Abstract
Background
Many studies have shown that abnormal circular RNA (circRNA) expression is associated with the malignant progression of breast cancer (BC), but the role of circ_0000732 in BC progression remains unclear.
Methods
The expression of circ_0000732, microRNA (miR)-1253 and collagen XI alpha 1 (COL11A1) was measured by quantitative real-time PCR. Cell proliferation, migration, invasion and stemness were assessed by cell counting kit 8 assay, Edu assay, transwell assay and sphere formation assay. Western blot analysis was used to determine protein expression. Dual-luciferase reporter assay was performed to assess the interaction between miR-1253 and circ_0000732 or COL11A1. The effect of circ_0000732 on BC tumor growth was confirmed by animal experiments.
Results
Circ_0000732 was overexpressed in BC tissues and cells, and its knockdown suppressed BC cell proliferation, metastasis and stemness. MiR-1253 could be sponged by circ_0000732, and anti-miR-1253 overturned the effects of circ_0000732 knockdown on BC cell progression. COL11A1 was targeted by miR-1253, and miR-1253 inhibited BC cell progression by targeting COL11A1. Circ_0000732 could sponge miR-1253 to upregulate COL11A1. Also, interference of circ_0000732 decreased BC tumor growth by miR-1253/COL11A1 pathway.
Conclusion
Our data showed that circ_0000732 might be a potential target for BC treatment, which could enhance BC malignant phenotype through miR-1253/COL11A1 axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is a malignant tumor that occurs in breast epithelium or ductal epithelium, which has caused serious harm to women’s health (Wormann 2017; Lovelace et al. 2019; Katsura et al. 2022). At present, it is believed that endocrine factors and genetic factors are the main causes of BC, and some gene mutations will increase the risk of BC (Litton et al. 2019; Gandhi and Das 2019). Targeted therapeutic drugs prepared for specific genes have achieved good results in the treatment of BC, such as docetaxel and trastuzumab (Nagini 2017; Bartsch 2020). Therefore, elucidating the effective targets that affect BC progression may provide theoretical basis for the development of new BC molecular targeted therapeutic drugs.
Circular RNA (circRNA) is a non-coding RNA (ncRNA) with circular structure formed by back-splicing in a non-classical splicing manner (Patop et al. 2019; Salzman 2016). As a new favorite in the field of ncRNA research, the important role of circRNA in human diseases has been clarified (Wang et al. 2022). CircRNA has extremely high stability, which provides advantages for it to become a potential therapeutic target for human diseases, especially cancer (Lei et al. 2019; Li et al. 2020). At present, many circRNAs have been found to participate in the regulation of BC progression. For example, circ_0061825 enhanced proliferation and metastasis in BC cells through upregulating TFF1 by sponging miR-326 (Pan et al. 2020). Also, the high expression of circ_0072995 in BC had been confirmed to promote cell proliferation and anaerobic glycolysis via the regulation of miR-149-5p/SHMT2 pathway (Qi et al. 2020). Additionally, circZNF609 was discovered to be a BC treatment biomarker, which could facilitate BC cell malignant phenotype through miR-145-5p/p70S6K1 (Wang et al. 2018). A past study screened the differentially expressed circRNA in BC tissues and normal tissue using human circRNA microarray, and pointed out that circ_0000732 was a significantly overexpressed circRNA in BC tissues (Liang et al. 2017). A recent study showed that circ_0000732 knockdown suppressed triple-negative BC cell proliferation and metastasis (Chen et al. 2022). Therefore, circ_0000732 might be an important regulator for BC progression, and its more functions and mechanisms in BC process deserve further exploration.
CircRNA can competitively bind to microRNA (miRNA) through miRNA response elements, thus removing the negative regulation of miRNA on target genes (Xiong et al. 2018; Dori and Bicciato 2019). In BC-related studies, miR-1253 had been discovered to be under-expressed in BC tissues, and it could hinder proliferation and metastasis in BC cells (Ding et al. 2021). COL11A1, a member of the fibrocollagen family, has recently been found to play an essential role in tumor progression (Vazquez-Villa et al. 2015; Zheng et al. 2021). COL11A1 has been confirmed to be a novel biomarker for BC prognosis and treatment (Shi et al. 2022; Luo et al. 2022). It was reported that COL11A1 accelerated BC cell proliferation and metastasis (Wang et al. 2020). Not only that, high COL11A1 expression in BC had been found to facilitate BC cell proliferation and reduce apoptosis (Gu et al. 2019). Here, we found that circ_0000732 had binding sites with miR-1253, and miR-1253 could COL11A1. Therefore, we proposed and confirmed the hypothesis that circ_0000732 regulated BC process through miR-1253/COL11A1 axis.
