Abstract
Early cancer metastases often occur in cervical cancer (CC) patients, resulting in poor prognosis and poor therapeutic outcome after resection of primary cancer. Hence, there is a compelling requirement for elucidating the molecular mechanisms underlying the CC cell invasiveness. Recently, the role of microRNAs (miRNAs) and pituitary tumor-transforming gene 1 (Pttg1) in the carcinogenesis of CC has been reported. Nevertheless, the relationship between miRNAs and Pttg1 remains ill-defined. Here, we showed that the levels of miR-3666 were significantly decreased and the levels of zinc finger E-box binding homeobox 1 (ZEB1) and Pttg1 were significantly increased in the CC specimens from patients, compared to the paired non-tumor tissue. Moreover, the levels of miR-3666 and ZEB1 inversely correlated. Bioinformatics analyses showed that miR-3666 targeted the 3′-untranslated region (3′-UTR) of ZEB1 messenger RNA (mRNA) to inhibit its translation, which was confirmed by luciferase reporter assay. Moreover, Pttg1 overexpression inhibited miR-3666 and subsequently increased ZEB1 and cell invasion, while Pttg1 depletion increased miR-3666 and subsequently decreased ZEB1 and cell invasion. Together, our data suggest that Pttg1 may increase CC cell metastasis, possibly through miR-3666-regulated ZEB1 levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer (CC) occurs in the cells lining the cervix at the lower part of the uterus of women. More than 90 % of all CCs are squamous cell carcinomas [1–4]. These cancers originate from cells in the exocervix and often in the transformation zone where the exocervix joins the endocervix [1–4]. Most of the other CCs are adenocarcinomas that develop from mucus-producing gland cells of the endocervix [1–4]. Long-lasting infections with certain types of human papillomavirus (HPV) account for the majority of all cases of CC, while CC can usually be cured when detected in the early stages [1–4]. However, some CC patients experience early metastases of the primary cancer, resulting in poor prognosis and poor therapeutic outcome [1–4]. Hence, it is critical to elucidate the molecular mechanisms underlying the CC cell invasiveness.
Epithelial-mesenchymal transition (EMT) plays a key in promoting cancer cell metastases. Many factors are known to trigger the EMT, including zinc finger E-box binding homeobox 1 (ZEB1), ZEB2, Snail1, Snail2, and E-cadherin [5–8]. The transcription factor ZEB1 possesses zinc finger clusters in both the N-terminal and C-terminal regions, and a homeodomain in the central region, and appears to be a key promoter of EMT initiation and a key factor for maintaining the adapted mesenchymal phenotype [9]. The zinc fingers of ZEB1 are essential for DNA binding, by specific binding to an E-box sequence CACCTG [9]. Interestingly, ZEB1 has been shown to bind to the promoter of E-cadherin to repress its expression [9].
MicroRNA (miRNA) is a class of non-coding small RNA that specifically recognizes the 3′-untranslated region (3′-UTR) of the messenger RNA (mRNA) of some genes through nucleotide pairing [10, 11]. miRNAs play a critical role in carcinogenesis of various tumors [12–14], as either tumor suppressor or enhancer. Among all miRNAs, miR-3666 has been rarely studied and reports on its involvement in cancer are lacking.
Pituitary tumor-transforming gene 1 (Pttg1) is a human homolog of yeast securin proteins, which prevent separins from promoting sister chromatid separation, and plays a critical role in mitosis [15–18]. Pttg1 has been shown to possess transforming activity in vitro and tumorigenic activity in vivo and is highly expressed in various tumors [19–21]. Interestingly, a very recent report showed that Pttg1 is negatively regulated by miR-494 in CC [22]. However, the molecular mechanisms underlying the effects of Pttg1 on CC cell invasion are undetermined.
Here, we showed that the levels of miR-3666 were significantly decreased and the levels of ZEB1 and Pttg1 were significantly increased in the CC specimens from patients, compared to the paired non-tumor tissue. Moreover, the levels of miR-3666 and ZEB1 inversely correlated. Bioinformatics analyses showed that miR-3666 targeted the 3′-UTR of ZEB1 mRNA to inhibit its translation, which was confirmed by luciferase reporter assay. Moreover, Pttg1 overexpression inhibited miR-3666 and subsequently increased ZEB1 and cell invasion, while Pttg1 depletion increased miR-3666 and subsequently decreased ZEB1 and cell invasion. Together, our data suggest that Pttg1 may increase CC cell metastasis, possibly through miR-3666-regulated ZEB1 levels.
