Abstract
Family with sequence similarity 83 (FAM83) members play important roles in carcinogenesis and tumor progression in several tumor types. However, the mechanism by which cancer cells regulate FAM83F still remains unclear. In this study, we found that the FAM84F protein and messenger RNA (mRNA) levels were consistently upregulated in esophageal squamous cell carcinoma (ESCC) tissues, which suggests that a post-transcriptional mechanism may be involved in the regulation of FAM83F. Since microRNAs (miRNAs) are powerful post-transcriptional regulators of gene expression, we performed bioinformatic analyses to search for miRNAs that could potentially target FAM83F. We identified the specific targeting site of miR-143 in the 3′-untranslated region (3′-UTR) of FAM83F and confirmed the inverse correlation between the levels of miR-143 and FAM83F protein and mRNA in ESCC tissue samples. By overexpressing or silencing miR-143 in ESCC cells, we experimentally validated that miR-143 directly binds to the 3′-UTR of the FAM83F transcript and degrades the FAM83F mRNA to regulate FAM83F expression. Furthermore, the biological consequences that miR-143 mediated by targeting FAM83F were examined using in vitro cell proliferation, apoptosis, migration, and invasion assays. We demonstrate that miR-143 exerted a tumor-suppressing effect by inhibiting the proliferation, migration, and invasion and inducing G1/G0 phase arrest of ESCC cells via the negative regulation of FAM83F expression. Taken together, our findings provide important evidence which supports the role of miR-143 as a tumor suppressor in ESCC via the inhibition of FAM83F expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal carcinoma is demonstrated to be one of the eight most common cancers and one of the six most common causes of cancer deaths worldwide. It can mainly be divided into two types, adenocarcinoma and squamous cell carcinoma [1]. Although esophageal adenocarcinoma (EAC) is becoming increasingly common in western countries, esophageal squamous cell carcinoma (ESCC) is still dominant in East Asia [2]. Thus far, although a number of alterations in genome and transcriptome of ESCC have been identified [3], the molecular mechanisms underlying ESCC remain to be elucidated.
Family with sequence similarity 83 (FAM83) members were recently demonstrated to exhibit oncogenic properties as well as significantly elevated levels of expression in multiple human tumor types [4]. In addition, evidence suggests that elevated expression of FAM83 members was associated with elevated tumor grade and decreased overall survival, which indicated that FAM83 members could play important roles in carcinogenesis and tumor progression.
MicroRNAs (miRNAs) are an endogenous conserved class of noncoding 20–22-nt small RNAs which modulate gene expression at the post-transcriptional level by mainly binding to 3′-UTR of target messenger RNAs (mRNAs), which leads to mRNA degradation or translation inhibition [5]. It has been proven that miRNAs regulate a variety of biological processes, including developmental timing, cell growth, and cell death [6]. Profiling has been applied to explore the potential role of miRNA genes which might be the downstream targets of activated oncogenic pathways or the target protein-coding genes involved in cancer [7]. In the ESCC model, miRNAs could modulate pivotal protein expressions in the signaling cascades and thus regulate tumor malignant behaviors [8]. We further propose that by overlapping the datasets of predictive miRNAs targeting FAM83F and the miRNA analysis, we might be able to identify the tumor-specific miRNAs regulating FAM83F expression in ESCC.
Hence, we identified miR-143 as a potential regulator of FAM83F through human ESCC miRNA profiling and bioinformatics analyses. The inverse correlation between miR-143 and FAM83F expressions was identified in ESCC tissues and paired noncancerous tissues. Furthermore, in this study, we experimentally confirmed the direct regulation of FAM83F by miR-143 and the biological role of the miR-143-mediated regulation of FAM83F expression in human ESCC.
Material and methods
Patients and clinicopathological data collection
Surgical tumor specimens were obtained from 40 patients, and their adjacent tissues were obtained. The study was approved by the hospital ethics committee, and written informed consent was obtained from all patients. None of the patients received radiotherapy or chemotherapy before surgery. The diagnosis was verified by clinical examination and histopathological analysis of the tissue specimens. The samples were collected and immediately snap frozen in liquid nitrogen and stored at −80 °C till further use. Clinical and pathological data including age, gender, pathological grading, tumor stage, and lymph node metastasis were acquired from the medical records.
