Abstract
Increasing results suggest microRNAs (miRNAs) could function important roles in malignant tumor progression. miR-30a-5p is downregulated in variety of cancers and acts as a cancer suppressing gene. The functions and molecular mechanisms of miRNA-30a-5p in hepatocellular carcinoma (HCC) remain unclear. In the present study, quantitative real-time PCR (qRT–PCR) was used to detect miR-30a-5p expression in 16 pairs of HCC and their adjacent non-cancerous tissues and HCC cell lines. By overexpression of miRNA-30a-5p, CCK-8 and colon formation assay were used to evaluate cell growth and flow cytometry to evaluate cell apoptosis. Western blot was used to test protein expression. And potential mechanisms were analyzed with luciferase activity assay. In vivo HepG2 tumor growth was observed with nude mice. Our results showed that miR-30a-5p expression in HCC tissues was significantly lower compared to adjacent non-cancerous liver tissues, and lower miR-30a-5p expression was also observed in HCC cell lines compared to normal liver cell. Luciferase assay showed that metadherin (MTDH) mRNA was a direct target of miR-30a-5p. A significant reverse correlation between miR-30a-5p and MTDH in liver cancer tissues was observed. miR-30a-5p overexpression in HCC cells significantly inhibited cell proliferation, colon formation, and induced apoptosis while MTDH overexpression reversed growth inhibition and apoptosis induction of miRNA-30a-5p in HCC cells. miRNA-30a-5p upregulated phosphatase and tensin homolog (PTEN) protein expression and thus inhibited AKT activating by targeting MTDH. miRNA-30a-5p also significantly inhibited HepG2 tumor growth in vivo. Our results suggest that underexpression of miR-30a-5p might function as a tumor suppressing miRNA by directly targeting MTDH in HCC and is therefore a potential candidate biomarker for HCC targeting therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are size of about 20 to 24 bases, single stranded, small non-coding RNAs. By completely or incompletely binding with the untranslated section of mRNA (3′-UTR), miRNAs inhibit protein expression [1]. Recently, more than 1000 miRNAs were found, and in the human body, at least 60 % protein expressions are regulated by miRNAs [2]. It is widely recognized that abnormal expression of miRNAs exists in a variety of cancers and, by importing the special miRNAs, could inhibit tumor growth [3]. Some miRNAs promoting cancer are frequently expressed higher in cancer tissues; conversely, some miRNAs inhibiting cancer are frequently reduced and in the whole to promote malignant tumor transformation [4]. It is also recognized that the structures of artificial miRNA and endogenous miRNAs have the similarity and will not cause endogenous reaction, and miRNA therapy for cancer is regarded as a potentially safe and promising treatment method [5].
Hepatocellular carcinoma (HCC) is the world’s fifth largest incidence of tumor with a situation still gradually increasing. When HCC diagnosis was made, only 30 % had surgery opportunity to remove the tumor; the postoperative 5-year survival rate was only about 30∼40 %, seriously threatening patient’s life and health [6]. Exploring the pathogenesis of liver cancer through miRNAs had achieved one important means to understand the occurrence and progression mechanisms in liver cancer, which expected to achieve promising prospects. For example, it was reported that some miRNAs such as miRNA122 and miRNA26 exist in the normal liver tissue but frequently downregulated in HCC tissues; exogenously importing corresponding miRNAs could significantly inhibit tumor proliferation [7]. On the contrary, some miRNAs, such as miRNA18b, expression were significantly upregulated in HCC tissues, and overexpression of such miRNAs could obviously promote HCC cells proliferation and migration.
In the process of liver embryogenesis, miRNA-30 family expressions gradually rise as the process of embryonic development [8]. Bud reported that microRNA-30 families frequently downregulated in HCC [9]. These results prompt the miRNA-30 families and low differentiation and hint that miRNA-30 families might function as a tumor suppressor in liver cancer. Recently, miR-30a-5p was found as a tumor suppressor in gastric cancer [10], and lower expression of miR-30a-5p in renal carcinoma was also found [11]. However, the contribution of miR-30a-5p in the tumor progress and potential mechanisms in HCC progression has not been fully clarified. To make clear the biological meaning of miR-30a-5p expression in HCC, we investigated the miR-30a-5p expression in HCC and their paired adjacent non-cancerous hepatic tissues in 16 HCC resected samples. Also, overexpression of miR-30a-5p in HepG2 cells on cell growth and apoptosis was observed, and potential mechanisms were analyzed.