Materials and methods
Patient tissue specimens
A total of 35 resected specimens from CC patients were collected for this study. CC specimens were compared with the paired non-tumor cervical tissue (NT) from the same patient. All specimens had been histologically and clinically diagnosed at the Department of Clinical Laboratory of the First Hospital of Jilin University from 2009 to 2014. For the use of these clinical materials for research purposes, prior patient’s consents and approval from the Institutional Research Ethics Committee were obtained.
Culture of human CC cell line
Human CC cell lines CaSki and HeLa S3 (H3) were used in the current study. CaSki is an epidermoid cervical cancer cell line prepared by R.A. Pattillo in 1977 [23], and H3 is a clonal derivative of the parent HeLa line, which was cloned in 1955 by T.T. Puck, P.I. Marcus, and S.J. Cieciura [24]. Both cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 15 % fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) in a humidified chamber with 5 % CO2 at 37 °C.
Plasmid transfection
The plasmids that contain transgene or shRNA for Pttg1 have been described before [19]. The constructs for miR-3666, miR-3666 antisense (as-miR-3666), were all purchased from GeneCopoeia (Rockville, MD, USA). Transfection was performed with 2 μg plasmids using the Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). The cells that were transfected with plasmid expressing a null sequence were used as control (null). Transfected cells expressing Pttg1, shPttg1, control scrambled sequence (scr), miR-3666, or as-miR-3666, or control null were purified by flow cytometry based on green fluorescent protein (GFP).
Western blot
The protein was extracted from the CC or paired normal tissue from the same patient, or from cultured cells, in RIPA lysis buffer (1 % NP40, 0.1 % sodium dodecyl sulfate (SDS), 100 μg/ml phenylmethylsulfonyl fluoride, 0.5 % sodium deoxycholate, in PBS) on ice. The supernatants were collected after centrifugation at 12,000×g at 4 °C for 20 min. Protein concentration was determined using a BCA protein assay kit (Bio-Rad, China), and whole lysates were mixed with 4× SDS loading buffer (125 mmol/l Tris-HCl, 4 % SDS, 20 % glycerol, 100 mmol/l DTT, and 0.2 % bromophenol blue) at a ratio of 1:3. Samples were heated at 100 °C for 5 min and were separated on SDS-polyacrylamide gels. The separated proteins were then transferred to a PVDF membrane. The membrane blots were first probed with a primary antibody. After incubation with horseradish peroxidase-conjugated second antibody, autoradiograms were prepared using the enhanced chemiluminescent system to visualize the protein antigen. The signals were recorded using X-ray film. Primary antibodies were rabbit anti-Pttg1, anti-ZEB1, and anti-α-tubulin (Cell Signaling, San Jose, CA, USA). Secondary antibody is HRP-conjugated anti-rabbit (Jackson ImmunoResearch Labs, West Grove, PA, USA). Blotting images were representative from five repeats. α-Tubulin was used as a protein loading control.
RT-qPCR
miRNA and total RNA were extracted from resected specimens from the patients or from cultured cells with miRNeasy mini kit, or RNeasy kit (Qiagen, Hilden, Germany), or Trizol (Invitrogen) for complementary DNA (cDNA) synthesis. cDNA was randomly primed from 2 μg of total RNA using the Omniscript reverse transcription kit (Qiagen). Reverse transcription quantitative PCR (RT-qPCR) was performed in duplicates with QuantiTect SYBR Green PCR Kit (Qiagen). All primers were purchased from Qiagen. Data were collected and analyzed using 2−△△Ct method for quantification of the relative mRNA expression levels. Values of genes were first normalized against α-tubulin and then compared to controls.
Luciferase reporter activity assay
Luciferase reporters were successfully constructed using molecular cloning technology. Target sequence for ZEB1 mRNA 3′-UTR clone was purchased from Creative Biogene (Shirley, NY, USA). miR-3666-modified CaSki and H3 cells were seeded in 24-well plates for 24 h, after which they were transfected with 1 μg of luciferase reporter plasmids per well. Luciferase activities were measured using the dual-luciferase reporter gene assay kit (Promega, Beijing, China), according to the manufacturer’s instructions.
Transwell cell migration assay
Cells (104) were plated into the top side of polycarbonate transwell filter coated with Matrigel in the upper chamber of the BioCoatTM Invasion Chambers (Becton-Dickinson Biosciences, Bedford, MA, USA) and incubated at 37 °C for 22 h. The cells inside the upper chamber with cotton swabs were then removed. Migratory and invasive cells on the lower membrane surface were fixed, stained with hematoxylin, and counted for 10 random 100× fields per well. Cell counts are expressed as the mean number of cells per field of view. Five independent experiments were performed, and the data are presented as mean ± standard deviation (SD).