Target gene prediction of miR-143
The target genes of miR-143 were predicted by bioinformatics analysis. The analysis was performed by the algorithms TargetScan 5.1 (http://targetscan.org/), PicTar (http://pictar.mdcberlin.de/), miRanda (http://microRNA.org), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), and DIANAmT (http://diana.cslab.ece.ntua.gr/microT/microT.php). Results of the forecast targets were intersected by miRWalk (http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/) website tools.
Cell culture
ECA-109 and KYSE-410 human esophageal cancer cell lines were primary purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). The cells were maintained in the Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10 % fetal bovine serum (Gibco, USA) in a humidified incubator at 37 °C and 5 % CO2. The medium was changed every 2–3 days, and the cells were trypsinized with trypsin when 80–90 % confluence was reached.
RNA extraction
miRNA-enriched RNA was successfully isolated from ESCC and matched adjacent tissues using miRNeasy Mini Kit (Qiagen, Denmark) following the manufacturer’s instructions. The eluted RNA was quantified spectrophotometrically and was stored at −80 °C till further use.
Quantitative real-time PCR
First-strand cDNA synthesis was carried out using miScript II RT Kit (Qiagen, Denmark) according to the manufacturer’s protocol. For analysis of the miR-143, real time-PCR was performed with miScript SYBR Green PCR Kit and miScript Primer PCR Assay (Qiagen, Germany). For analysis of the FAM83F mRNA expression, quantitative real-time PCR was performed using 2× SYBR Green PCR Mix (Aidlab, Beijing, China) following the procedure of the manufacturer. All reactions were performed on a Light Cycler480 Real-time PCR system (Roche Molecular Systems, Indianapolis, USA). The results of miRNA and mRNA real-time PCR were normalized to U6 and GAPDH, respectively, by using the 2−ΔΔCt method [9]. Each assay was performed in triplicate and repeated at least three times to calculate mean normalized gene expression. The primers were ordered from Kang-Chen Biotech (Kangcheng, Shanghai, China). Primer sequences are shown in Table 1.
miRNA overexpression and cell transfection procedures
The miR-143 inhibitor and scrambled oligonucleotide (negative inhibitor control, inhibitor-negative control (NC)) and miR-143 mimics and scrambled oligonucleotide (negative mimics control, mimics NC) used in this study were designed and synthesized by GenePharma (Shanghai, China). Cells were seeded on six-well plates at the density of 4 × 106/well, and the transfection was performed when the cells were approximately 70 % confluent. On the following day, miR-143 inhibitor, miR-143 mimics, or the NC was individually transfected using Lipofectamine 2000 transfection reagent (Invitrogen, USA) according to the manufacturer’s procedure. Transfection efficiency was measured by real-time reverse transcription (RT)-PCR and flow cytometry assay. Other experiments were carried out 48 h after transfection.
Plasmid construction and siRNA efficiency assay
A mammalian expression plasmid (pReceiver-M98-FAM83F) designed to specifically express the coding sequence (CDS) of human FAM83F without the 3′ untranslated region (UTR) was purchased from FulenGen (Guangzhou, China). An empty plasmid served as a NC. The small interfering RNA (siRNA) sequence targeting human FAM83F was designed and synthesized by GenePharma (Shanghai, China). A scrambled siRNA was used as a NC. The overexpression plasmid or siRNA was transfected into cell lines using Lipofectamine 2000 (Invitrogen) according to the manufacturer’ s instructions. Total RNA or protein was isolated 24 or 48 h after transfection. The FAM83F mRNA and protein expression levels were assessed via quantitative RT-PCR and Western blot, respectively.
Dual-luciferase assay
The 3′-UTR of human FAM83F mRNA was amplified and cloned into the site behind the firefly luciferase gene of a pGL3-Control Vector (Promega, USA) to generate a pGL3-FAM83F-3′-UTR-Wt vector (Wt vector). The sequences that interact with the seed region of miR-143 were mutated (from CAUCUCA to GUAGAGU for the miR-143 binding site), and the mutant FAM83F-3′-UTR was inserted into an equivalent luciferase reporter plasmid to generate the corresponding mutant plasmids (pGL3-FAM83F-3′-UTR-MT, MT vector). All the vectors were verified by DNA sequencing.