Materials and methods
Cell culture
Human HCC cell line (HepG2, SMCC-7721, Hu7, and MHCC-97H) and normal liver cell line LO2 were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). All these cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a 5 % CO2 incubator.
Tissue samples
Sixteen HCCs tissues and matched non-cancerous hepatic tissues were collected at the Department of Hepatibiliary Surgery in the Second Chongqing Medical Hospital, Chongqing, China between June 2013 and October 2014. The corresponding paraneoplastic tissues were taken at least 2 cm from the cancerous node. The tissues were immediately frozen in liquid nitrogen after surgical removal and stored at −80 °C until use. All the HCC tissues were 10 % formalin fixed, paraffin embedded, and cut into 3-μm thick slices for immunohistochemistry. All the recruited patients in this study had not a history of preoperative chemotherapy or radiotherapy. HCC diagnosis was based on the World Health Organization criteria. Liver function was assessed using the Child-Pugh scoring system. In this study, written informed consent was obtained from all patients, and the Chinese Medical Association Society of Medicine’s Ethics Committee approved all aspects of this study in accordance with the Helsinki Declaration.
Transfection of miRNA, siRNA, and cDNA
miR-30a-5p and negative control were purchased from RiboBio (Guang Zhou, China). MTDH siRNARNA and negative control were purchased from GenePharma (Shanghai, China). The transfection was performed in a final concentration of 100 nM using Lipofectamine TM 2000 (Invitrogen, Carlsbad, CA, USA) according to the instructions provided by the manufacturer. RNA transfection efficiency is approximately 70–80 %, and the overexpression of miRNA or siRNA persists for at least 48 h. For the MTDH cDNA plasmid without 3′-UTR (Addgene, Cambridge, USA) and miR-30a-5p mimics combination experiment, HepG2 and/or SMCC-7721 cells were first transfected with miR-30a-5p mimics (100 nM). Forty-eight hours later, these cells were co-transfected with MTDH cDNA plasmid (2 μg) and miR-30a-5p mimics (100 nM) for another 72 h.
Extraction of total RNA and q-PCR analysis of miR-30a-5p expression
Total RNA was extracted from cultured cells with TRIzol reagents (Life technologies, Carlsbad, CA, USA) according to the manufacturer protocol. RNA quality and concentration were detected by Nanodrop 2000 (Wilmington, DE 19810 USA). One microgram of total RNA which including miRNAs was reverse transcribed in a 15-μl reaction Prime Script RT reagent Kit (Takara, Dalian, China). And miR-30a-5p specific stem-looped primer (RiboBio, Guangzhou, China) for reverse transcription was used (made-to-order). SYBR green-based quantitative real-time PCR (qRT–PCR) analyses were performed with Applied Biosystems Step One Plus Real-Time PCR System (Life technologies, Carlsbad, CA, USA). U6 small RNA was used as an internal control for normalization and quantification of miR-30a-5p expression. The experiments were performed in triplicate. Relative expression for miR-30a-5p in HCC tissues to the matched non-cancerous liver tissues was calculated with the 2-ΔΔCT method.
Western blot analysis
Cells were collected and lysed with radio-immunoprecipitation assay buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 % (v/v) NP40, 0.1 % (w/v) SDS, 0.5 % (w/v) sodium deoxycholate) with protease inhibitors. Equal amounts of proteins (50 μg) were separated on 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). After blocking with 5 % non-fat milk, the membrane was incubated overnight at 4 °C with the primary MTDH antibody (Abcam, USA; 1:10000) or GAPDH (Cell Signal, USA; 1:1000), p-AKT (Cell Signal, USA; 1:1000) and AKT (Cell Signal, USA; 1:1000), phosphatase and tensin homolog (PTEN) (Cell Signal, USA; 1:1000), and then with horseradish peroxydase-coupled secondary antibody (Cell Signal, USA). Signal was detected with enhanced chemiluminescence (Millipore). And band signals were acquired in the linear range of the scanner and analyzed using QUANTITY ONE software (Bio-Rad, Hercules, CA, USA).