Materials and methods
Tissues samples
This study was approved by the Ethics Committee of People’s Hospital of Yangxin County. A total of 60 BC patients were recruited at People’s Hospital of Yangxin County. BC tumor tissues and adjacent normal tissues were obtained and stored at − 80 °C until use. Informed consent was obtained from all patients.
Cell culture and transfection
Human BC cells (MCF-7 and MDA-MB-231) and normal breast epithelial cells (MCF-10A) (ATCC, Manassas, VA, USA) were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco) and 1% Penicillin/Streptomycin (Sangon, Shanghai, China) at 37 °C in 5% CO2. Cell transfection could be carried out by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Circ_0000732 small interfering RNA (si-circ_0000732) or lentivirus short hairpin RNA (sh-circ_0000732), miR-1253 mimic or inhibitor (anti-miR-1253), pcDNA COL11A1 overexpression vector (pcDNA-COL11A1), and their controls were constructed by Ribobio (Guangzhou, China).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated by TRIzol reagent (Invitrogen), and cDNA was obtained by PrimeScript RT Reagent kit (Takara, Dalian, China). The synthesized cRNA was used for qRT-PCR with SYBR Premix Ex Taq (Takara) in PCR system. Primer sequences were listed in Table 1. The β-actin or U6 was used as internal control and the relative expression was calculated by 2–ΔΔCt method. In subcellular localization analysis, the nuclear RNA and cytoplasm RNA of BC cells were isolated by PARIS Kit (Invitrogen). Then, circ_0000732 expression in BC cell nuclear and cytoplasm was detected.
Cell counting kit 8 (CCK8) assay
After transfection, BC cells were harvested and re-seeded in 96-well plates. After culturing at indicated time periods, CCK8 solution (Beyotime, Shanghai, China) was added into each well and cultured for 4 h. Optical density (OD) value was analyzed by microplate reader at a wavelength of 450 nm.
Edu assay
Edu Cell Proliferation Kit (Sangon) was used in accordance with the kit instructions. Briefly, transfected BC cells were seeded into 24-well plates. The cells were stained with Edu solution, fixed with paraformaldehyde, treated with Triton X-100, and then hatched with DAPI solution. The fluorescence signal was captured under the fluorescence microscope, and relative Edu positive (Edu+) cells were counted.
Transwell assay
BC cells suspended with serum-free medium were plated into the upper of 24-well transwell chamber (Corning Inc., Corning, NY, USA) coated with or without Matrigel (for invasion and migration, respectively). The lower chambers were filled with complete medium. After 24 h, the migrated and invaded cells were fixed with paraformaldehyde and stained with crystal violet. Cell images were observed under a microscope (200 ×), and the number of cells was counted in 5 fields randomly.
Western blot (WB) analysis
Total protein was extracted by RIPA lysis buffer (Beyotime). After resolved by SDS-PAGE gel, proteins were transferred to PVDF membrane followed by blocked with skimmed-milk. Membrane was hatched with primary antibody and secondary antibody one by one, and then the protein blot was visualized using BeyoECL Moon (Beyotime). The antibodies (Abcam, Cambridge, MA, USA) were listed as below: anti-E-cadherin (1:5,000, ab40772), anti-N-cadherin (1:1,000, ab18203), anti-Vimentin (1:1,000, ab45939), anti-OCT4 (1:1,000, ab19857), anti-NANOG (1:2,000, ab109250), anti-CD133 (1:1,000, ab19898), anti-COL11A1 (1:500, ab64883), anti-β-actin (1:1,000, ab8227) and Goat Anti-Rabbit IgG (1:50,000, ab205718).
Sphere formation assay
In accordance with a previous study (Fang et al. 2021), BC cells were harvested and re-seeded into ultra-low attachment 6-well plates (4 × 103 cells/well) with serum-free medium containing 20 ng/mL EGF, 1:50 B27, and 10 ng/mL bFGF. 10 days later, the sphere state was observed under a microscope (100 ×), and the number of spheres (> 100 μm) was counted to calculate sphere formation efficiency.
