Abstract
hSNF2H partners with Rsf-1 to compose the Rsf complex to regulate gene expression. Recent studies indicated that hSNF2H was overexpressed in several human cancers. However, its expression pattern and biological mechanism in glioma remain unexplored. In this study, we found that hSNF2H was overexpressed in 32 % of glioma specimens. hSNF2H overexpression correlated with advanced tumor grade (p = 0.0338) and Rsf-1 positivity in glioma tissues (p = 0.016). Small interfering RNA (siRNA) knockdown was performed in A172 and U87 cell lines. MTT, colony formation assay, and cell cycle analysis showed that knockdown of hSNF2H inhibited cell proliferation, colony formation ability, and cell cycle transition. Matrigel invasion assay showed that hSNF2H depletion inhibited invasive ability of glioma cells. In addition, we demonstrated that hSNF2H depletion decreased temozolomide resistance of A172 and U87 cell lines and increased temozolomide induced apoptosis. Furthermore, hSNF2H depletion decreased cyclin D1, cyclin E, p-Rb, MMP2, cIAP1, Bcl-2 expression, and phosphorylation of IκBα and p65, suggesting hSNF2H regulates apoptosis through NF-κB pathway. Immunoprecipitation showed that hSNF2H could interact with Rsf-1 in both cell lines. To validate the involvement of Rsf-1, we checked the change of its downstream targets in Rsf-1 depleted cells. In Rsf-1 depleted cells, changes of cyclin E, Bcl-2, and p-IκBα were not significant using hSNF2H siRNA treatment. In conclusion, our study demonstrated that hSNF2H was overexpressed in human gliomas and contributed to glioma proliferation, invasion, and chemoresistance through regulation of cyclin E and NF-κB pathway, which is dependent on its interaction with Rsf-1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the most common malignant tumor in the central nervous system. Despite improvement in surgery and chemotherapy, the prognosis for glioma patients remains poor [1]. Overexpression of oncogene plays important roles during cancer development and progression. Thus identifying of new markers which play a part in tumor progression is very important for understanding biological characteristics of glioma and development of targeted therapy [2, 3].
Chromatin regulation is an important process of DNA replication, transcription, and DNA repair, and chromatin remodeling complexes play important roles in this process. The Imitation Switch (ISWI) is a chromatin remodeling complex family, which has many biological activities such as DNA dependent ATPase activity [4]. hSNF2H (human sucrose non-fermenting protein 2 homologue) belongs to ISWI chromatin remodeling complex and displays chromatin remodeling activity in development and cancer [5–7]. Previous reports showed that SNF2H expression was strong in proliferating population during brain development [8]. In a study using a SNF2H-null mice, blastocyst outgrowth experiments indicated that loss of SNF2H resulted in growth arrest and cell death of both the trophectoderm and inner cell mass. Reducing SNF2H level also inhibited CD34+ progenitors from undergoing cytokine-induced erythropoiesis in vitro [9, 10]. Recently, there are several studies indicating that hSNF2H was overexpressed in human cancers including gastric cancer, ovarian cancer, and prostate cancer [11–14]. It still remains unclear that hSNF2H was involved in development and progression of human glioma.
In the present study, we examined the expression pattern of hSNF2H in human glioma specimens. We also knocked down hSNF2H using small interfering RNA (siRNA) to explore its role in cell proliferation, invasion, and chemoresistance. In addition, we investigated the potential mechanism of hSNF2H during malignant cell progression.
Materials and methods
Specimens and immunohistochemistry
The study protocol was approved by the institutional reviewer board of China Medical University. All participants provided their written informed consent, and the investigation was conducted according to the principles expressed in the Declaration of Helsinki. Tumor specimens were obtained from 103 patients diagnosed with glioma who underwent resection in the First Affiliated Hospital and Shengjing Hospital of China Medical University between 2010 and 2013. Clinical and pathological data were obtained from medical records.