Statistical analysis
All statistical analyses were carried out using the SPSS 17.0 statistical software package. Bivariate correlations were calculated by Spearman’s rank correlation coefficients. All values are depicted as mean ± standard deviation and are considered significant if p < 0.05. All data were statistically analyzed using one-way ANOVA with a Bonferroni correction, followed by Fisher’s exact test for comparison of two groups.
Results
Altered levels of Pttg1, ZEB1, and miR-3666 in CC specimens
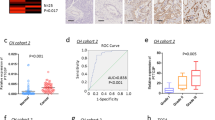
In the CC specimens from 35 patients, we detected significantly higher levels of Pttg1 (Fig. 1a) and significantly higher levels of ZEB1 (Fig. 1b), compared to the paired NT, by Western blot. Moreover, we detected significantly lower levels of miR-3666, compared to NT, by RT-qPCR (Fig. 1c). Then, we examined the relationship between miR-3666 and ZEB1 in CC specimens and detected a strong inverse correlation (Fig. 1d, ɤ = −0.65, p < 0.0001, N = 35). These data suggest the presence of a causal relationship between miR-3666 and ZEB1 in CC.
Altered levels of Pttg1, ZEB1, and miR-3666 in CC specimens (a–c). The levels of Pttg1 (a) and ZEB1 (b) in CC specimens by Western blot and the levels of miR-3666 by RT-qPCR (c), compared to the paired non-tumor cervical tissue (NT). d Correlation between miR-3666 and ZEB1. A strong inverse correlation was detected (ɤ = −0.65, p < 0.0001, N = 35). *p < 0.05. N = 35
miR-3666 targets 3′-UTR of ZEB1 mRNA to inhibit its translation in CC cells
Since our data suggest a relationship between miR-3666 and ZEB1 in CC cells, we checked whether miR-3666 may target ZEB1 mRNA. Based on bioinformatics analyses, we detected a miR-3666-binding site on the 3′-UTR of ZEB1 mRNA, ranged from 500th to 506th base site (Fig. 2a). In order to examine whether miR-3666 may regulate the translation of ZEB1 through base pairing in CC cells, we used two human CC cell lines: CaSki and H3. We either overexpressed miR-3666 or inhibited miR-3666 through transfection of the cells with a plasmid overexpressing miR-3666 or with a plasmid carrying as-miR-3666, respectively. The CaSki and H3 cells were also transfected with a plasmid carrying null sequence as a control (null). Co-expression of a GFP reporter in these plasmids allowed purification of transfected cells by flow cytometry. The overexpression or inhibition of miR-3666 in CaSki and H3 cells was confirmed by RT-qPCR (Fig. 2b). miR-3666-modified CaSki and H3 cells were then transfected with 1 μg of ZEB1-3′-UTR luciferase reporter plasmid. The luciferase activities were quantified in these cells, suggesting that miR-3666 targets 3′-UTR of ZEB1 mRNA to inhibit its translation (Fig. 2c).
miR-3666 targets 3′-UTR of ZEB1 mRNA to inhibit its translation. a Bioinformatics analyses of binding of miR-3666 to the 3′-UTR of ZEB1 mRNA. b We either overexpressed miR-3666 or inhibited miR-3666 through transfection of the cells with a plasmid overexpressing miR-3666 or with a plasmid carrying miR-3666 antisense (as-miR-3666), respectively. The CaSki and H3 cells were also transfected with a plasmid carrying null sequence as a control (null). Co-expression of a GFP reporter in these plasmids allowed purification of transfected cells by flow cytometry. The overexpression or inhibition of miR-3666 in CaSki and H3 cells was confirmed by RT-qPCR. c miR-3666-modified CaSki and H3 cells were then transfected with 1 μg of ZEB1-3′-UTR luciferase reporter plasmid. The luciferase activities were quantified. *p < 0.05. N = 5
miR-3666 decreases ZEB1 protein in CC cells without affecting ZEB1 mRNA
We found that modification of miR-3666 levels in CaSki and H3 cells did not alter the mRNA levels of ZEB1 (Fig. 3a). However, overexpression of miR-3666 significantly decreased ZEB1 protein levels in CaSki and H3 cells, while inhibition of miR-3666 significantly increased ZEB1 protein levels in CaSki and H3 cells, by Western blot (Fig. 3b). These data suggest that miR-3666 may inhibit ZEB1 protein translation in CaSki and H3 cells.