Cell were seeded in 24-well plates (1 × 105 cells per well) 24 h before transfections were performed. Cells with approximately 70 % confluent were co-transfected with matched recombinant plasmid (Wt vector or MT vector) (50 ng/well) and the matched miR-143 mimics or miR-143 inhibitor or scrambled (50 nmol/L, GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen). For luciferase assay, pRL-tk (50 ng/well) which encodes Renilla luciferase (Promega) was added to each group as an internal control for transfection efficiency. Luciferase activities were measured 24 h after transfection using the Dual-Luciferase Reporter Assay (Promega, USA) according to the manufacturer’s protocol. Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well.
Protein extraction and Western blot
The cells were scraped from the dishes and cellular lysate extracts prepared by homogenization in an ice-cold lysis buffer containing freshly protease inhibitor. After centrifugation at 12,000g for 10 min at 4 °C, supernatants were obtained and the total cell protein concentrations were determined using BCA protein assay kit (Pierce, USA). Equal amounts of protein, resolved and separated by SDS-PAGE, were transferred to a PVDF membrane at 320 mA for 2 h at a low temperature. After the membrane was blocked with 5 % fat-free milk with 0.05 % Tween 20 in PBS, the membrane was incubated with the primary antibodies (anti-FAM83Fand anti-GADPH) (Abcam, Cambridge, UK). After rinsing, the membranes were incubated in peroxidase-linked anti-rabbit IgG (Icllab, Shanghai, USA) and were finally visualized using chemiluminescence (Millipore, MA, USA) based on the manufacturer’s instructions. The blots were quantified with Bio-Rad Gel system (Bio-Rad, USA). Stained bands which were scanned, and pixel intensity was quantified using ImageJ based on NIH Image, and mean gray value of each tumor specimen was calculated.
Cell proliferation assays
Colony formation assay
The cells were seeded in six-well plate (500 cells/well) and were maintained under standard culture conditions for 14 days. The estimation of colonies was performed after the colonies were fixed with absolute methanol for 15 min and stained with crystal violet for 20 min.
CCK-8 assay
The viability of the cells was analyzed by CCK-8 (Kumamoto, Japan), conducted daily for 3 days. Viable cells were counted by absorbance measurements at 450 nm by auto microplate reader (infinite M200, Tecan, Austria).
Cell cycle analysis
Cells were seeded in a six-well plate and cultured for 16 h. Then, the cells were fixed with 70 % ethanol for 24 h. The cells were dyed with propidium iodide/RNase buffer (BD Biosciences, USA) for 30 min in the dark at room temperature. All samples were analyzed via flow cytometry (BD Biosciences, USA), and the data were processed using Cell Quest software (BD Biosciences, USA).
Tumor cell migration and invasion assay
The wound healing assay was performed to test cell migration ability. The artificial wounds were produced on the confluent cell monolayer with fetal bovine serum (FBS) free, using a 200-μL pipette tip at 24 h post-transfection. The images were respectively taken at 0, 12, 24, and 36 h after wound creation.
Cells’ migration and invasion abilities were assayed using a Millicell transwell chamber (Millipore, USA), with or without Matrigel (BD Biosciences, USA). For the invasion assay, a transwell chamber was placed into a 24-well plate that was precoated with a 5-mL mixture of BD Matrigel and DMEM (1:1, v/v) and was incubated for 40 min at 37 °C. In both transwell assays, 2 × 104 tumor cells in 0.1 mL of media without FBS were plated in the upper chamber. In the lower chamber, 0.6 mL of the medium with 10 % FBS was added. After 48 h of incubation, cells on the upper surface of the Millicell chambers, noninvasive cells, were scraped with a cotton swab. Tumor cells on the bottom surface of the membrane were fixed in 90 % ethanol for 20 min and stained with 0.1 % crystal violet for 15 min.
Statistical analysis
All experiments were performed at least three times with triplicate samples. Data are presented as the mean ± SD. SPSS19 Statistical software (USA) was used for all statistical analyses in this study. Differences between the miR-143 in tumor tissues and adjacent nontumor tissues were analyzed by the Wilcoxon matched pairs test. The unpaired two-group comparison and multiple comparisons were made with Student’s t test or one-way ANOVA, respectively. Bivariate correlation between miR-143 and FAM83F expressions was assessed by Pearson correlation coefficient and linear regression model. p < 0.05 (*) and p < 0.01 (**) were considered.