Surgically excised tumor specimens were fixed and embedded in paraffin, and 5-μm-thick sections were prepared. Immunostaining was performed using the S-P staining kit from Maixin (Ultrasensitive™, MaiXin, Fuzhou, China). After antigen retrieval in citrate buffer (pH 6.0) for 2 min in an autoclave. 0.3 % hydrogen peroxide was used for 15 min, and then the sections were incubated with goat serum. Tissue sections were incubated with hSNF2Hmouse monoclonal antibody (1:1000 dilution; Milipore), Rsf-1 (1:2000 dilution; Milipore). Mouse immunoglobulin (at the same concentration of the antigen specific antibody) was used as a negative control. Staining was performed at 4 °C overnight. Biotinylated goat anti-rabbit serum IgG was used as a secondary antibody. After washing, the sections were incubated with streptavidin–biotin conjugated complex with horseradish peroxidase, and the peroxidase reaction was developed with 3,3-diaminobenzidine tetrahydrochloride. Counterstaining with hematoxylin was performed and the sections were dehydrated in ethanol before mounting.
Two independent blinded investigators examined all tumor slides randomly. Positive nuclear/cytoplasmic staining was considered positive. Immunostaining of hSNF2H was scored on a semi-quantitative scale by evaluating staining intensity and percentage. We calculated the percentage of positively stained cells. The staining intensity was categorized as follows: 0, negative; 1, moderate; and 2, strong. The staining percentage of tumor specimens was scored as 0, 0 %; 1 1–5 %; 2, 6–25 %; 3, 26–75 %; and 4, 76–100 %. The scores of each tumor sample were multiplied to give a final score of 0 to 8, and the tumor samples with a final score of 4–8 were finally determined as hSNF2H overexpression.
Cell culture and transfection
A172 and U87 cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10 % fetal calf serum. Cells were grown on sterilized culture dishes and were passaged every 2 days with 0.25 % trypsin.
hSNF2H and Rsf-1 siRNA was bought from by Santa Cruz. DharmaFECT1 reagent was used for siRNA transfection (ThermoFisher, USA) according to the manufacturer’s instructions.
Western blot analysis
Total protein was extracted using Pierce lysis buffer (Pierce, Rockford, IL). Protein quantification was performed using the Bradford method. Fifty micrograms of protein was separated by SDS-PAGE and was transferred to PVDF membranes (Millipore, Billerica, MA, USA). Membranes were incubated at 4 °C overnight with antibody against hSNF2H (1: 1000, Millipore, USA), Bcl-2, MMP2, p-Rb, cIAP1, p-IκB, p-p65, cyclin D1, cyclin E, cleaved caspase3 (1:1000 dilution, Cell Signaling Technology, Boston, USA) and GAPDH (1:2000 dilution; Cell Signaling Technology, USA), after incubation with peroxidase-coupled anti-mouse or rabbit IgG antibody (1:1000 dilution, Cell Signaling Technology, USA) at 37 °C for 2 h. Target proteins on PVDF membrane were visualized using Pierce ECL kit and captured using a DNR BioImaging System (DNR, Jerusalem, Israel).
Immunoprecipitation
For immunoprecipitation, a sufficient amount of antibody was added to 200 μg protein and gently rotated at 4 °C overnight. The immunocomplex was captured by adding 25 μl protein A/G agarose beads (Beyotime, Jiangsu, China) and gently rotated at 4 °C for 3 h. Then, the mixture was centrifuged at 1500 g for 5 min at 4 °C, and the supernatant was discarded. The precipitate was washed three times with ice-cold RIPA buffer, resuspended in sample buffer, and boiled for 5 min to dissociate the immunocomplex from the beads. The supernatant was then collected by centrifugation and subjected to Western blot analysis.
Quantitative real-time PCR (SYBR Green method)
Total RNA was extracted from cells using Trizol (Invitrogen). Reverse transcription of 1 μg of RNA was done using the RT kit (TAKARA, Dalian, China) following the manufacturer’s instructions.
Real-time PCR was performed using SYBR Green master mix kit purchased from Applied Biosystems. PCR was performed using 7500 real-time PCR System (Applied Biosystems). β-actin was used as the endogenous control. Relative gene expression was calculated as ΔCt = Ct gene –Ct control. Relative gene expression was calculated by the 2-ΔΔCt method. The sequences of the primer pairs are as follows:
-
hSNF2H forward, 5′ TTGGCATCAATCTTGCGACT 3′
-
hSNF2H reverse, 5′ CCAATTCTATGTGCTCGGTCCA 3′
-
β-actin forward, 5′ ATAGCACAGCCTGGATAGCAACGTAC 3′
-
β-actin reverse, 5′ CACCTTCTACAATGAGCTGCGTGTG 3′
Colony formation and MTT assay
Colony formation assay: after transfection for 48 h, cells were plated into 6-cm cell culture dishes (about 2000 per dish). Then, cells were cultured for 12 days. Plates were washed with PBS and Giemsa staining was performed to visualize colony. The colonies with more than 50 cells were manually counted using a microscope.