Pttg1 decreases miR-3666 levels in CC cells
Then, we examined whether the levels of miR-3666 in CC cells may be regulated by Pttg1. We either overexpressed Pttg1 or inhibited Pttg1 in CC cells through transfection of the cells with a plasmid overexpressing Pttg1 or with a plasmid carrying short hairpin interfering RNA for Pttg1 (shPttg1), respectively. The CaSki and H3 cells were also transfected with a plasmid carrying a scrambled sequence as a control (scr). Co-expression of a GFP reporter in these plasmids allowed purification of transfected cells by flow cytometry. The overexpression or inhibition of Pttg1 in CaSki and H3 cells was confirmed by RT-qPCR (Fig. 4a). We found that overexpression of Pttg1 significantly decreased miR-3666 levels in CaSki and H3 cells, while inhibition of Pttg1 significantly increased miR-3666 levels in CaSki and H3 cells, by RT-qPCR (Fig. 4b). These data suggest that Pttg1 may suppress expression of miR-3666 levels in CC cells.
miR-3666 suppression by Pttg1 results in augment in ZEB1-mediated CC cell invasion
We found that overexpression of miR-3666 or depletion of Pttg1 resulted in decreases in cell invasiveness of CaSki and H3 cells in a transwell cell migration assay, shown by quantification (Fig. 5a) and by representative images (Fig. 5b). Similarly, depletion of miR-3666 or overexpression of Pttg1 resulted in increases in cell invasiveness of CaSki and H3 cells, shown by quantification (Fig. 5a) and by representative images (Fig. 5b). Together, these data suggest that Pttg1 may promote ZEB1-mediated CC cell invasion through miR-3666 suppression (Fig. 6).
Discussion
miRNAs play critical roles in EMT-associated CC growth, invasion, and metastases [5–8]. Their targets include many EMT-related proteins, e.g., ZEB1, ZEB2, Snail1, Snail2, and E-cadherin. Previous studies have shown that these regulations are essentially for tumorigenesis of various cancers [5–8]. However, a role of miR-3666 in the carcinogenesis has not been appreciated.
A recent study nicely showed that Pttg1 could be regulated by miR-494 in CC cells [22]. The authors detected significantly higher levels of Pttg1 and significantly lower levels of miR-494 in the resected CC specimens, compared with the adjacent normal cervical tissue from the same patient [22]. Interestingly, they found that Pttg1 levels inversely correlated with miR-494 levels. In vitro, Pttg1 levels positively affected CC cell invasiveness but were negatively regulated by miR-494 levels [22]. However, they found that miR-494 levels were not affected by Pttg1 levels, suggesting that miR-494 is upstream of Pttg1. Furthermore, miR-494 inhibited Pttg1 expression in CC cells, through directly binding and inhibition on 3′-UTR of Pttg1 mRNA [22]. This study is the first report for the regulation of oncogene Pttg1 in CC, whereas it left the downstream signaling of Pttg1 on CC cancer cell invasion unstudied.
In the current study, we addressed this remaining question, and we found that miR-3666 appeared to be a downstream target of Pttg1, and most importantly, miR-3666 was one potent suppressor of ZEB1 translation. We have analyzed all ZEB1-targeting miRNAs, including miR-3666, miR-429, miR-548, miR-23, miR-130, miR-301, miR-454, miR-21, miR-4295, miR-3666, miR-96, miR-507, and miR-1271, and only miR-3666 levels appeared to be regulated by Pttg1. Using two typical CC cell lines that represent different degrees of malignancy, we obtained similar results that demonstrate a model in which Pttg1 promotes ZEB1-mediated CC cell invasion through miR-3666 suppression.
Of note, we did not examine other EMT-associated genes that may be also regulated by Pttg1. Since those genes share some similar sequences that could be targeted by the same miRNA, it is possible that miR-3666 may have other target genes that also regulate EMT-associated cancer invasiveness. On the other hand, it may be possible that some other Pttg1-targeting miRNAs have a similar effect on EMT-associated factors other than ZEB1, to coordinate the cancer cell invasion and metastases with miR-3666/ZEB1 regulatory axis. Further experiments may be applied to approve this hypothesis.
To summarize, here we propose a model in which Pttg1 is not only regulated by miRNA in CC cells but also control the ZEB1-mediated cell invasion through miRNA. Hence, it is noteworthy that miRNAs may play a previously unappreciated role in the regulation of CC cell metastasis.
References
Kanayama T, Mabuchi S, Fujita M, Kimura T. Calcaneal metastasis in uterine cervical cancer: a case report and a review of the literature. Eur J Gynaecol Oncol. 2012;33:524–5.