Results
Patient characteristics
The surgical specimen used for qRT-PCR was obtained from 22 males and 18 females, and their details are shown in Table 2.
Downregulation of miR-143 in ESCC
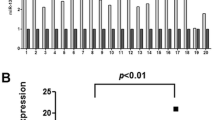
To further study the expression of miR-143 with ESCC, we detected the expression of miR-143 in 40 clinical samples of esophageal carcinoma and their matched normal tissues using qRT-PCR. Using the 2−ΔΔCt method and a twofold change criterion, the qRT-PCR results showed that the expression of miR-143 was downregulated in 87.5 % (35/40) of the clinical samples of ESCC (Fig. 1a). The expression level of miR-143 was significantly downregulated in ESCC compared to matched normal tissues (P < 0.01) as shown in Fig. 1b. We further explored the relationship between the clinical parameters and expression levels of miR-143. The mean fold changes of miRNAs in esophageal carcinoma samples and their possible connections with clinical parameters are presented in Table 3. Significant differences were observed in different T stage groups (Fig. 1c) (P = 0.005).
miR-143 expression in clinical samples. a miR-143 expression in 40 clinical patients. b miR-143 expression in tumor tissue and adjacent tissue. c The expression miR-143 was significantly associated with T stage. d Representative Western blot of paired tumor (ESCC) and normal adjacent tissues (NC). e Pearson’s correlation scatter plot of the fold change of miR-143 and FAM83F protein/mRNA. f Representations of the predicted target site for miR-143 in the FAM83F mRNA 3′-UTR
Upregulation of the FAM83F mRNA and protein in ESCC tissues
To further identify the correlation between the miR-143 levels and the FAM83F protein or FAM83F mRNA, the protein levels and mRNA levels in 40 paired ESCC and normal adjacent tissues were analyzed by Western blot and RT-PCR. We found that the expression level of FAM83F protein was significantly increased in ESCC tissues (Fig. 1d). Bivariate correlation analysis revealed that miR-143 and FAM83F protein were inversely correlated in ESCC tissues as opposed to the paired normal ones (Fig. 1e). Consistently, the FAM83F mRNA was downregulated in the ESCC tissue and inverse correlation was also observed (Fig. 1e). This inverse correlation strongly indicated that miR-143 might regulate the FAM83F negatively by decreasing the stability of mRNA.
miRNA target prediction
All five algorithms predicted the FAM83F as the target gene of miR-143. The predicted interactions between miR-143 and the target sites within the 3′-UTR of FAM83F are illustrated in Fig. 1f.
Validation of the 3′-UTR of FAM83F as a direct target of miR-143
The correlation between miR-143 and FAM83F expressions was further examined by evaluating FAM83F expression in the human ESCC cell lines (ECA109 and KYSE-410) after the overexpression and knockdown of miR-143.
First, the efficiency of overexpression (or knockdown) of miR-143 was demonstrated under inverted fluorescent microscopy and RT-PCR (Fig. 2a). Cellular miR-143 levels were significantly increased when ECA109 and KYSE410 cells were transfected with miR-143 mimics and dropped dramatically when treated with miR-143 inhibitor (Fig. 2a).
