Abstract
Kaposi’s sarcoma (KS) is a multicentric angioproliferative tumor of mesenchymal origin. The molecular and biologic aspects of KS are not fully understood. MicroRNAs are non-protein-coding small RNAs in the size range 19–25 nucleotides (nt) that play important roles in biological processes, including cellular differentiation, proliferation, and death. We performed a miRNA microarray analysis by detecting six paired KS and matched adjacent healthy tissues using the 7th generation of miRCURYTM LNA Array (v.18.0) (Exiqon) containing 3100 capture probes. We selected 10 significant differentially expressed miRNAs, which were confirmed by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in 18 paired KS and matched adjacent healthy tissue specimens. We also investigated the associations between clinical features and miRNA expression. Among the 3100 human miRNA probes in the microarrays, we identified 170 differentially expressed miRNAs (69 upregulated and 101 downregulated miRNAs) in KS versus adjacent healthy tissues. Among the most significantly upregulated miRNAs were miR-126-3p, miR-199a-3p, miR-16-5p, and the 13 KSHV-related miRNAs. The most significantly downregulated miRNAs included miR-125b-1-3p and miR-1183. Eight upregulated miRNAs, miR-181b-5p, miR-199a-3p, miR-15a-5p, miR-126-3p, miR-1297, kshv-miR-k12-12-3p, kshv-miR-k12-1-5p, and miR-16-5p, and two downregulated miRNAs, miR-125b-1-3p and miR-1183, were confirmed by qRT-PCR in 18 paired KS samples. The qRT-PCR results for 10 miRNAs were consistent with our microarray results. The miR-125b-1-3p and miR-16-5p had statistically significant associations with HHV-8 and HIV infections in KS. The results of miRNA profiling showed that KS appears to have unique expression patterns when compared with paired adjacent healthy tissues, suggesting that deregulation of miRNAs plays an important role in the progression of KS. These differentially expressed miRNAs may provide novel diagnostic and prognostic tools.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are small (19–25 nucleotides; nt), single stranded non-protein-coding RNAs that play a role in many biological processes [1]. MiRNAs regulate target mRNA expression by binding to complementary sequences of their target mRNA genes in the 3′-untranslated regions [2]. The functions of miRNAs include signal transduction, cell differentiation, cell proliferation, and apoptosis [3–6]. The role of miRNAs in cancer is important, and they include oncogenic/tumor suppressor miRNAs, and those involved in regulation of proliferation, angiogenesis, etc. MiRNAs generally have specific expression profiles in cancer [7], and therefore miRNAs can serve as markers for diagnosis and the response to therapy.
Kaposi’s sarcoma (KS) is a multicentric angioproliferative tumor of mesenchymal origin [8, 9]. The disease was first described in 1872 by Moritz Kaposi, and it mostly affects elderly men of Italian, Jewish, or Mediterranean origin [10, 11]. Xinjiang is a multiethnic gathering place in China with a population that includes Uyghur (45.7 %), Han (39.7 %), Kazakh (7 %), and Hui (4.5 %) ethnic groups [12]. More than 90 % of KS cases in China have occurred in Xinjiang [13–15]. A recent report described a clinical study of 105 KS cases between January 1997 and April 2013 in Xinjiang, China. The cases comprised 77 classic KS (CKS) and 28 AIDS-associated KS (AIDS-KS). Among them, 80 patients were Uyghur men and 11 patients were Uyghur women; the disease seldom occurred in Kazakh (eight men and three women), Hui (1 man), Xibo (1 man), and Han (1man) ethnic groups. The prevalence of human herpesvirus 8 (HHV-8) was very high (98.98 %) [16].
Many studies have demonstrated that HHV-8, also known as Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV), is the pathogen involved in KS [17]. Interestingly, the seroprevalence of HHV-8 in the general population of Uyghur and Kazakh people is not high, but KS is commonly seen among people of these ethnicities [17]. Owing to the lack of genomics research, the pathogenesis of KS in this population is unclear.
The particular geography, environment, and ethnic composition may have led to the specific characteristics of KS in Xinjiang, which may also be embodied in the miRNA expression profile in KS tissues. To discover diagnostic and therapeutic targets for KS, we first investigated the expression profile of miRNAs in six paired KS and matched adjacent healthy tissues. Subsequently, we verified the 10 obvious differentially expressed miRNAs using real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in 18 pairs of KS and matched adjacent healthy tissues. The associations between miRNA expression and clinical features were also analyzed.
