grove, IL) results were based on the 13th St. Gallen International Breast Cancer Conference [28]. Her2 immunohistochemistry was evaluated as positive when more than 10 % of tumor cells show homogeneous, dark circumferential staining. HER2 gene amplification by FISH was considered as positive when the ≥2.0 ratio of HER2 gene copy number to chromosome 17 copy number or ≥6.0 average HER2 signal per each tumor cell was observed. Histological grading was performed according to the Nottingham grading system [29]. The patients enrolled in this study received four to six cycles of dose-dense doxorubicin/cyclophosphamide (AC) followed by paclitaxel or combined docetaxel and cyclophosphamide (TC) regimen as adjuvant chemotherapy. Patients who had received neoadjuvant chemotherapy were excluded. Clinicopathological parameters and patient survival data were reviewed and collected via electronic medical records system. In the total cohort of 305 patients, we performed stratified analyses considering AJCC stage and therapeutic modality in order to assess their clinicopathological significance. This study was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. H-1511-085-720).
Immunohistochemistry and interpretation
Formalin-fixed paraffin-embedded tissues of whole sections were available for immunohistochemistry in all of the enrolled 305 cases. Representative tumor areas were selected, and a 4-mm-thick section for each block was stained immunohistochemically for the p53 antibody (1:500, DO-7; Dako) using a Benchmark automatic immunostaining device (Ventana). p53 staining was independently evaluated by three pathologists (MSJ, IAP, and HSR). Tumor cells with strong nuclear staining were chosen for evaluation. A case was considered positive for p53 expression if the staining proportion was greater than 10 % [30–32].
Cell culture
MDA-MB-231 and MDA-MB-468 were grown in Dulbecco’s Modified Eagle’s Medium (DMEM). Each medium was supplemented with 10 % FBS (GenDEPOT, Katy, TX, USA) and 1 % penicillin-streptomycin (Gibco, Grand Island, NY, USA). All cell lines were cultured at 37 °C in humidified atmosphere with 5 % CO2.
Western blot analysis
For intracellular protein extraction, transfected cells in a monolayer were lysed in ice-cold T-PER buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing a protease inhibitor cocktail (Roche, Basel, Switzerland). The isolated proteins were separated by polyacrylamide gel electrophoresis and were transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). After blocking with 5 % skim milk in Tris-buffered saline-Tween 20 (TBST), the membrane was incubated with various primary antibodies specific to p53 (1:500, DO-7; Dako) and β-actin (1:1000, C-2; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After incubation with primary antibodies, the membrane was washed in TBST once for 5 min and three times for 10 min each. The membrane was then incubated with secondary antibodies (horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit) for 1 h at room temperature. The membrane was washed again three times in TBST for 10 min each, and the proteins were then visualized using a luminol-based chemiluminescent detection kit (Elpis, Daejeon, Korea).
p53 siRNA and plasmid transfection
A small interfering RNA (Cosmo genetech, Seoul, Korea) specific for human p53 was used to knock down p53 in MDA-MB-231 and MDA-MB-468 cells harboring endogenous mutant p53 [18]. The sequence for the p53 siRNA was GACUCCAGUGGUAAUCUAC. A human wild-type p53 expression vector (pCMV-neo-BAM) and control vector (pCMV-tag2B) were kindly provided by Prof. Sang Hoon Kim (Kyung Hee University, Seoul, Korea). The cells were transfected with 50 nM of p53 siRNA using transfection reagent (Invitrogen, Carlsbad, CA, USA) with 1 μg of p53 plasmid using a transfection reagent (Poly Plus, Illkirch-Graffenstaden, France). The siRNA or plasmid-transfected cells were analyzed 48 h after transfection.