CCK-8 assay
Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) was used to measure the tumor cell proliferation. Cells were plated at a density of 5000 cells per well in 96-well plates with the complete media. At the end of transfection, 10 μl of the cell proliferation reagent WST-8 was added to each well and incubated for 2 h at 37 °C. The absorbance was measured at 450 nm with an ultraviolet spectrometer (Beckman Coulter, Brea, CA). The experiments were performed in quadruplicate and repeated in triplicate.
Colony formation assay
After transfection of mi-R30a-5p for 48 h in HepG2 and/or SMCC-7721 cells, 500 cells were plated into 6-well plates and continually cultured at 37 °C and at an atmosphere of 5 % CO2 for 10 days. The supernatants were then discarded and cells were rinsed in PBS for twice and fixed with 70 % ethanol for 15 min. The cells were stained with 0.1 % crystal violet for 10 min, and PBS was used to wash the rest of crystal violet for twice. The plates were dried at room temperature and the colony numbers containing more than 50 cells were microscopically counted. The experiments were triplicated.
Cell apoptosis assay
HepG2 and/or SMCC-7721 cells were transfected with 100 nM mi-R30a-5p mimics or negative control, and apoptosis assay was performed with Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit (Invitrogen) in 48 h after transfection according to the manufacturer’s instructions. The cell suspension (100 μl) was incubated with 5 μl of annexin V and 1 μl of propidium iodide at room temperature for 15 min. The stained cells were analyzed with fluorescent-activated cell sorting (FACS) using BD LSR II flow cytometry (BD Biosciences, San Diego, CA, USA). The percentage of early apoptotic cells located in the lower right quadrant (annexin V–FITC positive/PI negative cells), as well as late apoptotic cells located in the upper right quadrant (annexin V–FITC positive/PI positive cells) were determined.
Luciferase activity assay
The 3′-UTR of MTDH containing two putative miR-30a-5p-binding sites was amplified and cloned into pGL3 vector separately (Life technologies, Carlsbad, CA, USA). Two miR-30a-5p complementary sites with the sequence UGUUUACA in MTDH 3′-UTR were mutated, so that not complement to miR-30a-5p any more. HepG2 cells were cultured in 24-well plates and co-transfected with 100 ng of wild-type or mutated MTDH 3′-UTR constructs, or 100 nM of negative control or miR-30a-5p mimics. Luciferase activity was measured with the dual luciferase reporter assay system (Promega).
Immunohistochemical analysis
MTDH expression was detected in 3-μm thick HCC paraffin embedded slides with immunohistochemical staining method. Briefly, the slides were first dewaxed in xylol and rehydrated in 100, 95, and 85 % graded alcohol series, with antigen retrieval in 0.01 M sodium citrate solution 98 °C for 15 min. Endogenous peroxidase activity was blocked with 3 % H2O2-methanol, and normal goat serum was used to close non-specific binding. Anti-human MTDH rabbit monoclonal antibodies (EP4445, 1:100, Abcam Inc., Cambridge, CA) incubated in 4 °C for overnight. The next morning, the slides were washed three times with PBS and incubated with biotin labeled second antibody for 15 min. And DAB color-substrate solution was added for half a minute. Then, the slides were counterstained with hematoxylin for 1 min, dried in 60 °C for 2 h, dehydrated in 85, 95, and 100 % gradient ethanol, and sealed with neutral gum. The slides omitted first antibody used as negative controls and breast cancer tissues slides which had been confirmed to overexpress the MTDH protein as positive controls.
