Abstract
SMARCA5 partners with RSF-1 to compose the RSF complex, which belongs to the ISWI family of chromatin remodelers. Recent studies referred that SMARCA5 was overexpressed in some malignant tumors. However, expression pattern and biological roles of SMARCA5 in breast cancer have not been examined. In the present study, we found that SMARCA5 was overexpressed in breast cancer specimens by immunohistochemistry. Significant association was observed between SMARCA5 overexpression and TNM stage (p = 0.0199), tumor size (p = 0.0066), high proliferation index (p = 0.0366), and poor overall survival (p = 0.0141). SMARCA5 overexpression also correlated with Rsf-1 expression levels (p = 0.0120). Furthermore, colony formation assay and Matrigel invasion assay showed that knockdown of SMARCA5 expression in MDA-MB-231 and MDA-MB-435s cell lines with high endogenous expression decreased cell proliferation and cell invasion. Flow cytometry showed knockdown of SMARCA5-arrested cell cycle. Further analysis of cell cycle and invasion-related molecules showed that SMARCA5 downregulated cyclin A, MMP2 expression and upregulated p21 expression. In conclusion, our study demonstrated that SMARCA5 was overexpressed in human breast cancers and correlated with poor prognosis. SMARCA5 contributes to breast cancer cell proliferation and invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most commonly diagnosed cancer among women and the second most diagnosed type of cancer overall in the world [1, 2]. Although rapid development and great progress in prevention, surgical resection, and adjuvant chemoradiotherapy have been made in recent years, there are still some limitations in increasing longevity observed in oncology patients, especially due to breast cancer population [3–6]. Consequently, exploring new biomarkers to predict tumor progressions and potential target therapies is important [7–10].
The modification of chromatin structure is an important regulatory mechanism for many processes such as DNA replication, transcription and DNA repair, by factors involved in chromatin assembly or remodeling which mediate the inter-nucleosomal spacing of chromatin. The imitation switch (ISWI) in Drosophila belongs to the chromatin remodeling complexes, which are characterized by its DNA-dependent ATPase catalytic activity [11]. The human ISWI homologue SMARCA5 (also known as human sucrose nonfermenting protein 2 homologue), acting as ATPase, has been found in several structurally and functionally different remodeling complexes, such as CHRAC, ACF, WICH, NoRC, and RSF, which all display nucleosome remodeling activity in vitro [12–14].
The chromosome location that determined for the SMARCA5 gene, 4q31, is within a region where loss of heterozygosity (LOH) is frequently observed in hepatocellular carcinomas [15, 16]. Studies have showed that SMARCA5 is prevalent during the proliferative stage of neural development [17]. SMARCA5-null homozygous mice die early, SMARCA5-null cells implicate proliferation arrest and apoptosis of both inner cell mass and trophoblast-like structures, and SMARCA5 heterozygous mice exhibit a postnatal growth delay. In addition, SMARCA5 knock down in hematopoietic progenitors inhibited cytokine-induced proliferation in vitro [18, 19]. Recent studies referred that SMARCA5 was overexpressed in malignant tumors, such as gastric cancer, ovarian cancer, prostate cancer, and acute leukemia [20–23]. However, it remains unclear whether SMARCA5 is related with breast cancer and, if so, what roles does SMARCA5 play in tumor generation and development.
In this study, we evaluated the expression pattern of SMARCA5 in breast cancer and correlated it with multiple clinical pathological factors, including patients’ survival. In addition, SMARCA5-specific small interfering RNA (siRNA) was transfected into two breast cancer cell lines to investigate its effects on the biological behavior and to elucidate its possible mechanisms in malignant cell progression.
Materials and methods
Specimens and immunohistochemistry
The study protocol was approved by the institutional reviewer board of China Medical University. All participants provided their written informed consent, and the investigation was conducted according to the principles expressed in the Declaration of Helsinki. Tumor specimens were obtained from 110 patients diagnosed with invasive ductal carcinoma (IDC) who underwent resection in the First Affiliated Hospital of China Medical University between 2005 and 2008.