MTT assay: cells were plated in 96-well plates (approximately 3000 cells per well) and cultured for 5 days. Twenty microliters of 5 mg/ml MTT solution was added to well. After incubation for 4 h, the medium was removed and remaining MTT formazan was dissolved in 150 μl of DMSO. Solution was measured spectrophotometrically at 490 nm.
Flow cytometry for cell cycle analysis
Forty-eight hours after transfection, cells were harvested and fixed using 1 % paraformaldehyde. Then, cells were washed with PBS and stained in 5 mg/ml propidium iodide for 30 min at room temperature. Flow cytometry was performed using BD FACS Calibur flow cytometer systems (Becton Dickinson, USA).
Matrigel invasion assay
Cell invasion assay was performed using a 24-well Transwell chamber with a pore size of 8 μm (Costar, Cambridge, MA). The inserts were coated with 20 μl Matrigel (1:3 dilution, BD Bioscience, San Jose, CA, USA). After the transfection, cells were trypsinized, and 2 × 105 cells in 100 μl of serum-free medium were transferred to the upper Matrigel chamber and incubated for 18 h. Medium supplemented with 15 % FBS was added to the lower chamber. After incubation, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4 % paraformaldehyde and stained with hematoxylin.
Statistical analysis
SPSS version 11.5 for Windows was used for all statistical analyses. Chi-Square test was used to evaluate possible correlations between hSNF2H overexpression and clinicopathologic factors. t test was used to compare data between control and siRNA treated cells. All p values were based on the two-sided statistical analysis, and p < 0.05 was considered to be statistically significant in difference.
Results
Clinical significance of hSNF2H in glioma specimens
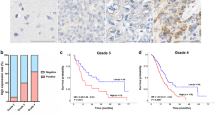
We examined the hSNF2H protein expression in a panel of 103 primary glioma specimens and 20 normal brain tissues using immunohistochemistry. We found that positive staining of hSNF2H protein was mainly localized in the nuclear compartment in glioma specimens. Normal glial cells showed negative hSNF2H expression (Fig. 1a). hSNF2H positivity was found in 33 out of 103 (32 %) glioma specimens (Fig. 1b–d). We analyzed the correlation between hSNF2H status and clinical parameters. We observed statistically significant correlation between hSNF2H positivity and advanced tumor grade (p = 0.0338). Positive rate of hSNF2H was 40.63, 21.21, and 0 % in advanced stage (stages III and IV), stage II, and stage I gliomas, respectively (Table 1). We found no difference between the hSNF2H status with age (p = 0.1659) and gender (p = 0.8365). We also examined expression 加of hSNF2H partner protein Rsf-1 in glioma tissues. The results showed that 54 cases scored positive for Rsf-1, which was also localized in nucleus of tumor cells (Fig. 1f). As shown in Table. 1, cases that had high level of hSNF2H expression tended to have strong Rsf-1 staining (p = 0.016) (Fig. 1e, f).
Expression of hSNF2H in human glioma. a Immunohistochemical staining of hSNF2H protein in glial cells was negative. b Negative hSNF2H staining in grade II glioma. c Positive nuclear hSNF2H staining in grade III glioma. d Strong nuclear hSNF2H staining in glioblastoma (grade IV). e, f Glioma samples with high hSNF2H expression (e) showed strong Rsf-1 nuclear expression (f). (Magnification ×200)
Knockdown of hSNF2H suppresses glioma cell proliferation and cell cycle progression
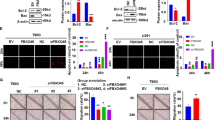
Expression level of hSNF2H was analyzed in three glioma cell lines by Western blot. High expression of hSNF2H protein was observed in A172 and U87 cell lines (Fig. 2a). To explore its biological function, hSNF2H specific siRNA was used to deplete its expression. hSNF2H knockdown efficiency was confirmed by Western blot and real-time PCR (Fig. 2b). Then, we tried to explored the effect of hSNF2H depletion on cell proliferation. Using MTT and colony formation assay, we found hSNF2H depletion decreased growth rate (Fig. 3a) and colony formation ability compared with control (A172 Con vs siRNA: 278 ± 8 vs 166 ± 11; U87 Con vs siRNA: 222 ± 18 vs 106 ± 11, p < 0.05) (Fig. 3b). Cell cycle analysis showed that S phase percentage was decreased in cells transfected with hSNF2H siRNA compared with control in both cell lines (Fig. 3c), suggesting hSNF2H downregulation could inhibit glioma cell proliferation by arresting cell cycle progression.