Materials and methods
Specimen collection
Diseased tissue samples from patients with KS and matched adjacent healthy tissues were obtained from surgical specimens immediately after resection, from January 2012 to September 2013 in the Department of Dermatology, People’s Hospital of the Xinjiang Uyghur Autonomous Region, China. The samples were flash frozen in liquid nitrogen and stored at −180 °C until RNA extraction. Among these samples, 6 KS and matched adjacent healthy tissues were used for miRNA microarray analysis and 18 pairs of KS and adjacent healthy tissues were using for qRT-PCR analysis. The tumor specimens underwent histological examination to confirm the diagnosis, establish the pathological stage, and detect HHV-8 infection. Written consent for tissue donation (for research purposes) was obtained from the patients before tissue collection, and the protocol was approved by the Institutional Review Board of the People’s Hospital of the Xinjiang Uyghur Autonomous Region.
miRNA microarray analysis
miRNA microarray
The 7th generation of miRCURYTM LNA Array (v.18.0) (Exiqon, Copenhagen, Denmark) contains 3100 capture probes, covering all human, mouse, and rat microRNAs annotated in miRBase 18.0, as well as all viral microRNAs related to these species. In addition, this array contains capture probes for 25 miRPlus™ human microRNAs.
RNA extraction
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and a miRNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. This efficiently recovered all RNA species, including miRNAs. The quality and quantity of RNA were measured using a Nanodrop spectrophotometer (ND-1000, Nanodrop Technologies), and RNA integrity was determined by gel electrophoresis.
RNA labeling
After isolation of RNA from the samples, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Copenhagen, Denmark) was used according to the manufacturer’s guidelines for miRNA labeling. One microgram of each sample was 3′-end-labeled with Hy3TM fluorescent label, using T4 RNA ligase and the following procedure. RNA in 2.0 μL of water was combined with 1.0 μL of CIP buffer and CIP (Exiqon). The mixture was incubated for 30 min at 37 °C, and was terminated by incubation for 5 min at 95 °C. Subsequently, 3.0 μL of labeling buffer, 1.5 μL of fluorescent label (Hy3TM), 2.0 μL of DMSO, and 2.0 μL of labeling enzyme were added to the mixture. The labeling reaction was incubated for 1 h at 16 °C, and terminated by incubation for 15 min at 65 °C.
Array hybridization
After stopping the labeling procedure, the Hy3TM-labeled samples were hybridized on the miRCURYTM LNA Array (v.18.0) (Exiqon) according to the array manual. The total 25-μL mixture from Hy3TM-labeled samples, with 25-μL hybridization buffer, was first denatured for 2 min at 95 °C, incubated on ice for 2 min and then hybridized to the microarray for 16–20 h at 56 °C in a 12-Bay Hybridization System (Hybridization System-Nimblegen Systems, Inc., Madison, WI, USA). Following hybridization, the slides were washed several times using a Wash buffer kit (Exiqon). The slides were scanned using an Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA).
Data analysis
Scanned images were imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged, and miRNAs with intensities ≥30 in all samples were chosen for calculation of the normalization factor. The data were normalized using the median normalization. Only those with a greater than twofold increase or twofold decrease in expression in two paired samples and with p < 0.05 were considered significantly changed [18, 19].
miRNA real-time RT-PCR quantification
The miRNA levels were determined by quantitative real-time reverse transcription polymerase chain reaction (qRT–PCR) using miScript SYBRTM Green mastermix (Exiqon) performed on complementary DNA (cDNA) generated from 1 μg of total RNA. Briefly, total RNA was extracted using TRIzol Reagent (Invitrogen) from clinical KS samples. The cDNAs were synthesized from total RNA using gene-specific primers. The reverse transcriptase reactions contained 700 ng RNA sample, 50 μM/μL stem–loop RT primer, 10 × RT buffer, 2.5 mmol/L each of the dNTPs, 200 U/μL MMLV reverse transcriptase, and 40 U/μL RNase inhibitor. The 20 μL reactions were incubated for 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C, and then held at 4 °C. The 10-μL PCR reactions included 2-μL RT product, 2 × PCR master mix, and 1-μL primers. Reactions were incubated in a 96-well optical plate at 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 1 min. The PCR reactions were run on a ViiA 7 Real-time PCR System (Applied Biosystems), and U6 small nuclear RNA was used as an internal control to normalize RNA input. The relative amounts of miRNAs were normalized against U6 snRNA, and the fold change for each miRNA was calculated by the 2–△△CT method [20]. The RT primer and qRT-PCR primer sequences were designed on the basis of the miRNA sequences obtained from the miRBase database (Supplementary data Tables 1 and 2). For the melting curve and amplification curve analyses, see Supplementary Figures 1 and 2.