Surgically excised tumor specimens were fixed with 10 % neutral formalin and embedded in paraffin, and 4-μm-thick sections were prepared. Immunostaining was performed using the avidin–biotin–peroxidase complex method (Ultrasensitive, MaiXin, Fuzhou, China). The sections were deparaffinized in xylene, rehydrated with graded alcohol, and then boiled in 0.01 M citrate buffer (pH 6.0) for 2 min in an autoclave. Hydrogen peroxide (0.3 %) was applied to block endogenous peroxide activity, and the sections were incubated with normal goat serum to reduce nonspecific binding. Tissue sections were incubated with SMARCA5 mouse monoclonal antibody (1:1000 dilution; Milipore), RSF-1(1:1000 dilution; Milipore), and Ki-67 antibody (MaiXin, Fuzhou, China). Mouse immunoglobulin (at the same concentration of the antigen-specific antibody was used as a negative control. Staining was performed at 4 °C overnight. Biotinylated goat anti-rabbit serum IgG was used as a secondary antibody. After washing, the sections were incubated with streptavidin–biotin-conjugated complex with horseradish peroxidase, and the peroxidase reaction was developed with 3,3-diaminobenzidine tetrahydrochloride. Counterstaining with hematoxylin was performed, and the sections were dehydrated in ethanol before mounting.
Two independent blinded investigators examined all tumor slides randomly. Positive nuclear/cytoplasmic staining was considered positive. Immunostaining of SMARCA5 was scored on a semiquantitative scale by evaluating staining intensity and percentage. We calculated the percentage of positively stained cells. The staining intensity was categorized as follows: 0, negative; 1, moderate; and 2, strong. The staining percentage of tumor specimens was scored as 0, 0 %; 1, 1–5 %; 2, 6–25 %; 3, 26–75 %; and 4, 76–100 %. The scores of each tumor sample were multiplied to give a final score of 0 to 8, and the tumor samples with a final score of 4–8 were finally determined as SMARCA5 overexpression.
Cell culture and transfection
MDA-MB-435s and MDA-MB-231 cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10 % fetal calf serum (Invitrogen), 100 IU/ml penicillin (Sigma-Aldrich), and 100 μg/ml streptomycin (Sigma-Aldrich). Cells were grown on sterilized culture dishes and were passaged every 2 days with 0.25 % trypsin (Invitrogen).
DharmaFECT1 reagent was used for siRNA transfection (Qiagen, Chicago, IL, USA) according to manufacturer’s instructions. The protein level was assessed 48 h later by western blotting. SMARCA5-siRNA was bought from Santa Cruz.
Western blot analysis
Total proteins from cells were extracted in lysis buffer (Pierce, Rockford, IL) and quantified using the Bradford method. Samples of 50 μg of protein were separated by SDS-PAGE. Samples were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) and incubated overnight at 4 °C with antibody against SMARCA5 (1:1000, milipore), cyclinA, cylinD1, cyclinB1, p21 (1:1000, Cell signaling, USA), MMP2, MMP9 (1:1000; San Diego, CA), and a mouse monoclonal antibody against beta-actin (1:1000; Santa Cruz). After incubation with peroxidase-coupled anti-mouse/rabbit IgG (Santa Cruz) at 37 °C for 2 h, bound proteins were visualized using ECL (Pierce) and detected using a BioImaging System (UVP Inc., Upland, CA, USA).
Quantitative real-time PCR (SYBR green method)
Total RNA was extracted from cells using Trizol (Qiagen). Reverse transcription of 1 μg of RNA was done using the high-capacity cDNA RT kit (Applied Biosystems) following the manufacturer’s instructions.
Quantitative real-time PCR was done using SYBR Green PCR master mix (Applied Biosystems) in a total volume of 20 μl on 7900HT fast Real-Time PCR system (Applied Biosystems) as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. The sequences of the primer pairs are as follows:
-
CyclinA forward, 5′-GCAGAGGCCGAAGACGAGA-3′,
-
CyclinA reverse, 5′-TCCAAGGAGGAACGGTGACA3-′.
-
p21 forward, 5′-CCTCATCCCGTGTTCTCCTTT-3′
-
p21 reverse, 5′-GTACCACCCAGCGGACAAGT-3′.
-
MMP2 forward, 5′-TGTGTTCTTTGCAGGGAATGAAT-3′,
-
MMP2 reverse, 5′-TGTCTTCTTGTTTTTGCTCCAGTTA-3′.
-
β-actin forward, 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′,
-
β-actin reverse, 5′-CACCTTCTACAATGAGCTGCGTGTG-3′.