hSNF2H knockdown inhibits cell proliferation and cell cycle progression. a MTT showed that siRNA treatment of hSNF2H decreased cell growth rate in A172 and U87 cells in comparison with cells transfected with negative siRNA. b hSNF2H depletion inhibited colony formation ability in both cell lines. c Cell cycle analysis showed that S phase percentage was decreased after hSNF2H depletion. *p < 0.05
hSNF2H depletion in glioma cells inhibits cell invasion and chemoresistance
Matrigel invasion assay was also performed to determine the role of hSNF2H on cell invasion. A significant decrease of cell invasion (A172: 58.7 %; U87: 44.1 %) was observed in hSNF2H depleted cells (Fig. 4a), suggesting that hSNF2H positively regulates glioma cell invasion. To investigate the involvement of hSNF2H in chemoresistance of glioma cells, temozolomide (TMZ) (100 μM) was used to treat A172 and U87 cells transfected with hSNF2H siRNA or negative control siRNA. We observed a remarkable decrease in cell viability after siRNA treatment using MTT assay (Fig. 4b). In addition, the degree of apoptosis was examined by AnnexinV/PI flow cytometry analysis. hSNF2H siRNA significantly increased apoptotic rate in TMZ treated glioma cells. We also found that the level of cleaved caspase 3 was significant upregulated after hSNF2H siRNA treatment in both cell lines. These results suggest that hSNF2H could mediate temozolomide resistance in glioma cells and its depletion enhanced temozolomide sensitivity.
hSNF2H knockdown inhibits cell invasion and chemoresistance. a Matrigel invasion assay showed that hSNF2H depletion decreased cell invasion in both A172 and U87 cell lines. b MTT assay showed a remarkable decrease in cell viability after siRNA treatment using MTT assay in temozolomide treated cells. c hSNF2H siRNA significantly increased apoptosis rate and caspase 3 cleavage in TMZ treated glioma cells. *p < 0.05
hSNF2H depletion downregulates cyclin D1, cyclin E, MMP2, Bcl-2, cIAP1 expression, and NF-κB signaling activity
To investigate the potential mechanism by which hSNF2H affected cell cycle, invasion, and chemoresistance, we examined the effect of hSNF2H depletion on cell cycle related proteins cyclin D1, cyclin E, and p-Rb. We also examined invasion related protein MMP2 and chemoresistance protein Bcl-2 and cIAP1. As shown in Fig. 5, Western blot analysis revealed that downregulation of hSNF2H decreased the protein levels of cyclin D1, cyclin E, p-Rb, MMP2, Bcl-2, and cIAP1. In addition, we found that the levels of NF-κB components p-IκBα and p-p65 were significantly decreased after hSNF2H depletion, suggesting hSNF2H regulates biological behaviors of glioma cells possibly through NF-κB pathway.
hSNF2H interacts with Rsf-1 to regulate NF-κB pathway
hSNF2H was previous regarded as Rsf-1 partner, which was reported to interact with cyclin E. To validate these, co-immunoprecipitation was performed to examine if there is an association between hSNF2H and Rsf-1. We immunoprecipitated hSNF2H and analyzed them by Western blotting for Rsf-1 binding. As shown in Fig. 6a, hSNF2H co-immunoprecipitated with Rsf-1 in glioma cells. To validate the role of Rsf-1 in hSNF2H mediated effects, we employed siRNAs to deplete Rsf-1 expression in A172 and U87 cells. Then, we examined if hSNF2H knockdown could decrease cyclin E, Bcl-2, and p-IκBα in Rsf-1 depleted cells. As shown in Fig. 6b, the effect of hSNF2H on cyclin E, Bcl-2, and p-IκBα was significantly blocked in Rsf-1 depleted cells.