HHV-8 immunohistochemistry
Fixed, paraffin-embedded tissue sections were examined immunohistochemically using the mouse monoclonal antibody to HHV-8 [LN35] (1:50; Abcam, Cambridge, UK). Tissue sections were stained with an automated immunostainer (BenchMark XT) using heat-induced epitope retrieval and a standard DAB detection kit (Ventana).
Statistical analysis
Analysis of the qRT-PCR data involved comparison of the CT values between tumor and normal tissue adjacent to tumors using the paired t test. Statistically significant associations between clinical features and the fold change in miRNA expression were evaluated using one-way analysis of variance or t test. A P value < 0.05 was considered statistically significant. All calculations were performed using Statistical Program for the Social Sciences (SPSS Inc., Chicago, IL, USA) software 17.0.
Results
Patient characteristics
The specimens for miRNA microarray analysis were all obtained from patients with CKS. The 18 specimens used for qRT-PCR included cases of CKS and AIDS-KS. Their details are shown in Table 1. Tumor specimens underwent histological examination to confirm the diagnosis and for HHV-8 staining (see Fig. 1). Data were recorded on patient age and gender; multifocal or single onset KS; how long since KS had been diagnosed; general area of lesion; tumor location; tumor stage; and HHV-8 infection. The male-to-female ratio was 17:1 (17/1). The mean age at the time of KS diagnosis was 60.1 ± 14.1 (range = 29–79) (CKS 67.7 ± 9.2, AIDS-KS 48.0 ± 11.9) years. The median age at onset of morbidity was 58.2 ± 14.0 (range = 29–78) (CKS 64.9 ± 11.0, AIDS-KS 47.6 ± 11.9) years. The duration of disease was 24.2 ± 33.8 (range = 1–132) (CKS 36.3 ± 39.1, AIDS-KS 5.3 ± 2.8) months. In the 11 cases of CKS, the viral (HHV-8) prevalence was 81.82 % (9/11). Among the seven cases of AIDS-KS, six patients were infected with HHV-8 (85.71 %). Three patients had KS accompanied by syphilis, and two patients had lymph node involvement.
Expression profiling of miRNAs in KS
To identify deregulated miRNAs associated with KS, we used miRNA microarray assay on paired tissues from six patients with KS. Using the miRNA microarray, we identified the miRNAs that were differentially expressed in the paired samples. Only miRNAs that were altered by at least twofold in at least two of the samples were considered significant candidates. Among the 3100 human miRNA probes in the microarrays, we identified 170 differentially expressed miRNAs (69 upregulated and 101 downregulated miRNAs) in KS versus normal skin adjacent to the tumors. The most significantly upregulated miRNAs were miR-126-3p, miR-199a-3p, and miR-16-5p. There were 25 KSHV-related miRNAs in the miRNA microarray. We detected 13 upregulated KSHV-related miRNAs, kshv-miR-K12-3-5p, kshv-miR-K12-9-3p, kshv-miR-K12-2-5p, kshv-miR-K12-12-3p, kshv-miR-K12-4-5p, kshv-miR-K12-10a-5p, kshv-miR-K12-11-3p, kshv-miR-K12-8-3p, kshv-miR-K12-7-3p, kshv-miR-K12-4-3p, kshv-miR-K12-5-3p, kshv-miR-K12-6-5p, and kshv-miR-K12-1-5p. Among the most significantly downregulated miRNAs were miR-125b-1-3p and miR-1183. Heat maps depict the differentially expressed miRNAs from six paired tissues from patients with KS (see Fig. 2). All raw and normalized miRNAs expression data are available from the GEO publicly accessible server (http://www.ncbi.nlm.nih.gov/geo/) with the accession number: GSE55625.