miR-143 regulates FAM83F expression by binding the 3′-UTR of FAM83F. a Representative microscopic images and quantitative RT-PCR analysis of the miR-143 levels in KYSE410 and ECA109 cells after transfection. b Sequence of the miR-143 binding sites within the human FAM83F 3′-UTR and a schematic diagram of the reporter constructs showing the FAM83F 3′-UTR sequence (WT) and the mutated sequence (MT). c Direct binding of the FAM83F 3′-UTR to miR-143 KYSE410 and ECA109 cells were co-transfected with a firefly luciferase reporter containing either wild-type (WT) or mutant (Mt) miR-143b binding sites in the FAM83F 3′-UTR and miR-143 mimics, mimics NC, miR-143 inhibitor or inhibitor NC. Twenty-four hours after transfection, the cells were assessed using a luciferase assay kit. The results are displayed as the ratio of firefly luciferase activity in the miR-143-transfected cells to that in the control cells. d Representative Western blot of FAM83F protein levels in KYSE410 and ECA109 cells after transfection. e Quantitative analysis of the FAM83F protein and mRNA levels in KYSE410 and ECA109 cells after transfection
To validate the 3′-UTR of FAM83F as a direct target of miR-143, the Wt vector (Fig. 2b) was co-transfected into ECA109 and KYSE-410 cells with either miR-143 mimics or mimics NC (miR-143 inhibitor or inhibitor NC). According to the result, the normalized luciferase activity was significantly decreased in the cells transfected with miR-143mimics compared with cells transfected with mimics NC. The significant decrease had also been observed in cells transfected with inhibitor NC compared with cells transfected with miR-143 inhibitor. Cells transfected with the MT vector were unaffected by the overexpression of miR-143 (Fig. 2b, c). This finding suggests that the putative miRNA binding sites of FAM83F were responsible for this miRNA-mRNA interaction.
The expression of FAM83F protein was significantly inhibited by the introduction of miR-143 mimics in two cell lines (Fig. 2d), while miR-143 inhibitor significantly increased the FAM83F protein level (Fig. 2d).
In order to determine the regulated level of miR-143, the expression level of FAM83F mRNA was also examined after transfection. The overexpression of miR-143 decreased the level of FAM83F mRNA significantly, and the knockdown of miR-143 increased the level of FAM83F mRNA inversely (Fig. 2e). These results demonstrated that miR-143 regulates FAM83F expression by decreasing the stability of mRNA.
miR-143 inhibits the proliferation and regulated the cell cycle of ESCC cells by targeting FAM83F
The roles of FAM83F in affecting cell proliferation and regulating the cell cycle were first detected by overexpression or silencing of FAM83F. The siRNA, which facilitates the degradation of FAM83F mRNA sequences, or plasmid, which expresses the FAM83F CDS, was transfected into KYSE410 and ECA109. The efficiency of overexpression and knockdown of FAM83F was demonstrated through Western blot (Fig. 3a). The results of colony formation assay after transfection are shown in Fig. 3b. Compared with control vector transfection, FAM83F vector transfection significantly increases the colony formation number. The proliferation was significantly dropped in cells transfected with FAM83F siRNA. To further understand the mechanisms by which cell proliferation is affected, flow cytometry was performed to analyze the cell cycle phase distribution. According to the result, ESCC cells transfected with FAM83F vector displayed an increased percentage of cells in S or G2/M phases and fewer cells in G1/G0 phase compared to cells transfected with control vector. Inversely, cells transfected with FAM83F siRNA showed decreased percentage of cells in S or G2/M phases and increased percentage of cells in G1/G0 phase compared to cells transfected with control siRNA (Fig. 3c).
The effect of FAM83F expression on the proliferation and cell cycle of ESCC cells. a Representative images of Western blot analysis of the FAM83F protein level in ESCC cells after transfection. b Representative images (left) and quantification (right) of ESCC cell colonies, as determined by colony formation assay after transfection. c Representative images (left) and quantification (right) of cell cycle in ESCC cells as analyzed by flow cytometry after transfection
Next, the effect of miR-143 on proliferation was detected. Cell lines transfected with miR-143 mimics exhibited reduced proliferation compared to mimics NC. In contrast, cells transfected with miR-143 inhibitor exhibited increased proliferation when compared to inhibitor NC (Fig. 4a). Besides, compared with mimics NC, cells transfected with miR-143 mimics showed an increased percentage of cells in G1/G0 phase and fewer cells in S or G2/M phase. Cells transfected with miR-143 inhibitor showed inverse results (Fig. 4b). These results suggested that the growth-suppressive effect of miR-143 was partly due to the G1/G0 phase arrest.