A dissociation procedure was performed to generate a melting curve for confirmation of amplification specificity. β-actin was used as the reference gene. The relative levels of gene expression were represented as ΔCt = Ct gene − Ct reference, and the fold change of gene expression was calculated by the 2-ΔΔCt Method. Experiments were repeated in triplicate.
Colony formation assay
For colony formation assay, cells were transfected for 48 h and then plated into three 6-cm cell culture dishes (1000 per dish) and incubated for 12 days. Plates were washed with phosphate-buffered saline (PBS) and stained with Giemsa stain. The number of colonies with more than 50 cells was counted. The colonies were manually counted using a microscope.
Flow cytometry for cell cycle analysis
Cells (500,000) were seeded into 6 cm tissue culture dishes. Twelve hours later, cells were transfected with SMARCA5-specific siRNA plasmid or control. Forty-eight hours after transfection, cells were harvested, fixed in 1 % paraformaldehyde, washed with PBS, and stained with 5 mg/ml propidium iodide in PBS supplemented with RNase A (Roche, Indianapolis, IN) for 30 min at room temperature. Data were collected using BD systems. One-parameter histogram was plotted according to the distribution of nuclear DNA content in each cell detected by flow cytometer. Cells in each individual phase of the cell cycle were determined based on their DNA ploidy profile.
Matrigel invasion assay
Cell invasion assay was performed using a 24-well Transwell chamber with a pore size of 8 μm (Costar, Cambridge, MA). The inserts were coated with 20 μl Matrigel (1:3 dilution; BD Bioscience, San Jose, CA, USA). Forty-eight hours after the transfection, cells were trypsinized and 3 × 105 cells in 100 μl of serum-free medium were transferred to the upper Matrigel chamber and incubated for 16 h. Medium supplemented with 15 % FBS was added to the lower chamber. After incubation, the noninvaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4 % paraformaldehyde and stained with hematoxylin.
Statistical analysis
SPSS version 11.5 for Windows was used for all statistical analyses. Chi-square test was used to evaluate possible correlations between SMARCA5 overexpression and clinicopathologic factors. Student’s t test was used to compare data between control and transfected cells. All p values were based on the two-sided statistical analysis, and p < 0.05 was considered to be statistically significant in difference.
Results
Expression and localization of SMARCA5 in breast cancer specimens
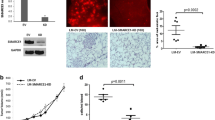
We investigated the SMARCA5 protein expression in a panel of 110 primary breast cancer samples as well as paired autologous normal mammary gland by immunohistochemistry. We used immunoglobulin as negative control which showed negative staining. The immunohistochemical results showed that positive staining of SMARCA5 protein was mainly localized in the nuclear compartment in breast cancer specimens. SMARCA5 was showed negative expression in normal breast tissue (Fig. 1a). Negative/weak nuclear expression was found in ductal carcinoma in situ (DCIS) (Fig. 1b). We found overexpression of SMARCA5 in 60 out of 110 (54.5 %) IDC specimens. Weak/negative SMARCA5 staining was considered as normal expression (Fig. 1c). Strong nuclear staining with a final score of ≥4 was considered a SMARCA5 overexpression (Fig. 1d).
Expression pattern of SMARCA5 in breast cancers and its correlation with survival. a Weak staining of SMARCA5 in normal breast tissue. b Negative/weak nuclear staining of SMARCA5 in ductal carcinoma in situ (DCIS). c Weak/negative SMARCA5 staining in invasive ductal carcinoma (IDC). d Strong nuclear staining in IDC. e The overall survival and relapse-free survival analyses showed that patients with high SMARCA5 expression had lower RFS and 5-year OS than those with low SMARCA5 expression (both p < 0.05)
The clinical significance of SMARCA5 protein expression in breast cancer
To evaluate the role of SMARCA5 protein in breast cancer progression, the correlation between SMARCA5 expression and clinical features of patients and biological markers was analyzed. As summarized in Table 1, no statistical difference was found between the SMARCA5 overexpression and age (p = 0.4785) and lymph node metastasis (p = 0.2362). However, there was significant correlation between SMARCA5 overexpression and advanced TNM stage (p = 0.0199), which was higher in advanced stage (stages II–III) breast cancers than in early stage (stage I) cases. In addition, overexpression of SMARCA5 showed a correlation with the tumor size (p = 0.0066) (Table 1).