Immunohistochemical results evaluation
Semi-quantitative assessment method which combines staining intensity and the percentage of positive cells was used to evaluate MTDH dyeing results. The staining intensity was scored as 0, negative; 1, weak; 2, moderate; and 3, strong. The percentage of positive cells was scored as <10 % = 1, 10–34 % = 2, 35–70 % = 3; and >70 % = 4. The final staining score was calculated by a staining index (SI) of MTDH ranged from 0 to 12 was finally determined as the product of staining intensity and proportion of immunopositive cancer cells according to previously published criteria. A total score equal or less than 5 was considered to be of low MTDH expression, while a score greater than 5 indicated high MTDH expression. All the sections were independently assessed by two experienced pathologists, and inconsistent immunohistochemical results were reviewed again by the two pathologists and to obtain the final pathological diagnosis.
In vivo tumorigenicity
For the HCC xenograft model, 4-week-old BALB/c nude mice were housed in a pathogen-free environment and used for experiment. Medium (0.2 ml) containing 5 × 106 mi-R30a-5p mimics or negative control transfected HepG2 cells were injected subcutaneously into the left posterior flank regions of each mouse. And tumor growth was examined every week. Mice were sacrificed after tumor inoculation for 6 weeks, and the volume of each tumor were calculated. Tumor volume was calculated using the formula: L × S/2, where L is the longest tumor diameter and S is the shortest tumor diameter. All experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by our institutional ethical guidelines for animal experiments.
Statistical analysis
All the data were analyzed with SPSS 17.0 software. Measurement data were presented as means ± SD from at least three separate experiments. Two-sided Student’s t test was used to analyze the differences of HepG2 cell proliferation, colony formation number, percent of cell cycle and apoptotic rates. The miR-30a-5p-5p expression and the MTDH protein immunohistochemical scores were evaluated by Spearman’s rank correlation coefficients. Statistically significance was considered when P <0.05.
Results
Expression of miR-30a-5p was significantly downregulated in HCC cell lines and tissues
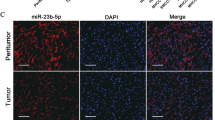
To investigate the roles of miR-30a-5p-5p in HCC progression, we first used SYBR green-based qRT-PCR assays to detect the expression level of miR-30a-5p-5p in HCC cells HepG2, SMCC-7721, Hu7, MHCC-97H, and normal liver cell line LO2. We found that miR-30a-5p expression in HCC cells was significantly downregulated compared to normal liver cell line LO2 (Fig. 1a). The relative expression of miR-30a-5p in SMCC-7721, HepG2, Hu7, and MHCC-97H were 0.43 ± 0.13, 0.32 ± 0.05, 0.40 ± 0.10, and 0.16 ± 0.07, respectively, compared to the expression level of miR-30a-5p in LO2 cell. We also determined the expression level of mature miR-30a-5p in 16 pairs of HCC tissues and matched non-cancerous liver tissues by qRT-PCR assays. And 13 (81.25 %) cases showed lower expression of miR-30a-5p in HCC tissues compared with matched non-cancerous liver tissues (Fig. 1b). Thus, for the first time, we showed that expression level of miR-30a-5p was significantly decreased in both HCC tissues and cells. These results suggested that miR-30a-5p might have important roles in HCC pathogenesis.
Decrease expression of miR-30a-5p in HCC cells and tissues. a Expression of miR-30a-5p in normal liver cell line LO2 and 4 HCC cell lines detected by stem-loop real-time PCR. b The relative expression of mature miR-30a-5p was further downregulated in HCC tissues (HCC) compared to matched non-cancerous liver tissues (NAT). U6 RNA was used as an internal control
MTDH is a direct target of miR-30a-5p in HCC
To further explore the molecular mechanism of miR-30a-5p-mediated pathological functions in HCC, we used three independently bioinformatics algorithms: TargetScan, PicTar, and miRanda to analyze the miR-30a-5p targeted genes. We focused on MTDH protein which functions to promote cancer cell proliferation, invasion, and frequently upregulated in HCC. The miRNA: mRNA alignment analysis showed that the 3′-UTR of MTDH mRNA sequence contained two putative binding sites for miR-30a-5p located at 287–294 nt and 1569–1577 nt (Fig. 2a). Then, we cloned each of the putative binding sites of miR-30a-5p individually into the pGL3 vector to validate direct targeting of MTDH by miR-30a-5p in HepG2 cells. As indicated in Fig. 2b, apparently, co-transfection of HepG2 with miR-30a-5p mimics and the pGL3-wt1 3′-UTR vector showed a significantly decreased luciferase activity compared to those cells transfected with the negative control. Decreased luciferase activity was also observed when co-transfection of HepG2 with miR-30a-5p mimics and the pGL3-wt2 3′-UTR vector. But co-transfection of HepG2 with miR-30a-5p mimics and the pGL3-MU1 3′-UTR vector or pGL3-MU2 3′-UTR vector that with several nucleotide substitutions in the core binding sites had little affection on luciferase activity.