Invasion assay
Invasion assays were performed using BioCoatTM Matrigel® Invasion Chamber (Corning Inc., Corning, NY, USA) according to the protocol provided by the manufacturer. Cells with p53 knockdown or with p53 overexpression were used. After pre-heating the invasion chamber, the cells were collected in serum-free Opti-MEM (Gibco) and plated at 5 × 105 cells per well in the top chamber. DMEM or RPMI containing 10 % FBS was used as a chemo-attractant. After 48 h of incubation, the non-invading cells on the upper surface of membrane were removed with a cotton swab, and the invading cells on the lower surface of membrane were washed and then fixed with absolute ethanol for 10 min, followed by HE staining. Stained cells were counted using a phase-contrast microscope (Nikon Eclipse 80i, Japan), and pictures were taken using a Digital Sight DS-Fi1 camera (Nikon) attached to a microscope.
Statistical analysis
Pearson’s chi-square test and Fisher exact test were used to analyze the statistical significance of the association between LNM and categorical parameters. To identify independent prognostic factors for the risk of LNM, multivariate logistic regression analysis was performed. Kaplan-Meier survival curves with log-rank tests were used to analyze the time to disease progression and patients’ survival. Multivariate analysis was performed using Cox proportional hazards model. P values <0.05 were considered statistically significant. Two-tailed Student’s t test was performed to compare means among different groups for in vitro studies. Data was statistically analyzed using SPSS 21.0 for Windows (IBM SPSS INC., Armonk, NY, USA).
Results
Relationship between p53 expression and LNM in TNBC patients based on immunohistochemistry
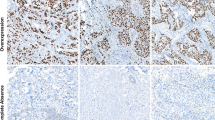
Comparisons of categorical variables predicting LNM using Pearson chi-square test in 305 TNBC patients are shown in Table 1. The number of cases positive for p53 was 172 (56.4 %). Photomicrographs of the histological findings and immunohistochemistry for p53 protein are presented in Fig. 1. TNBC showing p53 expression was significantly associated with decreased LNM (P < 0.001). Three clinicopathological variables including pT stage (P = 0.004, OR = 5.680), lymphovascular invasion (P < 0.001, OR = 3.446), and p53 protein expression (P < 0.001, OR = 0.361) were independent factors predicting LNM in multivariate analysis (Table 2).
Histologic and p53 immunohistochemical features of two representative cases of triple-negative breast cancer. Left panels, stained with hematoxylin and eosin (a) and showing entirely negative expression of corresponding p53 staining (c). Right panels, stained with hematoxylin and eosin (b) and showing diffuse and strong nuclear p53 staining of corresponding section (d) (a–d, original magnification ×4 and ×400 (insets))
In additional stratified analyses according to different pT stages, p53 expression revealed a significantly decreased likelihood of LNM in both univariate and multivariate regression analyses (Supplementary Table 1 and Supplementary Table 2).
Effect of p53 protein expression on survival prediction in TNBC patients
We evaluated the relationship between clinicopathological variables and survival in TNBC patients (Table 3 and Fig. 2). Univariate survival analysis demonstrated that higher p53 expression was associated with longer disease-free survival (DFS) (P = 0.012) and overall survival (OS) (P = 0.008). A trend for favorable survival in p53-positive patients was revealed on multivariate analysis (DFS, OR = 0.629, P = 0.076 and OS, OR = 0.570, P = 0.057; Table 4).
Kaplan-Meier disease-free survival (upper, a–e) and overall survival (lower, f–j) analyses according to AJCC stage and treatment modality. Total cohort (a, f), early breast cancer group (b, g), localized advanced breast cancer group (c, h), adjuvant chemotherapy group (d, i), and no adjuvant chemotherapy group (e, j)
In the stratified analysis, elevated p53 protein expression was associated with increased DFS and OS in univariate analysis in TNBC patients with early stage (DFS, P = 0.020 and OS, P = 0.005) and those who received adjuvant chemotherapy (DFS, P = 0.016 and OS, P = 0.012; Supplementary Table 3 and Supplementary Table 4). In multivariate analysis, although no statistically significant correlation was observed in the early stage disease group or patients who received adjuvant chemotherapy, there was a strong association between p53 expression and patient survival (data not shown, Supplementary Table 5).