The effect of miR-143 on the proliferation and cell cycle of ESCC cells. a Representative images (left) and quantification (right) of ESCC cell colonies, as determined by colony formation assay after transfection. b Representative images (left) and quantification (right) of cell cycle in ESCC cells as analyzed by flow cytometry after transfection
Finally, the biological consequences of the miR-143-mediated suppression of FAM83F expression in ESCC cells were detected. ECA109 and KYSE410 cells were co-transfected with FAM83F vector or control vector and miR-143 mimics or mimics NC. The Western blot demonstrated that the protein level in the overexpression sample was apparently higher than that of the NC even under the suppression of miR-143 (Fig. 5a). According to the results of colony formation number (Fig. 5b), the miR-143-resistant FAM83F expression rescued the suppression of FAM83F expression by miR-143 and attenuated the anti-proliferative effect of miR-143. To further demonstrate the viability of ESCC cells, the CCK-8 assay was conducted. The results of CCK-8 assay are presented in the Fig. S1, which were consistent with that of colony formation assay.
miR-143 inhibits proliferation and regulates cell cycle of ESCC cells by targeting FAM83F. a Representative images of Western blot analysis of the FAM83F protein level in ESCC cells after transfection. b Representative images (left) and quantification (right) of ESCC cell colonies, as determined by colony formation assay after transfection. c Representative images (left) and quantification (right) of cell cycle in ESCC cells as analyzed by flow cytometry
Furthermore, according to the result of cell cycle analysis, when ESCC cells were co-transfected with miR-143 mimics and the FAM83F overexpression plasmid, the G1/G0 phase arrest effect of miR-143 was significantly attenuated (Fig. 5c).
miR-143 inhibits the migration and invasion of ESCC cells by targeting FAM83F
The effect of FAM83F in promoting cell migration and invasion was first detected by scratching assay and transwell assay. Both cell lines transfected with FAM83F vector exhibit significantly increased migration ability when compared with cells transfected with vector control at 12, 24, and 36 h after scratching. Cells transfected with FAM83F siRNA showed decreased migration ability when compared with cells transfected with siRNA control (Fig. S2). For transwell result, the number of migrating and invasive cells per field in cell lines transfected with FAM83F vector was significantly higher than those transfected with control vector (Fig. 6a, b).
The effects of miR-143 on cell migration and invasion were also analyzed. Cell lines transfected with miR-143 mimics were distinctively less migratory than mimics NC at 12, 24, and 36 h after scratching (Fig. S2). According to the result of transwell assay, the number of migrating and invasive cells per field in cell lines transfected with miR-143 mimics was dramatically decreased compared to the cells transfected with mimics NC. In contrast, cells transfected with miR-143 inhibitor exhibited increased invasiveness compared to inhibitor NC (Fig. 7a, b).
Next, we investigated the restoration effect of FAM83F in miR-143mimics-transfected cells. According to the result, the cells co-transfected with miR-143 and the FAM83F overexpression plasmid exhibited a significantly increased migration and invasion ability than cells co-transfected with miR-143 mimics and control vector. This indicated that the restoration of FAM83F rescued the suppression of FAM83F expression mediated by miR-143 and leads to an activation of invasion and migration (Fig. 8, Fig. S2).
Discussion
FAM83F proteins were recently identified as a novel oncogene family that overexpressed in diverse tumor types including breast cancer, lung cancer, and ovarian cancer. The FAM83 family could promote epithelial cell transformation and act as critical mediators of CRAF/MAPK signaling and EGFR/RAS signaling [4, 10].
In this study, we predicted the FAM83F as a target of miR-143, a miRNA that has been reported to be downregulated and to function as a tumor suppressor in several tumors [11], such as glioblastoma [12], colorectal cancer [11], and prostate cancer [13]. For esophageal cancer, miR-143 was found to be downregulated and its overexpression had been found to play an important role in inhibiting cell mobility [14]. Recent studies reported that the miR-143 could act as a negative regulator of TNF-related apoptosis in ESCC cells [15]. In addition, miR143 could affect the risk of ESCC through regulating fascin homolog 1 (FSCN1) [15]. In this study, we found an alternative mechanism through which the FAM83F was regulated by miR-143 at the post-transcriptional level in ESCC.
First, we detected the expression of miR-143 in 40 patients and found that the expression of miR-143 was significantly downregulated in most of the cancer sample (35/40) compared with adjacent normal tissue. Also, it should be noticed that this ratio would change according to the cutoff value (cutoff = 2) that we chose, and when it was set higher, there could be relatively fewer patients with miR-143 downregulated in cancer tissues. On the other hand, significant decrease was also (p < 0.01) observed in the expression level of miR-143 in tumor tissue compared with the adjacent normal tissue (Fig. 1). However, there was still 12.5 % of the tumor tissue that had high miR-143 expression, and this might be due to the individual difference among patients. Some of the individuals might have had different gene expression patterns during tumorigenesis or cancer progression.