MTDH is a direct target of miR-30a-5p. a Sequence alignment of miR-30a-5p with 3′-UTR of MTDH. There are two direct recognition sites of MTDH 3′-UTR by miR-30a-5p independently. b 520 bp sequence from the MTDH 3′-UTR containing the miR-30a-5p target sequence (position of 287–294 for UTR-wt1 or position 1569–1576 of for UTR-wt2), or identically insert with a mutated MTDH 3′-UTR constructs (UTR-mut1 or UTR-mut2) were cloned into the pGL3 vector separately (Life technologies, Carlsbad, CA, USA)
Reverse relation between miR-30a-5p and MTDH expression in liver cancer tissues
MTDH immunohistochemical scores in 16 included HCC tissues and the relative miR-30a-5p expression were analyzed with linear regression. As shown in Fig. 3a, MTDH was differently expressed in HCC tissues, and the positive expression rate of MTDH in HCC tissues was 68.75 % (11/16). MTDH expression mainly located in the cytoplasm and cell membrane, and MTDH was also found in the nucleus in HCC tissues. And Western blot results confirmed that the expression of MTDH protein in HCC tissues was significantly higher compared to matched non-cancerous hepatic tissues (Fig. 3b). The linear regression results showed that patients with high miR-30a-5p expression were MTDH negative while patients with low miR-30a-5p expression were MTDH positive. The reverse relation of MTDH and miR-30a-5p in HCC tissues was statistically significant. (Spearman rank test r = −0.5584; P = 0.0013) (Fig. 3c). All these data indicated that miR-30a-5p could regulate MTDH protein in HCC and thus affect prognosis of HCC patients.
Reverse relationship between MTDH and miR-30a-5p in HCC tissues. a Representative MTDH microphotographs of immunohistochemistry. b Western blot was used to compare MTDH protein expression in HCC tissues (HCC) and matched non-cancerous liver tissues (NAT). c Reverse relationship of MTDH immunohistochemistry scores and relative miR-30a-5p expression in HCC tissues and matched non-cancerous liver tissues
Overexpression of miR-30a-5p has the same effect as knockdown MTDH on HCC cells proliferation, colony formation, and apoptosis
HepG2 and/or SMCC-7721 cell lines overexpressing miR-30a-5p mimics were generated. CCK-8 assay was used to test the growth inhibiting effect of miR-30a-5p in HepG2 and/or SMCC-7721 cells. As shown in Fig. 4a, b, overexpression of miR-30a-5p significantly inhibited HepG2 and SMCC-7721 cell proliferation at 48, 72, and 96 h compared to the negative control group. And knockdown MTDH expression had the same growth inhibiting effect as overexpression of miR-30a-5p in HCC cells.