Wild-type TP53 gene restoration suppresses invasion ability in human TNBC cell lines
Because of the strong association between p53 expression and LNM in TNBC patients observed in this study, we investigated the functional consequences of wild-type p53 gain and how the loss of mutant p53 contributes to cancer cell invasion in two human TNBC cell lines with endogenous mutant p53. We observed that silencing of p53 mutant expression mediated by p53-siRNA in MDA-MB-231 and MDA-MB-468 cells significantly reduced the number of migrating cells compared to those with mock and nonsense-siRNA treatment (Fig. 3). Additionally, plasmid transfection of the cell lines with wild-type p53 revealed significantly reduced cell transmigration ability (Fig. 3).
Mutant TP53 gene silencing and its effect on tumor cell invasion. a, b Alteration of p53 protein expression in MDA-MB-231 and MDA-MB-468 cells transfected with p53-siRNA and wild-type p53 plasmid vector is noted. The number of transmigrated invasive cells in two cell lines (MDA-MB-231 and MDA-MB-468) transfected with p53-siRNA (a) and p53 plasmid vector (b) is significantly reduced compared to mock and nonsense-siRNA or empty vector groups in invasion assay (c). All experiments are performed in triplicate
Discussion
Over the past decade, the emerging role of p53 in the regulation of cancer invasion in TNBC has been documented in several reports on TNBC; these studies suggested the oncogenic role of the mutant p53 gene in tumor progression using transgenic mice models or cancer cell lines [21, 23–25, 33, 34].
In our study, strong p53 protein expression appears to be an independent predictor for metastasis to axillary lymph nodes, which is comparable to the results of the conventional predictive variables including lymphatic invasion and tumor size [35], both in distinctive cohorts re-classified according to pathologic tumor size as well as in the cohort including all patients. This is the largest cohort study showing an inverse correlation between p53 protein expression and LNM in TNBC patients. Although comparative analysis in our study was limited since different methods were used, previous reports suggest that p53 expression increases the invasion and migration abilities of breast cancer cells [19–23], which is in direct contrast with our results.
Additionally, in this study, increased p53 expression in TNBC was found to have a statistically significant or strong correlation with better survival outcome in both the entire cohort as well as the stratified patient groups based on AJCC stage and the use of adjuvant chemotherapy. Although there are only a few studies that have evaluated the relationship between LNM and p53, numerous studies have found that p53 expression is associated with clinical outcome in TNBC patients. Although the significance of these results remains controversial, several previous studies showed either decreased survival in patients with p53 protein overexpression [14, 36–38] or no significance [15, 39], which is in contrast with the results of our study. However, the oncosuppressive effect of p53 through the miRNA pathway, particularly in TNBC, has been demonstrated in preclinical studies [16, 40]. Coates et al. [17] also showed that p53-positive expression on immunohistochemistry was associated with increased DFS and OS among TNBC patients, which is concordant with the results of our study.
We used immunohistochemistry to evaluate p53 expression levels, which is one of the most widely used diagnostic tools in investigating target agents and prediction of clinical outcome [41]. Although next generation sequencing (NGS) allows whole genomic evaluation, the high cost and complex interpretation required for this option make it impractical for most clinical laboratories. Immunohistochemistry is still considered a valuable diagnostic tool with easy accessibility and well-established efficacy in examining the functional unit in cancer that can overcome discrepancies between mRNA and protein expression by transcriptional and translational regulation [42, 43].
It remains controversial whether p53 immunohistochemistry reflects the mutant or wild-type p53. According to Lacroix et al. [18], p53 protein translated from the mutant TP53 gene is typically not observed by immunohistochemical staining because p53 protein exists as a truncated structure with reduced stability. However, the wild-type TP53 gene shows strong immunoreactivity because the gene is commonly overexpressed as a compensatory mechanism to repair DNA damages that occur during tumorigenesis [18]; this may explain why our results demonstrated a strong association between p53 protein expression and reduced LNM as well as decreased risks of recurrence and death.