Setoodeh R, Hakam A, Shan Y. Cerebral metastasis of cervical cancer, report of two cases and review of the literature. Int J Clin Exp Pathol. 2012;5:710–4.
Peters P, Bandi H, Efendy J, Perez-Smith A, Olson S. Rapid growth of cervical cancer metastasis in the brain. J Clin Neurosci. 2010;17:1211–2.
Chen B, Zhang C, Dong P, Guo Y, Mu N. Molecular regulation of cervical cancer growth and invasion by VEGFA. Tumour Biol. 2014;35:11587–93.
Hu Q, Tong S, Zhao X, Ding W, Gou Y, Xu K, et al. Periostin mediates TGF-beta-induced epithelial mesenchymal transition in prostate cancer cells. Cell Physiol Biochem. 2015;36:799–809.
Sa Y, Li C, Li H, Guo H. Timp-1 induces alpha-smooth muscle actin in fibroblasts to promote urethral scar formation. Cell Physiol Biochem. 2015;35:2233–43.
Lan A, Qi Y, Du J. Akt2 mediates TGF-beta1-induced epithelial to mesenchymal transition by deactivating GSK3beta/snail signaling pathway in renal tubular epithelial cells. Cell Physiol Biochem. 2014;34:368–82.
Teng Y, Zhao L, Zhang Y, Chen W, Li X. Id-1, a protein repressed by mir-29b, facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human ovarian cancer cells. Cell Physiol Biochem. 2014;33:717–30.
Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–66.
Di Leva G, Croce CM. MiRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11.
Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–9.
Mei Q, Li F, Quan H, Liu Y, Xu H. Busulfan inhibits growth of human osteosarcoma through miR-200 family microRNAs in vitro and in vivo. Cancer Sci. 2014;105:755–62.
Wang F, Xiao W, Sun J, Han D, Zhu Y. MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via suppressing akt phosphorylation in glioblastoma. Tumour Biol. 2014;35:8653–8.
Liu G, Jiang C, Li D, Wang R, Wang W. MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in gastric cancer. Tumour Biol. 2014;35:9801–6.
Yu R, Ren SG, Horwitz GA, Wang Z, Melmed S. Pituitary tumor transforming gene (PTTG) regulates placental JEG-3 cell division and survival: evidence from live cell imaging. Mol Endocrinol. 2000;14:1137–46.
Caporali S, Alvino E, Levati L, Esposito AI, Ciomei M, Brasca MG, et al. Down-regulation of the pttg1 proto-oncogene contributes to the melanoma suppressive effects of the cyclin-dependent kinase inhibitor pha-848125. Biochem Pharmacol. 2012;84:598–611.
Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–86.
Tfelt-Hansen J, Kanuparthi D, Chattopadhyay N. The emerging role of pituitary tumor transforming gene in tumorigenesis. Clin Med Res. 2006;4:130–7.
Huang S, Liao Q, Li L, Xin D. Pttg1 inhibits SMAD3 in prostate cancer cells to promote their proliferation. Tumour Biol. 2014;35:6265–70.
Zhang G, Zhao Q, Yu S, Lin R, Yi X. Pttg1 inhibits TGFbeta signaling in breast cancer cells to promote their growth. Tumour Biol. 2015;36:199–203.
Lin YH, Tian Y, Wang JS, Jiang YG, Luo Y, Chen YT. Pituitary tumor-transforming gene 1 regulates invasion of prostate cancer cells through mmp13. Tumour Biol. 2015. doi:10.1007/s13277-015-3796-1.
Chen B, Hou Z, Li C, Tong Y. MiRNA-494 inhibits metastasis of cervical cancer through Pttg1. Tumour Biol. 2015. doi:10.1007/s13277-015-3440-0.
Pattillo RA, Hussa RO, Story MT, Ruckert AC, Shalaby MR, Mattingly RF. Tumor antigen and human chorionic gonadotropin in caski cells: a new epidermoid cervical cancer cell line. Science. 1977;196:1456–8.
Puck TT, Marcus PI, Cieciura SJ. Clonal growth of mammalian cells in vitro; growth characteristics of colonies from single hela cells with and without a feeder layer. J Exp Med. 1956;103:273–83.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, L., Han, LY., Yu, M. et al. Pituitary tumor-transforming gene 1 enhances metastases of cervical cancer cells through miR-3666-regulated ZEB1. Tumor Biol. 37, 15567–15573 (2016). https://doi.org/10.1007/s13277-015-4047-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4047-1