miRNAs deregulated in KS as detected by microRNA microarray. Six pairs of KS and matched normal tissues were analyzed by the 7th generation of miRCURY™ LNA Array (v.18.0). Unsupervised hierarchical cluster analysis of miRNA expression in six patients with KS. Rows: miRNAs; columns: cases. The analysis showed that two chips in each case were consistent. For each miRNA, red represents higher expression and blue represents lower expression than the average. XJ-100, sample 1; XJ-111, sample 2; XJ-113, sample 3; XJ-114, sample 4; XJ-115, sample 5; XJ-117, sample 6. M: KS tumor; C: matched adjacent healthy tissues. Red words represent the 10 verified miRNAs
qRT-PCR validation
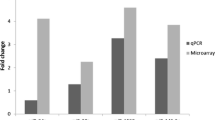
To validate the results of the microarray data, we first examined the expression of eight upregulated miRNAs (miR-181b-5p, miR-199a-3p, miR-15a-5p, miR-126-3p, miR-1297, kshv-miR-k12-12-3p, kshv-miR-k12-1-5p, miR-16-5p) and two downregulated miRNAs (miR-125b-1-3p and miR-1183) by qRT-PCR in the same six paired KS tissues used for the microarray. Further, we validated the 10 miRNAs by qRT-PCR in the fresh tissues from additional patients, comprising 12 case–control pairs. The total validation rate was 100 % when comparing the qRT-PCR with the microarray. The Supplementary Table 3 shows the results of the microarray and qRT-PCR. The levels of miR-181b-5p, miR-199a-3p, miR-15a-5p, miR-126-3p, miR-1297, kshv-miR-k12-12-3p, kshv-miR-k12-1-5p, and miR-16-5p were upregulated and those of miR-125b-1-3p and miR-1183 were downregulated in the tissues from KS specimens when compared with matched normal tissues (p < 0.05). The qRT-PCR results for the 10 miRNAs were consistent with our microarray results (see Fig. 3). As shown in Figure 3, the expression levels of the 10 miRNAs were significantly different in KS and matched adjacent healthy tissues. We also compared the results between qRT-PCR and microarray for the 10 significant differentially expressed miRNAs, using the same six pairs of tissue samples. This also showed the consistency of the results between qRT-PCR and microarray (see Supplementary Figure 3).
The levels of expression of 10 miRNAs measured using miRNA microarray and qRT-PCR. Expression patterns of miR-125b-1-3p (a), miR-1183 (b), kshv-miR-K12-12-3p (c), miR-1297 (d), miR-199a-3p (e), miR-15a-5p (f), miR-126-3p (g), miR-16-5p (h), miR-181b-5p (i), and kshv-miR-K12-1-5p (j) in tumor and matched adjacent healthy tissues. The left most two bars of each panel show miRNA validated by quantitative reverse-transcriptase PCR (qRT-PCR) in 18 pairs of KS tissues. C: matched adjacent healthy tissues; T: KS tumor. Data are means ± SE; *p < 0.05 compared with matched adjacent healthy tissues
Associations between levels of the 10 miRNAs and clinical data from patients with KS
In order to investigate the clinical correlates of the 10 miRNAs, we analyzed the relationships between the levels of expression of the 10 miRNAs in KS tissues and clinical data on the patients with KS (see Table 2). As shown in Table 2, there was no statistically significant difference in expression of miR-125b-1-3p and miR-16-5p in association with the clinical data, except for HHV-8 and HIV infections (see Fig. 4). The other eight miRNAs showed no statistically significant difference in expression in association with the clinical data.
Discussion
In recent years, miRNAs have become known as important regulators of protein expression. Being involved in the expression of many genes, miRNAs are related to the initiation and progression of various cancers [21]. Many studies have indicated that some miRNAs are disordered in human cancer, which demonstrates that miRNAs may play an important role in tumorigenesis [22]. Some miRNAs have tissue-specific or disease-specific expression profiles [7, 23]. One study reported six pregnancy-associated miRNAs as biomarkers for the diagnosis and prognostication of fetal neural tube defects [19]. In this case, a diagnosis made in the first trimester allows early intervention. In general, the identification of biomarkers for diagnosis and therapy is very important.