Because we found a distinctive prognostic role of p53 protein expression that differs from what is generally accepted, we attempted to minimize bias and error as follows. First, immunohistochemical results for p53 were interpreted by three well-trained pathologists who were blinded to the clinicopathological information, and consistent results were obtained among the investigators. Second, we performed statistical analyses according to treatment modalities and AJCC stage, which showed a significant predictive ability of p53 for LNM and clinical outcome, with the exception of the stratified cohort without the event of death. Third, we analyzed p53 expression using whole section slides to reduce tumor heterogeneity and overcome area limitations of the tissue microarray. Fourth, we determined the function of p53 in tumor invasion in multiple human TNBC cell lines that had been genetically manipulated; invasion ability was suppressed after introduction of wild-type p53 or substitution of endogenous mutant TP53 for wild-type TP53.
In conclusion, we evaluated p53 protein expression using whole-section slides in a large cohort with TNBC. We found that increased expression of p53 was significantly associated with decreased LNM as well as improved patient survival in stratified patient groups according to pathologic tumor size, AJCC stage, and adjuvant chemotherapy as well as in the entire cohort. Additionally, re-introduction of wild-type TP53 or repression of mutant p53 on the genomic level significantly diminished the invasion ability of human TNBC cell lines. We therefore, suggest that strong immunohistochemical expression of p53 protein in TNBC reflects the production of the wild-type TP53 gene, not of the mutant one. These results suggest that application of p53-targeting agents may in fact inhibit the normal function of p53 and that its use should be reconsidered in clinical practice.
Currently, molecular target agents in breast cancer have been developed based on the results interpreted via immunohistochemistry. In addition, several ongoing clinical trials targeting specific biomarkers including PARP1 are being validated using immunohistochemistry. Since clinical trials evaluating the significance of p53 as a novel candidate for target therapy are currently underway, the different clinicopathological behaviors of p53 in TNBC must be elucidated, particularly because TNBC shows the highest frequency of p53 mutation among all of the intrinsic subtypes of breast cancer. Once immunohistochemical evaluation of p53 is validated, p53 may serve as a novel predictive marker along with ER and HER2. Further validation combining mutational analyses to validate the significance of p53 immunohistochemistry is necessary.
References
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Kumar V, Abbas AK, Aster JC. Robbins and Cotran pathologic basis of disease. 9th ed. Philadelphia: Elsevier Saunders; 2014. p. 293–6.
Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–40.
Roger L, Jullien L, Gire V, Roux P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J Cell Sci. 2010;123:1295–305.
Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–18.
Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, et al. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–901.
Roger L, Gadea G, Roux P. Control of cell migration: a tumour suppressor function for p53? Biol Cell. 2006;98:141–52.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–41.
Desmedt C, Voet T, Sotiriou C, Campbell PJ. Next-generation sequencing in breast cancer: first take home messages. Curr Opin Oncol. 2012;24:597–604.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Duffy MJ, Synnott NC, McGowan PM, Crown J, O'Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40:1153–60.
Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20:3569–80.
Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets. 2014;15:80–9.
Maeda T, Nakanishi Y, Hirotani Y, Fuchinoue F, Enomoto K, Sakurai K, et al. Immunohistochemical co-expression status of cytokeratin 5/6, androgen receptor, and p53 as prognostic factors of adjuvant chemotherapy for triple negative breast cancer. Med Mol Morphol. 2015. doi:10.1007/s00795-015-0109-0.
Kashiwagi S, Yashiro M, Takashima T, Aomatsu N, Ikeda K, Ogawa Y, et al. Advantages of adjuvant chemotherapy for patients with triple-negative breast cancer at Stage II: usefulness of prognostic markers E-cadherin and Ki67. Breast Cancer Res. 2011;13:R122.
Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6:458–72.
Coates AS, Millar EK, O'Toole SA, Molloy TJ, Viale G, Goldhirsch A, et al. Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX. Breast Cancer Res. 2012;14:R143.
Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325.