Restored expression of MTDH in HCC cells reverses proliferation and colon formation inhibition effect of miR-30a-5p. Overexpression of miR-30a-5p had the same proliferation and colon formation inhibiting effect as knockdown MTDH expression in HepG2 and SMMC-7721 cells. While restored MTDH expression could reverse miR-30a-5p proliferation a, b and colon formation (c) inhibiting effect in HepG2 and SMMC-7721 cells. The results were analyzed with Student’s t test (***P < 0.001) (d)
Colony formation assay also showed that overexpression of miR-30a-5p could significantly inhibit HepG2 and/or SMCC-7721 colony formation compared to the negative control group. And knockdown MTDH expression had the same colon formation inhibiting effect as overexpression of miR-30a-5p in HCC cells (Fig.4c, d). And the cell apoptosis was determined by FACS. Compared to the negative control group, significant increase in cell apoptosis was inducted by overexpression of miR-30a-5p in HepG2 and/or SMCC-7721 cells (Fig. 5a, b). Knockdown MTDH expression had the same apoptosis induction effect as overexpression of miR-30a-5p in HCC cells. These results strongly supported the potential meaning that miR-30a-5p anti-cancer activity in HCC by targeting MTDH.
miR30a-5p regulated apoptosis in HCC cells by targeting MTDH/PTEN/AKT pathway. Annexin V–FITC/PI assay for determination of apoptosis of pre-miRNA transfected and 100 nM MTDH siRNA transfected cells with flow cytometer. MiR-30a-5p overexpression had the same apoptosis induction effect as knockdown MTDH expression in HepG2 and SMMC-7721 cells. While restored expression of MTDH in HCC cells reverses apoptosis induction effect of miR-30a-5p a The apoptosis results were analyzed with Student’s t test (***P < 0.001) (b). MiR-30a-5p overexpression had the same p-AKT inhibition and PTEN activation effect as knockdown MTDH expression in HepG2 and SMMC-7721 cells while restored MTDH expression could re-activate PTEN/AKT pathway (c)
Restoration of MTDH prevents miR-30a-5p induced inhibition of HCC cell proliferation and promotion of HCC cell apoptosis
To further determine whether MTDH overexpression in HCC cells could reverse miR-30a-5p induced inhibition of HCC growth, colony formation, and apoptosis induction effect, we transfected miR-30a-5p overexpressed HCC cells with a MTDH expression vector devoid of the 3′-UTR. Briefly, HepG2 and/or SMCC-7721 stable cells (with or without miR-30a-5p overexpression) were transfected with a MTDH expression vector (MTDH in pcDNA3) or a control vector (pcDNA3). MTDH overexpression prevented miR-30a-5p induced inhibition of cell proliferation, colon formation (Fig. 4a–c), and apoptosis promotion effect (Fig. 5a, b).
miR-30a-5p inhibits PTEN/AKT signaling pathway by targeting MTDH
PTEN is tightly controlled by various non-genomic mechanisms. It is already demonstrated that MTDH could decrease PTEN expression by inhibiting PTEN transcription. Our results confirmed that miR-30a-5p could inhibit PTEN expression, and overexpression of MTDH could reverse this effect. Consistent with the documented inhibition of Akt phosphorylation by PTEN, we also observed stable overexpression of miR-30a-5p enhanced PTEN protein expression and inhibited Akt phosphorylation; these effects were reversed by overexpression of MTDH (devoid of the 3′-UTR) (Fig. 5c). These results indicated that through targeting MTDH, miR-30a-5p upregulated PTEN protein and inhibited activation of AKT. Thus, miR-30a-5p functioned as a tumor suppressor gene in HCC.
miR-30a-5p inhibits HepG2 tumor growth in vivo
Nude mice bearing miR-30a-5p or control xenografts were sacrificed after 6 weeks of inoculation. Tumors were excised and measured (Fig. 6a). The tumor volume of the mice bearing miR-30a-5p was obviously smaller than that of the mice bearing negative control tumors (Fig. 6b). And immunohistochemical results showed that overexpression of miR-30a-5p significantly inhibited PCNA expression compared to negative control tumors. Furthermore, the tumor volume (Fig. 6c) and tumor weight of the miR-30a-5p overexpression group was smaller than of the negative control tumors (Fig. 6d).