Dual-luciferase reporter assay
Starbase3.0 software and Targerscan software were used to predict the binding sites between miR-1253 and circ_0000732 or COL11A1 3’UTR. As the previous study described (Ding et al. 2022), the circ_0000732-WT/MUT vectors and COL11A1 3’UTR-WT/MUT vectors were generated using pmirGLO vectors. The vectors were transfected with miR-1253 mimic/miR-NC into BC cells using Lipofectamine 3000. 48 h later, Dual-luciferase Reporter Gene Assay Kit (Beyotime) was used for analyzing relative luciferase activity.
Mice xenograft models
As the previous study described (Ding et al. 2022), BALB/c nude mice (n = 5/group; Vital River, Beijing, China) were subcutaneously injected with MCF-7 cells (2 × 106) stably transfected with sh-NC or sh-circ_0000732. Every 7 days, tumor volume was calculated until 35 days. After the mice were sacrificed, the tumor tissues were weighted. Then, the tumor tissue was used to prepare paraffin sections for Ki67 immunohistochemical (IHC) staining. Animal experiments were approved by the Animal Ethics Committee of People’s Hospital of Yangxin County.
Statistical analysis
Results were denoted as the mean ± SD. All experiments were performed in triplicate, with each independent experiment set 3 times to generate an average value. GraphPad Prism 7.0 software was used for statistical analysis. Differences between groups were analyzed by Student’s t-test or ANOVA. Pearson correlation analysis was used to analyze the linear correlation. P < 0.05 was considered as statistically significant.
Results
The high expression of circ_0000732 was found in BC tissues and cells
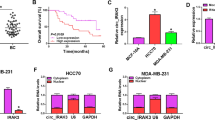
First, we revealed the expression of circ_0000732 in BC tissues and cells to speculate the role of circ_0000732 in BC process. Through measuring circ_0000732 expression in BC tumor tissues and adjacent normal tissues, we discovered that circ_0000732 was remarkably upregulated in BC tumor tissues (Fig. 1A). Also, circ_0000732 expression was higher in BC cells (MCF7 and MDA-MB-231) than in MCF-10A cells (Fig. 1B). The results of subcellular localization analysis suggested that circ_0000732 was mainly distributed in the cytoplasm of BC cells (Fig. 1C, D). This suggested that circ_0000732 might be mainly involved in post-transcriptional regulation.
The expression of circ_0000732 in BC tissues and cells. A The circ_0000732 expression in BC tumor tissues and adjacent normal tissues was measured by qRT-PCR. B QRT-PCR was used to detect circ_0000732 expression in BC cells and MCF-10A cells. C, D Subcellular localization analysis was used to determine the distribution of circ_0000732 in BC cell cytoplasm and nuclear. *P < 0.05
Interference of circ_0000732 suppressed BC cell proliferation, metastasis and stemness
Given the high expression of cc, we speculated that circ_0000732 might facilitate BC progression. To confirm circ_0000732 roles in BC cell progression, si-circ_0000732 was transfected into MCF7 and MDA-MB-231 cells to silence circ_0000732 expression (Fig. 2A). As presented in Fig. 2B–D, circ_0000732 knockdown repressed BC cell viability and Edu+ cells, confirming that circ_0000732 promoted BC cell proliferation. Also, circ_0000732 silencing could restrain the number of migrated and invaded cells, decrease the N-cadherin and Vimentin protein expression, while increase the E-cadherin protein expression (Fig. 2E–G). These confirmed that circ_0000732 enhanced BC cell metastasis. Furthermore, silenced circ_0000732 also inhibited the sphere formation efficiency of BC cells and reduced the protein expression of cancer stem cell markers (OCT4, NANOG and CD133) (Fig. 2H, I), revealing that circ_0000732 facilitated BC cell stemness.