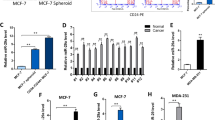
Furthermore, no significant correlation was observed between SMARCA5 and ER, PR, and Her2 status. However, we were able to show that the positive rate of SMARCA5 protein was higher in cancer cases with high Ki-67 expression (Fig. 2f) compared with those with low Ki-67 expression. We also detected RSF-1 expression in breast cancer tissues. The results showed that 71 cases scored positive for RSF-1 overexpression, which was localized in nuclear compartments of tumor cells (Fig. 2b). Cases that had high levels of SMARCA5 expression tended to have high RSF-1 labeling (p < 0.001) (Table 1).
Correlation of SMARCA5 expression with RSF-1 and Ki-67. a, b Cases that had a high level of SMARCA5 expression showed a high RSF-1 expression. c, d Cases showed negative staining of SMARCA5 and RSF-1 protein. e, f Cases that had a high level of SMARCA5 expression showed a high Ki-67 expression. g, h Cases showed negative staining of SMARCA5 and Ki-67 protein
We followed up on 110 patients and divided them into two groups according to SMARCA5 expression. Using Kaplan–Meier survival analysis, we found that patients with low expression of SMARCA5 had a statistically significantly longer survival rate than those with high expression of SMARCA5. To further substantiate the importance of high SMARCA5 expression in breast cancer progression, we analyzed relapse-free survival (RFS) and 5-year overall survival (OS) of 110 breast cancer cases and found that patients with high SMARCA5 expression had lower RFS and 5-year OS than those with low SMARCA5 expression (both p < 0.05) (Fig. 1e). These data suggested that SMARCA5 could be a valuable prognostic factor in breast cancer.
Knockdown of SMARCA5 suppressed breast cancer cells proliferation by regulation of cyclin A and p21 protein
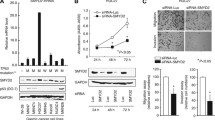
Expression of SMARCA5 was analyzed in six breast cancer cell lines by western blot. We found high levels of SMARCA5 in MDA-MB-435s and MDA-MB-231 cell lines (Fig. 3a). In order to explore the biological function of SMARCA5 in breast cancer, we employed siRNA to deplete SMARCA5 expression. Western blot confirmed the SMARCA5 knockdown efficiency in both of the two cell lines (Fig. 3a). Colony formation assay was employed to characterize the role of SMARCA5 on cell proliferation. We found that treatment of SMARCA5-specific siRNA resulted in a decrease in colony formation ability compared with control siRNA (231 Con vs siRNA, 873 ± 46 vs 357 ± 39; 435s Con vs siRNA, 423 ± 25 vs 305 ± 16) (p < 0.05) (Fig. 3c). Cell cycle analysis demonstrated that for the cells transfected with SMARCA5-specific siRNA, the percentage of cells in S phase was decreased compared with that for the control groups (Fig. 3d). These findings suggest that downregulation of SMARCA5 can inhibit cell proliferation by arresting cell cycle progression in breast cancer cells. To investigate the mechanism by which SMARCA5 affected the cell cycle, we examined the effect of SMARCA5 depletion on cyclin D1, cyclin A, cyclin B1, and p21 expression levels. As shown in Fig. 4, analysis revealed that downregulation of SMARCA5 decreased the protein and RNA levels of cyclin A and upregulated p21 expression. These results suggest that overexpression of SMARCA5 promotes lung cancer cell growth, likely mediated by regulation of cyclin A and p21.