miRNA-30a-5p suppresses HepG2 tumorigenicity in vivo. HepG2 cells were transfected with NC or miR-30a-5p. At 48 h after transfection, the cells were collected. a Photographs of dissected tumors from nude mice. b PCNA expression was tested with immunohistochemistry. c Tumor growth curves measured after subcutaneous injection of HepG2 cells were transfected with NC or miR-30a-5p. The tumor volume was calculated for 6 weeks (**P < 0.01, ***P < 0.001; Student’s t test). d The tumor tissues from the animal were weighted (***P < 0.001, Student’s t test)
Discussion
It is well known that during cancer progression, multiply onco-proteins upregulated; on the contrary, multiple tumor-suppressing proteins downregulated [12]. The mechanisms of special protein expression change are complicated, and miRNAs are recognized to play an important role in epigenetic changes. By binding with target mRNA 3′-UTR, microRNAs could effectively regulate protein expression. Recently, many reports have found miRNAs dysregulation in HCC, and obviously, miRNAs participated in malignant change of HCC [13]. Some miRNAs promoting hepatic tumorigenesis were continued to be upregulated and silenced such that miRNA could upregulate tumor suppressing gene expression. For example, blocking miR-221 could prompt HCC survival [14]. On the contrary, some miRNAs functioning as tumor suppressors were continued to be downregulated. For example, downregulated miR199a in HCC promoted cancer progress, and miR199a could regulate many onco-proteins expression such as mTOR, c-Met, and HIF-1ɑ [15]. MiR148a could target Met and regulate many onco-proteins such as MSK1, Bcl-2, and IGF-IR in HCC [16]. Mir27a could inhibit drug-resistant hepatocellular carcinoma cell BEL-7402/5-fluorouracil (BEL/5-FU) proliferation through downregulating multiple drug resistance (MDR) protein expression [17]. These findings indicated that miRNAs correlated intimately with HCC malignant progress process.
In our study, we showed that miR-30a-5p expression was significantly decreased both in HCC cell lines compared to normal liver cell line LO2 and in HCC cancer tissues compared to matched non-cancerous liver tissues. Our results also showed that overexpression of miR-30a-5p could significantly inhibit HepG2 cell proliferation, colon formation, and induce cell apoptosis. These results suggested that miR-30a-5p might function as a tumor suppressor in HCC. However, with the small size of tissue samples in this study, a further large population will be necessary to testify the prognostic meaning of miR-30a-5p expression in HCC.
Although we found that miR-30a-5p could function as a tumor suppressor in HCC, the detailed mechanisms of miR-30a-5p function in HCC cells remain unclear. To understand the molecular mechanisms of miR-30a-5p-mediated pathological functions in HCC, we used bioinformatics algorithms: TargetScan, PicTar, and miRanda to analyze the target genes. We found that miR30a-5p could target MTDH 3′-UTR, and ectopic miR-30a-5p expression with special mimics could degrade its protein and mRNA. MTDH is a single-pass transmembrane protein composed of 582 amino acids with a gene located at chromosome 8q22.5 [18]. Overexpression of MTDH is frequently observed in melanoma, breast cancer, prostate cancer, and esophagus cancer and is correlated with poor clinical outcomes [19]. MTDH could inhibit cancer cell apoptosis and increasing invasiveness and metastasis. And many signaling pathways are regulated by MTDH such as nuclear factor-kappaB, Wnt/β-catenin, MAPK/ERK, and PI3K/AKT [20].
In HCC patients, it was also found that the clinical outcome was consistently poorer for the high MTDH expression group than for the low MTDH expression group [21]. High MTDH expression in HCC is positive with tumor microvascular invasion, tumor grade and stage, and high recurrence rate [22]. MTDH overexpression in HCC enhances anchorage-independent growth, in vivo tumorigenicity, and angiogenesis through the enhancement of multiple signal pathway including PI3K/Akt, MAPK, and Wnt/β-catenin pathways [23]. It is also reported that MTDH could inhibit PTEN protein expression and thus promote malignant tumor growth [24].