Effects of si-circ_0000732 on BC cell progression. BC cells were transfected with si-NC or si-circ_0000732. A The circ_0000732 expression was detected by qRT-PCR. CCK8 assay (B, C), Edu assay (D) and transwell assay (E, F) were used to measure cell proliferation, migration and invasion. G The protein expression of N-cadherin, Vimentin and E-cadherin was examined by WB analysis. H Sphere formation assay was performed to assess cell stemness. I The protein expression of OCT4, NANOG and CD133 was tested by WB analysis. *P < 0.05
MiR-1253 could interact with circ_0000732
To further uncover the potential molecular mechanisms by which circ_0000732 regulated BC progression, we performed bioinformatics analysis. The Starbase3.0 software was used to predict the targeted miRNA for circ_0000732, and miR-1253 was found to be complementary to circ_0000732 (Fig. 3A). MiR-1253 mimic was confirmed to promote miR-1253 expression in BC cells (Fig. 3B). After miR-1253 mimic and the circ_0000732-WT/MUT reporter vectors were co-transfected into BC cells, we discovered that miR-1253 mimic reduced the luciferase activity of circ_0000732-WT vector, confirming the interaction between circ_0000732 and miR-1253 (Fig. 3C, D). The results of Fig. 3E showed that circ_0000732 knockdown markedly increased miR-1253 expression. In addition, miR-1253 expression was discovered to be lower in BC tumor tissues and cells than that in adjacent normal tissues and MCF-10A cells, respectively (Fig. 3F, G).
MiR-1253 could interact with circ_0000732. A The sequences of circ_0000732-WT/MUT vectors were shown. B The transfection efficiency of miR-1253 mimic was measured by qRT-PCR. C, D Dual-luciferase reporter assay was used to assess RNA interaction. E MiR-1253 expression was determined by qRT-PCR in BC cells transfected with si-NC or si-circ_0000732. F The miR-1253 expression was examined by qRT-PCR in BC tumor tissues and adjacent normal tissues. G QRT-PCR was used to measure miR-1253 expression in BC cells and MCF-10A cells. *P < 0.05
Circ_0000732 regulated BC cell proliferation, metastasis and stemness by miR-1253
In view of the above results, we speculated that circ_0000732 might target miR-1253 to mediate BC progression. To determine whether miR-1253 participated in the regulation of circ_0000732 on BC progression, we performed the rescue experiments. The anti-miR-1253 indeed reduced miR-1253 expression in BC cells (Fig. 4A). In BC cells co-transfected with si-circ_0000732 and anti-miR-1253, we confirmed that the inhibitory effects of si-circ_0000732 on BC cell viability, Edu+ cells, and the numbers of migrated and invaded cells were revoked by miR-1253 inhibitor (Fig. 4B–F). Besides, the decreased N-cadherin and Vimentin protein expression, and the increased E-cadherin protein expression induced by si-circ_0000732 also were reversed by miR-1253 inhibitor (Fig. 4G). As shown in Fig. 4H, I , anti-miR-1253 also abolished the suppressive effect of si-circ_0000732 on BC cell sphere formation efficiency and the protein expression of OCT4, NANOG and CD133.
Effects of si-circ_0000732 and anti-miR-1253 on BC cell progression. A The transfection efficiency of anti-miR-1253 was assessed by qRT-PCR. B–I BC cells were transfected with si-circ_0000732 or anti-miR-1253. Cell proliferation, migration and invasion were determined by CCK8 assay (B, C), Edu assay (D) and transwell assay (E, F). G WB analysis was performed to measure the protein expression of N-cadherin, Vimentin and E-cadherin. H Cell stemness was evaluated by sphere formation assay. I The protein expression of OCT4, NANOG and CD133 was analyzed using WB analysis. *P < 0.05
COL11A1 was targeted by miR-1253
In order to reveal the downstream target of miR-1253, bioinformatics analysis was performed. We used Targetscan and GEPIA software to jointly predict the target genes of miR-1253, and found that COL11A1 was the target of miR-1253 (Fig. 5A). In the GEPIA software, we confirmed that COL11A1 had increased expression in BC tumor tissues compared to normal tissues (Fig. 5B). According to the binding sites between COL11A1 and miR-1253, we constructed the COL11A1 3’UTR-WT/MUT vectors (Fig. 5C). We found that the luciferase activity driven by the COL11A1 3’UTR-WT vector could be reduced by miR-1253 mimic (Fig. 5D, E). In BC cells transfected with miR-1253 mimic or inhibitor, COL11A1 mRNA and protein expression was decreased and increased, respectively (Fig. 5F, G). Additionally, we discovered that COL11A1 was highly expressed in BC tumor tissues and cells at the mRNA and protein levels (Fig. 5H–K).