SMARCA5 siRNA knockdown inhibited breast cancer cell proliferation and invasion. a Endogenous expression of SMARCA5 and RSF-1 was examined in the normal breast epithelial cell and six breast cancer cell lines by western blot. SMARCA5 siRNA treatment markedly decreases its levels in 231 and 435s cells in comparison with cells transfected with negative control. b Colony formation assay was performed in cells transfected with SMARCA5 siRNA and negative control. A decrease in colony formation was observed in the groups with SMARCA5 siRNA compared with the control. c MTT assay showed marked decrease of penetrated cells in 231 and 435s cells transfected with SMARCA5 siRNA. d Cell cycle analysis showed that SMARCA5 siRNA transfection decreased S phase percentage and increased G1 phase percentage
SMARCA5 depletion in breast cancer cells inhibited invasion with MMP2 downregulation
The role of SMARCA5 on breast cell invasion was determined by Matrigel invasion assay. It showed that a significant decrease of cell invasion (231, 52.5 %; 435s, 49.3 %) was observed in cells with SMARCA5 knockdown compared with scramble controls (Fig. 3b), demonstrating that SMARCA5 could regulate invading ability in breast cancer cells. To explore the potential mechanism of SMARCA5 on the invading ability of cancer cells, we used real-time PCR to analyze expression change of MMP family which is related to cancer cell invasion. We observed a remarkable decrease in MMP2 expression at both mRNA and protein levels after siRNA treatment while MMP9 expression was not changed (Fig. 4). These results suggest that SMARCA5 could modulate MMP2 transcription, thus regulating cell invasion.
Discussion
Given the crucial roles of chromatin remodeling factors in biology, it comes as no surprise that defects in, or aberrant expression of, chromatin remodeling proteins are associated with various developmental disorders and cancer [24]. Earlier studies reported SMARCA5 expressed higher in gastric cancer samples than in normal mucosa. Recently, SMARCA5 was verified to directly interact and co-express with RSF-1 in ovarian cancer [23]. However, SMARCA5 expression pattern and biological roles in breast cancer have not been examined. In this study, we performed immunohistochemistry to detect the staining of SMARCA5 protein in breast cancer tissue. We found that IHC staining of SMARCA5 was mainly localized in the nuclear compartment of cancer cells, which was in accord with previous reports [22]. The expression level of SMARCA5 in invasive ductal carcinoma was higher than that in normal breast tissues. It was reported that SMARCA5 partnered with RSF-1to compose the RSF complex, which belongs to the ISWI family of chromatin remodelers [25]. RSF have been reported to participate in several biological processes and may serve as a survival signal. Previous studies showed that SMARCA5 promoted the function of RSF-1in tumorigenesis [23]. In our study, we also demonstrated that high expression of SMARCA5 correlated with RSF-1 overexpression in breast cancer tissues. Both SMARCA5 and RSF-1 staining were localized in nucleus. According to the findings above, we speculate that SMARCA5 protein level also might be used as an early diagnostic indicator of tumour.
Despite previous studies demonstrated SMARCA5 protein was overexpressed in the tumor [20–23], there have been few reports of SMARCA5 protein expression-based outcomes in tumor patients. In our study, we confirmed the clinical characters of SMARCA5 by analyzing the association of its expression levels with clinical pathological parameters. As a result, there were markedly correlations between SMARCA5 upregulation and tumor stage, and nodal metastasis, suggesting SMARCA5 might associate with breast cancer progression. Ki-67 is a nuclear antigen present throughout the whole cell cycle (G1, S, G2, and M phases) except during the rest phase (G0 phase) or in the early G1 phase [26]. Ki-67 nuclear staining has been related to biological aggressiveness and prognosis in several cancers, and a greater proliferative index indicates more aggressive behavior of the neoplasia [27, 28]. Therefore, we examined the expression of Ki-67 to investigate the relationship between SMARCA5 positivity and proliferative activity of cancer tissues by immunohistochemistry. The result showed that cases that had a high level of SMARCA5 expression showed a high Ki-67 expression. More importantly, we found that the patients with negative expression of SMARCA5 protein had better prognosis than those with positive expression. There was also a significant difference in relapse survival rate (p = 0.0141), suggesting that SMARCA5 may be a prognostic indicator for breast cancer patients.