Discussion
Chromatin remodeling factors play important roles in diverse biological process. Aberrant expression of chromatin remodeling complex was reported to associate with many diseases and cancers [15]. hSNF2H was reported to be overexpressed and to interact with Rsf-1 in ovarian cancer [14]. However, clinical significance of hSNF2H and its biological roles in human glioma have not been explored. To address this issue, immunohistochemistry was performed to detect the staining of hSNF2H protein in glioma tissues. We found that hSNF2H was overexpressed in 32 % of glioma tissues compared with normal glial cells. hSNF2H protein was mainly localized in the nucleus of cancer cells and correlated with advanced grade. This was in accord with previous study suggesting hSNF2H was a potential oncoprotein in glioma and might associate with glioma progression [13]. It was reported that hSNF2H partnered with Rsf-1 to form an ISWI chromatin remodeling complex [16]. Rsf-1 served as a survival signal in many biological activities and was overexpressed in many human cancers [17–19]. Previous studies demonstrated that hSNF2H interacted with Rsf-1 and promoted the function of Rsf-1 during cancer development and progression [14, 20]. In the present study, we showed that hSNF2H colocalized with Rsf-1 in the nuclear compartment of glioma tissues, suggesting Rsf-1 may play a part in the biological roles of hSNF2H in glioma.
Immunohistochemical staining suggested potential roles of hSNF2H in glioma progression. We depleted hSNF2H expression by using siRNA in A172 and U87 cell lines with high endogenous expression and examined the biological roles of hSNF2H. MTT and colony formation assay were performed, and we observed a remarkable decrease of proliferation rate and colony formation ability in hSNF2H depleted glioma cells. Cell cycle analysis indicated that hSNF2H depletion might arrest G1-S transition to inhibit cell proliferation. Accordingly, analysis of cell cycle related proteins showed that hSNF2H siRNA significantly decreased cyclin D1, cyclin E, and p-Rb, which was in accordance with cell cycle inhibition caused by siRNA treatment. In addition, Matrigel invasion assay was performed and the results showed that hSNF2H depletion significantly decreased invading ability of glioma cells, with downregulation of MMP2 protein expression. Thus, hSNF2H regulates both proliferation and invading ability of glioma cells, which correlated well with the fact that hSNF2H positively associated with advanced glioma grade.
The involvement of hSNF2H in cancer chemoresistance was not clear. To explore this issue, we examined cell viability in hSNF2H depleted glioma cells with TMZ treatment. We found that hSNF2H sensitized cancer cells to TMZ treatment in A172 and U87 cell lines. In addition, the rate of apoptosis and the level of caspase 3 cleavage were upregulated after hSNF2H knockdown in TMZ treated cells. These data suggested that hSNF2H overexpression in glioma induced temozolomide resistance and its depletion increased drug sensitivity. The mechanism of hSNF2H related temozolomide sensitivity was unclear. We examined the level of apoptosis related proteins and found that Bcl-2 and cIAP1 expression were downregulated after hSNF2H depletion in both cell lines. Bcl-2 and cIAP1 were important inhibitors of cell apoptosis and played an important role in the development of TMZ resistance [21, 22]. In addition, we found that the level of NF-κB components p-IκBα and p-p65 were significant inhibited after hSNF2H knockdown. Bcl-2 and cIAP1 were both downstream targets of NF-κB signaling pathway [23–25]. These results suggested that hSNF2H upregulated Bcl-2 and cIAP1 expression to enhance TMZ resistance of A172 and U87 cells through NF-κB pathway.
Rsf-1 was previously reported to interact with cyclin E and induce cell proliferation [26]. Rsf-1 was also shown to interact with hSNF2H in ovarian cancer cells [20]. Since hSNF2H is a Rsf-1 partner protein, we examined if there was an association between hSNF2H and Rsf-1. Using immunoprecipitation, we confirmed that hSNF2H could interact with Rsf-1. To further confirm the involvement of Rsf-1 in hSNF2H induced effect, we employed siRNA to deplete both hSNF2H and Rsf-1 expression in A172 and U87 cells. The effect of hSNF2H on cyclin E, Bcl-2, and p-IκBα was significantly blocked in Rsf-1 depleted glioma cells. These results indicated that Rsf-1 plays an important role in hSNF2H mediated biological effect.