COL11A1 was targeted by miR-1253. A Targetscan and GEPIA software was used to jointly predict the target genes of miR-1253. B COL11A1 expression in BC tumor tissues and normal tissues was analyzed by GEPIA software. C The sequences of COL11A1 3’UTR-WT/MUT vectors were presented. D, E Dual-luciferase reporter assay was performed to evaluate RNA interaction. F, G The mRNA and protein expression of COL11A1 was determined by qRT-PCR and WB analysis in BC cells transfected with anti-miR-1253 or miR-1253 mimic. (H, I) The COL11A1 mRNA and protein expression was examined by qRT-PCR and WB analysis in BC tumor tissues and adjacent normal tissues. (J, K) QRT-PCR and WB analysis were used to measure COL11A1 mRNA and protein expression in BC cells and MCF-10A cells. *P < 0.05
MiR-1253 inhibited BC cell progression via targeting COL11A1
In view of the above results, we speculated that miR-1253 might target COL11A1 to regulate BC progression. The following experiments were used to confirm that miR-1253 targeted COL11A1 to regulate BC progression. The pcDNA-COL11A1 was found to enhance the mRNA and protein expression of COL11A1 in BC cells (Fig. 6A, B). Then, miR-1253 mimic and pcDNA-COL11A1 were co-transfected into BC cells. The results revealed that miR-1253 overexpression inhibited BC cell viability, Edu+ cells, the numbers of migrated and invaded cells, while these effects were overturned by COL11A1 overexpression (Fig. 6C–G). Also, miR-1253 mimic reduced the protein expression of N-cadherin and Vimentin, while promoted the protein expression of E-cadherin. However, this effect also was reversed by pcDNA-COL11A1 (Fig. 6H). In addition, overexpressed COL11A1 revoked the inhibition of miR-1253 mimic on BC cell sphere formation efficiency and the protein expression of OCT4, NANOG and CD133 (Fig. 6I, J).
Effects of miR-1253 and COL11A1 on BC cell progression. A, B The transfection efficiency of pcDNA-COL11A1 was assessed by qRT-PCR and WB analysis. C–J BC cells were transfected with miR-1253 mimic or pcDNA-COL11A1. CCK8 assay (C, D), Edu assay (E) and transwell assay (F, G) were utilized for measuring cell proliferation, migration and invasion. H The protein expression of N-cadherin, Vimentin and E-cadherin was analyzed by WB analysis. I Sphere formation assay was performed to measure cell stemness. J WB analysis was performed to determine the protein expression of OCT4, NANOG and CD133. *P < 0.05
Circ_0000732 regulated COL11A1 via sponging miR-1253
The above results confirmed that circ_0000732 sponged miR-1253, and miR-1253 targeted COL11A1. Therefore, we speculated that circ_0000732 might regulate miR-1253 to mediate COL11A1 expression. Circ_0000732 knockdown significantly decreased COL11A1 mRNA and protein expression, and this effect could be reversed by miR-1253 inhibitor (Fig. 7A, B). In BC tumor tissues, we found that miR-1253 expression was negatively correlated with circ_0000732 and COL11A1 expression, and circ_0000732 expression was positively correlated with COL11A1 expression (Fig. 7C–E). These results confirmed that circ_0000732 sponged miR-1253 to positively regulate COL11A1.
Circ_0000732 regulated COL11A1 via sponging miR-1253. A, B The COL11A1 mRNA and protein expression was determined by qRT-PCR and WB analysis in BC cells transfected with si-circ_0000732 or anti-miR-1253. C, E Pearson correlation analysis was used to assess the correlations among circ_0000732, miR-1253 and COL11A1 expression in BC tumor tissues. *P < 0.05
Circ_0000732 knockdown reduced BC tumorigenesis
To further confirm our conclusions, we conducted in vivo experiments. In animal experiments, we found that the tumor volume and weight were decreased after circ_0000732 knockdown compared to the control group (Fig. 8A, B). In the tumor tissues of the sh-circ_0000732 group, circ_0000732 expression was indeed reduced (Fig. 8C). Also, miR-1253 expression was increased, while COL11A1 mRNA and protein expression was inhibited in the tumor tissues of the sh-circ_0000732 group (Fig. 8D–F). Moreover, IHC staining was used to detect the Ki67 positive cell rate in the tumor tissues of each group, and the results showed that the Ki67 positive cell rate also was repressed by circ_0000732 knockdown (Fig. 8G).