Currently, the pathogenesis of KS is not very clear. In this study, we identified differences in miRNA profiles between six KS tissues and their matched adjacent healthy tissues, and we also analyzed the associations between the level of expression of 10 dysregulated miRNAs and the clinical data from patients with KS.
The results clearly identified 170 differentially expressed miRNAs, of which 69 were upregulated and 101 downregulated between KS and matched adjacent healthy tissues. Among the obviously differentially upregulated miRNAs were miR-126-3p, miR-199a-3p, miR-16-5p, and the 13 KSHV-related miRNAs. The downregulated miRNAs included miR-125b-1-3p and miR-1183. In further validation by qRT-PCR, the expression of 10 miRNAs (miR-181b-5p, miR-199a-3p, miR-15a-5p, miR-126-3p, miR-1297, kshv-miR-k12-12-3p, kshv-miR-k12-1-5p, miR-16-5p, miR-125b-1-3p, and miR-1183) agreed with that shown by the microarray assay.
In this study, our miRNA microarray results partially agree with the findings of Ene [24] and O’Hara [25]. O’Hara detected the levels of 84 mature miRNAs using a qRT-PCR-based miRNA array. The results showed that miR-143/145 was a novel KS tumor-upregulated miRNA biomarker and that miR221/222, miR-155, miR-220, and let-7 family were downregulated in KS. However, in our study, miR-221, miR-143, miR-200a-3p, miR-200c-3p, let-7g-5p, let-7d-5p, and let-7f-5p were upregulated in KS. The levels of miR-200 and let-7 in O’Hara’s study were contrary to our results. In addition, we did not detect miR-145, miR-222, and miR-155.
The results of Ene’s study [24] demonstrated 185 differentially expressed miRNAs, among which 76 were upregulated and 109 were downregulated, between 17 formalin-fixed paraffin-embedded KS samples and three KSHV-negative normal FFPE samples. The authors used a miRNA Microarray Kit V2 platform (containing 723 human and 76 human viral miRNAs from the Sanger database, v.10.1). The most significantly downregulated miRNAs were miR-99a, the miR-200 family (miR-200a, b, c, miR-141, and miR-429), miR-199b-5p, miR-100, miR-335, and the let-7 family (let-7a to let-7i); kshv-miR-K12-4-3p, kshv-miR-K12-1, kshv-miR-K12-2, kshv-miR-K12-4-5p, kshv-miR-K12-8, kshv-miR-k12-6-3p, kshv-miR-6-5p, kshv-miR-6-3p, kshv-miR-K12-6-5p, kshv-miR-K12-3, kshv-miR-K12-11, and kshv-miR-K12-7 were significantly upregulated. In our study, miR-221, miR-199b-5p, miR-200a-3p, miR-200c-3p, let-7g-5p, let-7d-5p, let-7f-5p, and 13 KSHV-related miRNAs were upregulated. Moreover, we did not detect miR-99a, miR-222, miR-100, miR-200b, miR-141, miR-429, miR-335, and some let-7 family members. The level of expression of nine KSHV-related miRNAs in our study was identical to Ene’s results. Possible reasons for the similarities and differences between studies are differences in the number of miRNAs analyzed, differences in the patient population, and the different miRNA platforms. The miRNA expression profile in patients with KS from the Xinjiang area in China may be significantly altered and show apparent regional features. Our results provide new understanding of the pathogenic mechanism of KS in the Xinjiang area.
We selected 10 miRNAs for qRT-PCR validation. MiR-181b-5p, miR-199a-3p, miR-15a-5p, miR-1297, and miR-16-5p were upregulated, and miR-125b-1-3p and miR-1183 were downregulated in KS samples compared with controls. Many studies have indicated that these miRNAs are involved in the development, progression, treatment, prognosis, or suppression of angiogenesis in some tumors [26–31]. In Li’s [32] study, the overexpression of miR-1297 promoted carcinoma cell proliferation. Moreover, it was found that phosphatase and tensin homologue gene (PTEN) is a novel and crucial tumor suppressor target gene of miR-1297. PTEN is expressed in KS and contributes to the activation of the PI3KAkt-mTOR pathway and reduces colony formation in KS [33].