After measuring the expression levels of FAM83F in paired human ESCC and noncancerous tissue, we detected an inverse correlation between the miR-143 levels and the FAM83F protein levels. The downregulated effects of miR-143 to FAM83F were then verified by evaluating FAM83F mRNA and protein expression in the human ESCC cell lines ECA109 and KYSE-410 after the overexpression and knockdown of miR-143. Then, we demonstrated the 3′-UTR of FAM83F mRNA as a direct target of miR-143 by dual-luciferase experiment. Furthermore, by overexpressing or silencing miR-143 in ESCC cells, we experimentally validated that miR-143 regulates FAM83F expression by decreasing the stability of mRNA at the post-transcriptional level.
This study observed a statistically significant association between the expression of miR-143 and the ESCC invasion stage. These results suggest that alterations of miR-143 could be involved in ESCC cancer progression. Studies in vitro confirmed speculation that the miR-143 can negatively regulate the cell proliferation, migration, and invasion. On the other hand, the role of FAM83F in promoting cell proliferation, migration, and invasion had also been observed. Finally, we showed that the restoration of FAM83F expression reverses miR-143-induced cell effects including inhibition of cell proliferation through G1/G0 phase arrest and suppression of migration and invasion. These results delineate a novel regulatory network by which miR-143 and FAM83F regulate relative biological process and further affect tumor progression.
Conclusion
In conclusion, our expression and functional studies suggest that the miR-143 was downregulated and could act as a tumor suppressor through inhibiting proliferation, migration, and invasion and inducing G1/G0 phase arrest by directly targeting FAM83F in ESCC. Thus, our study provides basic information to better understand ESCC tumor biology and could help to facilitate the development of miRNA-based anti-tumor strategies.
References
Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17(1):38–44. doi:10.1016/j.semradonc.2006.09.007.
Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15(19):6192–200. doi:10.1158/1078-0432.CCR-09-1467.
Huang J, Zhang SY, Gao YM, Liu YF, Liu YB, Zhao ZG, et al. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 2014;47(4):277–86.
Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res. 2014;12(8):1156–65. doi:10.1158/1541-7786.MCR-13-0289.
Roush SF, Slack FJ. Micromanagement: a role for microRNAs in mRNA stability. ACS Chem Biol. 2006;1(3):132–4. doi:10.1021/cb600138j.
Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–4.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi:10.1038/nrc1997.
Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–5. doi:10.1038/nature13176.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-8. doi:10.1006/meth.2001.1262
Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122(9):3211–20. doi:10.1172/JCI60498.
Li JM, Zhao RH, Li ST, Xie CX, Jiang HH, Ding WJ, et al. Down-regulation of fecal miR-143 and miR-145 as potential markers for colorectal cancer. Saudi Med J. 2012;33(1):24–9.
Koo S, Martin GS, Schulz KJ, Ronck M, Toussaint LG. Serial selection for invasiveness increases expression of miR-143/miR-145 in glioblastoma cell lines. BMC Cancer. 2012;12:143. doi:10.1186/1471-2407-12-143.
Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350(1-2):207–13.
Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF, Huang Q, et al. miRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J Gastroenterol. 2011;17(1):79–88.
Zhou K, Yan Y, Zhao S. Esophageal cancer-selective expression of TRAIL mediated by MREs of miR-143 and miR-122. Tumour Biol. 2014;35(6):5787–95. doi:10.1007/s13277-014-1768-5.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We declare that all human and animal studies have been approved by the hospital ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We declare that all patients gave informed consent prior to inclusion in this study.
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s13277-016-5470-7.
Rights and permissions
About this article
Cite this article
Mao, Y., Liu, J., Zhang, D. et al. miR-143 inhibits tumor progression by targeting FAM83F in esophageal squamous cell carcinoma. Tumor Biol. 37, 9009–9022 (2016). https://doi.org/10.1007/s13277-015-4760-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4760-9