Circ_0000732 knockdown reduced BC tumorigenesis. MCF7 cells transfected with sh-NC or sh-circ_0000732 were injected into cells. Tumor volume (A) and tumor weight (B) were detected. C, D The expression of circ_0000732 and miR-1253 in the mice tumor tissues was measured by qRT-PCR. E, F The COL11A1 mRNA and protein expression in the mice tumor tissues was assessed by qRT-PCR and WB analysis. G IHC staining was performed to assess Ki67 positive cell rate in the mice tumor tissues. *P < 0.05
Discussion
CircRNA has been proven to regulate the functions of many molecules, thereby regulating the physiological and pathological activities of cells (Patop and Kadener 2018; Kristensen et al. 2018). In this, we investigated the function of circ_0000732 in BC progression. Through qRT-PCR, we confirmed that circ_0000732 had elevated expression in BC, which was consistent with the reported study (Liang et al. 2017; Chen et al. 2022). In loss-of-function experiments, silenced circ_0000732 restrained BC cell proliferation, metastasis and stemness, as well as suppressed BC tumor growth. These results confirmed the role of circ_0000732 in promoting the malignant phenotype of BC, revealing that circ_0000732 might be a potential therapeutic target for BC.
Further analysis suggested that circ_0000732 sponged miR-1253. MiR-1253 is a key miRNA regulating the malignant progression of tumors. A recent study suggested that miR-1253 functioned an anti-tumor role in colon cancer, which could reduce cell growth and metastasis (Yang and Zhang 2021). Studies had indicated that miR-1253 had an inhibition on osteosarcoma cell proliferation and invasion (Huang et al. 2018). Also, miR-1253 was found to suppress medulloblastoma cell cycle and proliferation to inhibit tumor progression (Kanchan et al. 2020). Similar to previous results (Ding et al. 2021), our data confirmed the low miR-1253 expression in BC, and demonstrated that miR-1253 restrained BC cell proliferation, metastasis and stemness. These data verified that miR-1253 acted a tumor suppressor role in BC. Also, we revealed that circ_0000732 targeted miR-1253 to decrease miR-1253 expression, thereby promoting BC cell progression.
COL11A1 is thought to be a proto-oncogene that promotes cancer progression, such as lung cancer (Tu et al. 2021), colorectal cancer (Chen et al. 2020) and ovarian cancer (Wu et al. 2014). COL11A1 was considered to be an independent prognostic factor for breast ductal carcinoma in situ (Toss et al. 2019). Consistent with previous study (Wang et al. 2020; Gu et al. 2019), we confirmed that COL11A1 was overexpressed in BC tissues and cells. Besides, we pointed out that COL11A1 could be targeted by miR-1253. Functional experiments indicated that miR-1253 targeted COL11A1 to suppress its expression and then to inhibit BC cell proliferation, metastasis and stemness. Additionally, our data suggested that circ_0000732 knockdown reduced COL11A1 expression by sponging miR-1253, confirming the existence of circ_0000732/miR-1253/COL11A1 pathway.
To sum up, our research pointed to a new circRNA that regulated BC malignant behaviors. Our results showed that circ_0000732 promoted BC proliferation, metastasis and stemness, which was mainly achieved by targeting miR-1253 to mediate COL11A1 expression. According to our test results, we believed that clinically targeted inhibition of circ_0000732 might be an effective way to treat BC.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Bartsch R (2020) Trastuzumab-deruxtecan: an investigational agent for the treatment of HER2-positive breast cancer. Expert Opin Investig Drugs 29(9):901–910
Chen D, Qin Y, Dai M, Li L, Liu H, Zhou Y et al (2020) BGN and COL11A1 regulatory network analysis in colorectal cancer (CRC) reveals that bgn influences CRC cell biological functions and interacts with miR-6828-5p. Cancer Manag Res 12:13051–13069
Chen D, Wang M, Zhang H, Zhou S, Luo C (2022) Estrogen receptor beta2 (ERbeta2)-mediated upregulation of hsa_circ_0000732 promotes tumor progression via sponging microRNA-1184 in triple-negative breast cancer (TNBC). Inflamm Res 71(2):255–266