Jasar D, Smichkoska S, Kubelka K, Filipovski V, Petrushevska G. Expression of p53 protein product in triple negative breast cancers and relation with clinical and histopathological parameters. Prilozi. 2015;36:69–79.
Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB, et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2013;32:2992–3000.
Ali A, Shah AS, Ahmad A. Gain-of-function of mutant p53: mutant p53 enhances cancer progression by inhibiting KLF17 expression in invasive breast carcinoma cells. Cancer Lett. 2014;354:87–96.
Arjonen A, Kaukonen R, Mattila E, Rouhi P, Hognas G, Sihto H, et al. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Invest. 2014;124:1069–82.
Noll JE, Jeffery J, Al-Ejeh F, Kumar R, Khanna KK, Callen DF, et al. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene. 2012;31:2836–48.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98.
Boudreau HE, Casterline BW, Burke DJ, Leto TL. Wild-type and mutant p53 differentially regulate NADPH oxidase 4 in TGF-beta-mediated migration of human lung and breast epithelial cells. Br J Cancer. 2014;110:2569–82.
Romond EH, Perez EA, Bryant J, Suman VJ, Geyer Jr CE, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84.
Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–18.
Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–10.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–47.
Zlobec I, Steele R, Michel RP, Compton CC, Lugli A, Jass JR. Scoring of p53, VEGF, Bcl-2 and APAF-1 immunohistochemistry and interobserver reliability in colorectal cancer. Mod Pathol. 2006;19:1236–42.
Yamashita H, Toyama T, Nishio M, Ando Y, Hamaguchi M, Zhang Z, et al. p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast Cancer Res. 2006;8:R48.
Kurshumliu F, Gashi-Luci L, Kadare S, Alimehmeti M, Gozalan U. Classification of patients with breast cancer according to Nottingham prognostic index highlights significant differences in immunohistochemical marker expression. World J Surg Oncol. 2014;12:243.
Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet Jr RJ, Broxmeyer HE, Kopelovich L, et al. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene. 2007;26:3329–37.
Heinlein C, Krepulat F, Lohler J, Speidel D, Deppert W, Tolstonog GV. Mutant p53(R270H) gain of function phenotype in a mouse model for oncogene-induced mammary carcinogenesis. Int J Cancer. 2008;122:1701–9.
Gajdos C, Tartter PI, Bleiweiss IJ. Lymphatic invasion, tumor size, and age are independent predictors of axillary lymph node metastases in women with T1 breast cancers. Ann Surg. 1999;230:692–6.
Wu M, Wei W, Xiao X, Guo J, Xie X, Li L, et al. Expression of SIRT1 is associated with lymph node metastasis and poor prognosis in both operable triple-negative and non-triple-negative breast cancer. Med Oncol. 2012;29:3240–9.
Zhang J, Wang Y, Yin Q, Zhang W, Zhang T, Niu Y. An associated classification of triple negative breast cancer: the risk of relapse and the response to chemotherapy. Int J Clin Exp Pathol. 2013;6:1380–91.
Biganzoli E, Coradini D, Ambrogi F, Garibaldi JM, Lisboa P, Soria D, et al. p53 status identifies two subgroups of triple-negative breast cancers with distinct biological features. Jpn J Clin Oncol. 2011;41:172–9.
Wang J, Zhang C, Chen K, Tang H, Tang J, Song C, et al. ERbeta1 inversely correlates with PTEN/PI3K/AKT pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res Treat. 2015;152:255–69.
Mahamodhossen YA, Liu W, Rong-Rong Z. Triple-negative breast cancer: new perspectives for novel therapies. Med Oncol. 2013;30:653.
Gossner W. A brief history of the Society for Histochemistry: its founders, its mission and the first 50 years. Histochem Cell Biol. 2002;118:91–4.
Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–73.
Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Jin, MS., Park, I.A., Kim, J.Y. et al. New insight on the biological role of p53 protein as a tumor suppressor: re-evaluation of its clinical significance in triple-negative breast cancer. Tumor Biol. 37, 11017–11024 (2016). https://doi.org/10.1007/s13277-016-4990-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4990-5