In our study, miR-30a-5p overexpression in HCC cells could significantly downregulate MTDH protein expression. Luciferase activity assay showed that miR-30a-5p could directly bind the sequence of MTDH 3′-UTR in HepG2 cell. And relation analysis showed a reverse relation of MTDH level and miR-30a-5p in HCC tissues. miR-30a-5p effectively increased PTEN protein expression which relates to decreased MTDH protein expression. All these results indicated that miR-30a-5p might act as a tumor suppressor by regulating MTDH expression in HCC. For the first time, our results showed that miR-30a-5p functioned as a tumor suppressor at least partly through inhibiting MTDH expression in HCC.
In conclusion, our results found miR-30a-5p expression was decreased in HCC tissues compare to non-cancerous hepatic tissues. Further in vitro cell experiments demonstrated that miR-30a-5p could inhibit liver cancer cell HepG2 proliferation, colon formation, and induced apoptosis. Mechanism analysis indicated that MTDH was regulated by miR-30a-5p in HCC. All these results suggest that miRNA-30a-5p closely related to the HCC malignant development and has potentially important value in the HCC treatment.
References
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschi T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346(6209):608–13.
Heyns M, Kovalchuk O. Non-coding RNAs including miRNAs, piRNAs, and tRNAs in human cancer. Oncotarget. 2015;6(27):23055–7.
Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread micro-RNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50.
Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–10172.
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55.
Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–83.
Hand NJ, Master ZR, EauClaire SD, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136(3):1081–90.
Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907.
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33(6):2915–23.
Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459(2):234–9.
Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74(24):7430–41.
Murakami Y, Tamori A, Itami S, Tanahashi T, Toyoda H, Tanaka M, et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade and malignancy and prognosis. BMC Cancer. 2013;13:99.
Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–16.
Callegari E, Elamin BK, D’Abundo L, Falzoni S, Donvito G, Moshiri F, et al. Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. Plos One. 2013;8(9), e73964.
Zhang JP, Zeng C, Xu L, Gong J, Fang JH, Zhuang SM. MicroRNA-148a suppresses the epithelial–mesenchymaltransition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2014;33(31):4069–76.
Zhaolin C, Ma T, Huang C, Zhang L, Lv X, Xu T, et al. MiR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/β-catenin pathway in hepatocellular carcinoma cells. Cell Signal. 2013;25(12):2693–701.
Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15(18):5615–20.
Huang Y, Li LP. Progress of cancer research on astrocyte elevated gene-1/Metadherin (Review). Oncol Lett. 2014;8(2):493–501.
Gong Z, Liu W, You N, Wang T, Wang X, Lu P, et al. Prognostic significance of metadherin overexpression in hepatitis B virus-related hepatocellular carcinoma. Oncol Rep. 2012;27(6):2073–9.
Zhu K, Dai Z, Pan Q, Wang Z, Yang GH, Yu L, et al. Metadherin promotes hepatocellular carcinoma metastasis through induction of epithelial mesenchymal transition. Clin Cancer Res. 2011;17(23):7294–302.
Ahn S, Hyeon J, Park CK. Metadherin is a prognostic predictor of hepatocellular carcinoma after curative hepatectomy. Gut and Liver. 2013;7(2):206–12.
Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119(3):465–77.
Du C, Yi X, Liu W, Han T, Liu Z, Ding Z, et al. MTDH mediates trastuzumab resistance in HER2 positive breast cancer by decreasing PTEN expression through an NF-kB dependent pathway. BMC Cancer. 2014;14:869.
Acknowledgments
This research was funded by the National Natural Science Foundation of China, projects No: 81272570, and the Health Bureau of Chongqing City, projects No: 2012-1-035.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Human and animal rights and informed consent
In this study, written informed consent was obtained from all patients, and the Chinese Medical Association Society of Medicine’s Ethics Committee approved all aspects of this study in accordance with the Helsinki Declaration. All experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by our institutional ethical guidelines for animal experiments.
Rights and permissions
About this article
Cite this article
Li, Wf., Dai, H., Ou, Q. et al. Overexpression of microRNA-30a-5p inhibits liver cancer cell proliferation and induces apoptosis by targeting MTDH/PTEN/AKT pathway. Tumor Biol. 37, 5885–5895 (2016). https://doi.org/10.1007/s13277-015-4456-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4456-1