MiR-126 regulates vascularization, and is encoded by intron 7 of the epidermal growth-factor-like domain 7 (Egfl7) gene [34]. It was discovered to be downregulated in lung, breast, oral, pancreatic, stomach, cervical, bladder, and prostate cancers and has been considered as a suppressor gene; the mechanism involves repression of VEGF-A expression [35]. Moreover, VEGF has an important role in KSHV pathogenesis and is a molecular biomarker of KS lesions [36]. However, in our study, the expression of miR-126 was upregulated, and its function has not been determined in KS. It may promote the growth of KS by increasing VEGF-A expression.
KSHV-miR-K12-11-3p (23.1-fold increase) and kshv-miR-K12-1-5p (19.6-fold increase) were the genes with the most significant differential expression in our study. K12 (the Kaposin gene) is a latent gene that is activated during lytic replication of KSHV and maintains latent KSHV infection during KSHV-induced oncogenesis [37]. KSHV-miR-K12-11 provides a unique model for studying tissue-specific gene regulatory networks involved in viral infection and pathogenesis. It has been shown to have similar seed sequences and functional targets to miR-155 [38, 39].
We identified 10 disordered miRNAs in 18 KS case–control tissue pairs and investigated the clinical associations of the 10 miRNAs. Only miR-125b-1-3p and miR-16-5p had statistically significant associations with HHV-8 and HIV infections in KS. The other eight miRNAs showed no statistically significant difference in miRNA expression in association with the clinical data. It may be that HHV-8 and HIV infections promote the level of expression of miR-125b-1-3p and miR-16-5p.
In summary, the current study has identified differentially regulated miRNAs that are involved in the pathogenesis of KS. We verified 10 dysregulated miRNAs in 18 KS case–control tissue pairs and confirmed the results of the microarray assay. These miRNAs may be useful biomarkers for KS. The miR-125b-1-3p and miR-16-5p showed statistically significant associations between HHV-8, HIV infection, and KS. Although the current results have revealed the miRNA profile of KS, further investigations are necessary to understand the regulatory mechanisms of these miRNAs in KS.
References
Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25(46):6156–62.
Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–9.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
Farh KK-H, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–21.
Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5(2):351–8.
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6.
Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–30.
Hengge UR, Ruzicka T, Tyring SK, Stuschke M, Roggendorf M, Schwartz RA, et al. Update on kaposi’s sarcoma and other hhv8 associated diseases. Part 1: epidemiology, environmental predispositions, clinical manifestations, and therapy. Lancet Infect Dis. 2002;2(5):281–92.
Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36(7):687–93.
Kaposi M. Idiopathic multiple pigmented sarcoma of the skin. Arch Dermatol Syphilol. 1872;4(6):265–73.
Kaloterakis A, Papasteriades C, Filiotou A, Economidou J, Hadjiyannis S, Stratigos J. Hla in familial and nonfamilial mediterranean Kaposi’s sarcoma in Greece. Tissue Antigens. 1995;45(2):117–9.
Dilnur P, Katano H, Wang ZH, Osakabe Y, Kudo M, Sata T, et al. Classic type of Kaposi’s sarcoma and human herpesvirus 8 infection in Xinjiang, China. Pathol Int. 2001;51(11):845–52.
Pu X-M, Wu W-D, Ju H-E. The detection of hhv-8 in the serum of Kaposi’s sarcoma before and after the therapy with interferon. J Clin Dermatol. 2004;33(2):87–8.
He F, Wang X, He B, Feng Z, Lu X, Zhang Y, et al. Human herpesvirus 8: serovprevalence and correlates in tumor patients from Xinjiang, China. J Med Virol. 2007;79(2):161–6.
Wang X, He B, Zhang Z, Liu T, Wang H, Li X, et al. Human herpesvirus-8 in northwestern China: epidemiology and characterization among blood donors. Virol J. 2010;7(1):1–7.
Wu XJ, Pu XM, Kang XJ, Halifu Y, An CX, Zhang DZ, Yakeya B, Mijit J. One hundred and five kaposi sarcoma patients: A clinical study in xinjiang, northwest of china. J Eur Acad Dermatol Venereol. 2014.
Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in aids-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–9.
Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4(5):350–7.
Gu H, Li H, Zhang L, Luan H, Huang T, Wang L, et al. Diagnostic role of microRNA expression profile in the serum of pregnant women with fetuses with neural tube defects. J Neurochem. 2012;122(3):641–9.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative ct method. Nat Protoc. 2008;3(6):1101–8.
Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci Signal. 2007;2007(367):re1.
Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14.
Ene AMC, Borze I, Guled M, Costache M, Leen G, Sajin M, et al. MicroRNA expression profiles in Kaposi’s sarcoma. Pathol Oncol Res. 2014;20(1):153–9.
O’Hara AJ, Wang L, Dezube BJ, Harrington WJ, Damania B, Dittmer DP. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113(23):5938–41.
Chen L, Yang Q, Kong WQ, Liu T, Liu M, Li X, et al. MicroRNA-181b targets camp responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64(7):628–35.
Nurul S, Yoke-Kqueen C, Sabariah AR, Shiran MS, Singh A, Learn-Han L. Differential microRNA expression and identification of putative miRNA targets and pathways in head and neck cancers. Int J Mol Med. 2011;28(3):327–36.
Ratert N, Meyer H-A, Jung M, Mollenkopf H-J, Wagner I, Miller K, et al. Reference miRNAs for miRNAome analysis of urothelial carcinomas. PLoS One. 2012;7(6):e39309.
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. Mirna-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor epb41l3. Mol Cancer Res. 2011;9(7):824–33.
He J, Jing Y, Li W, Qian X, Xu Q, Li F-S, et al. Roles and mechanism of mir-199a and mir-125b in tumor angiogenesis. PLoS One. 2013;8(2):e56647.
Emmrich S, Henke K, Hegermann J, Ochs M, Reinhardt D, Klusmann J-H. MiRNAs can increase the efficiency of ex vivo platelet generation. Ann Hematol. 2012;91(11):1673–84.
Li X, Wang H-l, Peng X, Zhou H-l, Wang X. Mir-1297 mediates pten expression and contributes to cell progression in LSCC. Biochem Biophys Res Commun. 2012;427(9):254–60.
Roy D, Dittmer DP. Phosphatase and tensin homolog on chromosome 10 is phosphorylated in primary effusion lymphoma and Kaposi’s sarcoma. Am J Pathol. 2011;179(4):2108–19.
Parker LH, Schmidt M, Jin S-W, Gray AM, Beis D, Pham T, et al. The endothelial-cell-derived secreted factor egfl7 regulates vascular tube formation. Nature. 2004;428(6984):754–8.
Meister J, Schmidt MHH. Mir-126 and mir-126*: new players in cancer. Sci World J. 2010;10(9):2090–100.
Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, et al. The Kaposi’s sarcoma-associated herpes virus g protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1 alpha. Cancer Res. 2000;60(17):4873–80.
Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102(15):5570–5.
Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, et al. A viral microRNA functions as an orthologue of cellular mir-155. Nature. 2007;450(7172):1096–9.
Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of mir-155. J Virol. 2007;81(23):12836–45.
Acknowledgments
This work was supported by a grant from the Natural Science Foundation of China (Grant No. 81260311) and Natural Science Foundation of Xinjiang Uygur Autonomous Region (Grant No. 2014211A059). We are most grateful to all the families who have so willingly participated in this study and made this study possible. We also acknowledge the support of the Clinical Dermatology Institute of Xinjiang Uygur Autonomous Region and National Clinical key specialty construction projects.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiu-Juan Wu and Xiong-Ming Pu contributed equally to this work and should be considered joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOCX 14 kb)
Supplementary Table 2
(DOCX 18 kb)
Supplementary Table 3
(DOCX 18 kb)
Supplementary Figure 1
(DOCX 412 kb)
Supplementary Figure 2
(DOCX 357 kb)
Supplementary Figure 3
(DOCX 238 kb)
Rights and permissions
About this article
Cite this article
Wu, XJ., Pu, XM., Zhao, ZF. et al. The expression profiles of microRNAs in Kaposi’s sarcoma. Tumor Biol. 36, 437–446 (2015). https://doi.org/10.1007/s13277-014-2626-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2626-1