In conclusion, this study demonstrates that hSNF2H is overexpressed in glioma tissues and correlates with advanced grade. hSNF2H interacts with Rsf-1 and regulates cell proliferation, invasion, and chemoresistance through NF-κB signaling. Our findings indicate that hSNF2H might serve as a therapeutic target of glioma.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29.
Krupkova Jr O, Loja T, Redova M, Neradil J, Zitterbart K, et al. Analysis of nuclear nestin localization in cell lines derived from neurogenic tumors. Tumour Biol. 2011;32:631–9.
Zadran S, Amighi A, Otiniano E, Wong K, Zadran H. ENTPD5-mediated modulation of ATP results in altered metabolism and decreased survival in gliomablastoma multiforme. Tumour Biol. 2012;33:2411–21.
Kokavec J, Podskocova J, Zavadil J, Stopka T. Chromatin remodeling and SWI/SNF2 factors in human disease. Front Biosci. 2008;13:6126–34.
LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–4.
Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–87.
Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, et al. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–900.
Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001;77:1145–56.
Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39:614–22.
Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100:14097–102.
Gigek CO, Lisboa LC, Leal MF, Silva PN, Lima EM, et al. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29:162–6.
Stopka T, Zakova D, Fuchs O, Kubrova O, Blafkova J, et al. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14:1247–52.
Reis ST, Timoszczuk LS, Pontes-Junior J, Viana N, Silva IA, et al. The role of micro RNAs let7c, 100 and 218 expression and their target RAS, C-MYC, BUB1, RB, SMARCA5, LAMB3 and Ki-67 in prostate cancer. Clinics (Sao Paulo). 2013;68:652–7.
Sheu JJ, Choi JH, Yildiz I, Tsai FJ, Shaul Y, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68:4050–7.
Sheu JJ, Guan B, Choi JH, Lin A, Lee CH, et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285:38260–9.
Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23:6759–68.
Li H, Zhang Y, Zhang Y, Bai X, Peng Y, et al. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker. Tumour Biol. 2014;35:5771–6.
Lin CY, Tian YF, Wu LC, Chen LT, Lin LC, et al. Rsf-1 expression in rectal cancer: with special emphasis on the independent prognostic value after neoadjuvant chemoradiation. J Clin Pathol. 2012;65:687–92.
Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumour Biol. 2012;33:1485–91.
Choi JH, Sheu JJ, Guan B, Jinawath N, Markowski P, et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69:1407–15.
Qi XC, Xie DJ, Yan QF, Wang YR, Zhu YX, et al. LRIG1 dictates the chemo-sensitivity of temozolomide (TMZ) in U251 glioblastoma cells via down-regulation of EGFR/topoisomerase-2/Bcl-2. Biochem Biophys Res Commun. 2013;437:565–72.
Wagner L, Marschall V, Karl S, Cristofanon S, Zobel K, et al. Smac mimetic sensitizes glioblastoma cells to Temozolomide-induced apoptosis in a RIP1- and NF-kappaB-dependent manner. Oncogene. 2013;32:988–97.
Liu S, Shen H, Xu M, Liu O, Zhao L, et al. FRP inhibits ox-LDL-induced endothelial cell apoptosis through an Akt-NF-{kappa}B-Bcl-2 pathway and inhibits endothelial cell apoptosis in an apoE-knockout mouse model. Am J Physiol Endocrinol Metab. 2010;299:E351–363.
Chen GG, Liang NC, Lee JF, Chan UP, Wang SH, et al. Over-expression of Bcl-2 against Pteris semipinnata L-induced apoptosis of human colon cancer cells via a NF-kappa B-related pathway. Apoptosis. 2004;9:619–27.
Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, et al. IKKepsilon-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep. 2013;3:724–33.
Sheu JJ, Choi JH, Guan B, Tsai FJ, Hua CH, et al. Rsf-1, a chromatin remodelling protein, interacts with cyclin E1 and promotes tumour development. J Pathol. 2013;229:559–68.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (No. 81000824, 81473285, and 81101402).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Zhao, XC., An, P., Wu, XY. et al. Overexpression of hSNF2H in glioma promotes cell proliferation, invasion, and chemoresistance through its interaction with Rsf-1. Tumor Biol. 37, 7203–7212 (2016). https://doi.org/10.1007/s13277-015-4579-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4579-4