Evidences may suggest SMARCA5 a potential role in carcinogenesis. However, the potential mechanism by which SMARCA5 regulates breast cancer cell proliferation and invasion is unclear at the present time. By immunohistochemistry, we were able to demonstrate that expression of SMARCA5 protein correlated with the proliferation index, suggesting its potential role in promoting cell proliferation. In support of the immunohistochemical results, we blocked SMARCA5 function by using siRNA treatment in the MDA-MB-231 and MDA-MB-435s cell line which has high endogenous SMARCA5 expression. We found that SMARCA5 depletion caused an obvious decrease in the proliferation rate and invading ability. In addition, cell cycle analysis showed that the percentage of cells in S phase was decreased in the cells with knockdownof SMARCA5 compared with control cells, suggesting that SMARCA5 might arrest G1 to S phase transition to promote cell growth. We thereafter examined a series of cell cycle-related factors in the cells SMARCA5-specific siRNA and noted apparently inhibit expression of cyclinA and enhanced expression of p21. These results correlated well with the cell cycle alternation after SMARCA5 knockdown. After examining in the SMARCA5 depletion cell lines the expression of MMP family, which was closely related with cells migration, we found the protein and RNA levels of MMP2 was decreased. The data in the present study suggested that SMARCA5 impact cell proliferation via regulated expression of cyclin A and p21 and migration with MMP2 expression level in breast cancer cell lines, but the exact machinery by which SMARCA5 modulates cyclin and MMPs needs to be further elucidated in future studies.
In conclusion, this study with expression level and biological effect of SMARCA5 in breast cancer demonstrated that it existed high expressed in tumor tissues and correlated with tumor size, p-TNM stage, tumor cell proliferation, invasion, and poor survival of the patients. Given these findings, SMARCA5 might be a potential therapeutic target in the treatment of breast cancer, and future studies are warranted to investigate these possibilities.
References
Chavarri-Guerra Y et al. Breast cancer in Mexico: a growing challenge to health and the health system. Lancet Oncol. 2012;13(8):e335–43.
Jemal A et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Siegel R et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–41.
Ambrogi F et al. Molecular subtyping of breast cancer from traditional tumor marker profiles using parallel clustering methods. Clin Cancer Res. 2006;12(3 Pt 1):781–90.
Charpentier M, Martin S. Interplay of stem cell characteristics, EMT, and microtentacles in circulating breast tumor cells. Cancers (Basel). 2013;5(4):1545–65.
Cuppone F et al. Magnitude of risks and benefits of the addition of bevacizumab to chemotherapy for advanced breast cancer patients: meta-regression analysis of randomized trials. J Exp Clin Cancer Res. 2011;30:54.
Ahn S et al. The prognostic significance of tumor-associated stroma in invasive breast carcinoma. Tumour Biol. 2012;33(5):1573–80.
Kabbage M et al. Expression of the molecular chaperone alpha B-crystallin in infiltrating ductal breast carcinomas and the significance thereof: an immunohistochemical and proteomics-based strategy. Tumour Biol. 2012;33(6):2279–88.
Elfagieh M et al. Serum tumour markers as a diagnostic and prognostic tool in Libyan breast cancer. Tumour Biol. 2012;33(6):2371–7.
Kurbel S. In search of triple-negative DCIS: tumor-type dependent model of breast cancer progression from DCIS to the invasive cancer. Tumour Biol. 2013;34(1):1–7.
Kokavec J et al. Chromatin remodeling and SWI/SNF2 factors in human disease. Front Biosci. 2008;13:6126–34.
LeRoy G et al. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282(5395):1900–4.
Poot RA et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19(13):3377–87.
Strohner R et al. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–900.
Aihara T et al. Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet Cell Genet. 1998;81(3–4):191–3.
Buetow KH et al. Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1989;86(22):8852–6.
Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001;77(4):1145–56.
Chong S et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39(5):614–22.
Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100(24):14097–102.
Gigek CO et al. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29(2):162–6.
Stopka T et al. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14(7):1247–52.
Reis ST et al. The role of micro RNAs let7c, 100 and 218 expression and their target RAS, C-MYC, BUB1, RB, SMARCA5, LAMB3 and Ki-67 in prostate cancer. Clinics (Sao Paulo). 2013;68(5):652–7.
Sheu JJ et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68(11):4050–7.
Sheu JJ et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285(49):38260–9.
Loyola A et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23(19):6759–68.
Gerdes J et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–5.
Brown DC, Gatter KC. Monoclonal antibody Ki-67: its use in histopathology. Histopathology. 1990;17(6):489–503.
Cher ML et al. Cellular proliferation in prostatic adenocarcinoma as assessed by bromodeoxyuridine uptake and Ki-67 and PCNA expression. Prostate. 1995;26(2):87–93.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jin, Q., Mao, X., Li, B. et al. Overexpression of SMARCA5 correlates with cell proliferation and migration in breast cancer. Tumor Biol. 36, 1895–1902 (2015). https://doi.org/10.1007/s13277-014-2791-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2791-2