Ding X, Zheng J, Cao M (2021) Circ_0004771 accelerates cell carcinogenic phenotypes via suppressing miR-1253-mediated DDAH1 inhibition in breast cancer. Cancer Manag Res 13:1–11
Ding D, Yang F, Chen Z, Ying J (2022) Circ_0007385 regulates cell proliferation, apoptosis and stemness via targeting miR-493-3p/RAB22A axis in non-small cell lung cancer. Thorac Cancer 13(4):571–581
Dori M, Bicciato S (2019) Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes (basel) 10(9):642
Fang G, Chen T, Mao R, Huang X, Ji L (2021) Circular RNA circ_0089153 acts as a competing endogenous RNA to regulate colorectal cancer development by the miR-198/SUMO-specific peptidase 1 (SENP1) axis. Bioengineered 12(1):5664–5678
Gandhi N, Das GM (2019) Metabolic reprogramming in breast cancer and its therapeutic implications. Cells 8(2):89
Gu SQ, Luo JH, Yao WX (2019) The regulation of miR-139-5p on the biological characteristics of breast cancer cells by targeting COL11A1. Math Biosci Eng 17(2):1428–1441
Huang L, Chen M, Pan J, Yu W (2018) Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun 500(2):511–517
Kanchan RK, Perumal N, Atri P, Chirravuri Venkata R, Thapa I, Klinkebiel DL et al (2020) MiR-1253 exerts tumor-suppressive effects in medulloblastoma via inhibition of CDK6 and CD276 (B7–H3). Brain Pathol 30(4):732–745
Katsura C, Ogunmwonyi I, Kankam HK, Saha S (2022) Breast cancer: presentation, investigation and management. Br J Hosp Med (lond) 83(2):1–7
Kristensen LS, Hansen TB, Veno MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37(5):555–565
Lei B, Tian Z, Fan W, Ni B (2019) Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 16(2):292–301
Li J, Sun D, Pu W, Wang J, Peng Y (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6(4):319–336
Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL (2017) Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7(7):1566–1576
Litton JK, Burstein HJ, Turner NC (2019) Molecular testing in breast cancer. Am Soc Clin Oncol Educ Book 39:e1–e7
Lovelace DL, McDaniel LR, Golden D (2019) Long-term effects of breast cancer surgery, treatment, and survivor care. J Midwifery Womens Health 64(6):713–724
Luo Q, Li J, Su X, Tan Q, Zhou F, Xie S (2022) COL11A1 serves as a biomarker for poor prognosis and correlates with immune infiltration in breast cancer. Front Genet 13:935860
Nagini S (2017) Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med Chem 17(2):152–163
Pan G, Mao A, Liu J, Lu J, Ding J, Liu W (2020) Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif 53(2):e12720
Patop IL, Kadener S (2018) circRNAs in cancer. Curr Opin Genet Dev 48:121–127
Patop IL, Wust S, Kadener S (2019) Past, present, and future of circRNAs. EMBO J 38(16):e100836
Qi C, Qin X, Zhou Z, Wang Y, Yang Q, Liao T (2020) Circ_0072995 promotes cell carcinogenesis via up-regulating miR-149-5p-mediated SHMT2 in breast cancer. Cancer Manag Res 12:11169–11181
Salzman J (2016) Circular RNA Expression: its potential regulation and function. Trends Genet 32(5):309–316
Shi W, Chen Z, Liu H, Miao C, Feng R, Wang G et al (2022) COL11A1 as an novel biomarker for breast cancer with machine learning and immunohistochemistry validation. Front Immunol 13:937125
Toss MS, Miligy IM, Gorringe KL, Aleskandarany MA, Alkawaz A, Mittal K et al (2019) Collagen (XI) alpha-1 chain is an independent prognostic factor in breast ductal carcinoma in situ. Mod Pathol 32(10):1460–1472
Tu H, Li J, Lin L, Wang L (2021) COL11A1 Was involved in cell proliferation, apoptosis and migration in non-small cell lung cancer cells. J Invest Surg 34(6):664–669
Vazquez-Villa F, Garcia-Ocana M, Galvan JA, Garcia-Martinez J, Garcia-Pravia C, Menendez-Rodriguez P et al (2015) COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumour Biol 36(4):2213–2222
Wang S, Xue X, Wang R, Li X, Li Q, Wang Y et al (2018) CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res 10:3881–3890
Wang H, Ren Y, Qian C, Liu J, Li G, Li Z (2020) Over-expression of CDX2 alleviates breast cancer by up-regulating microRNA let-7b and inhibiting COL11A1 expression. Cancer Cell Int 20:13
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan Q et al (2022) The influence of circular RNAs on autophagy and disease progression. Autophagy 18(2):240–253
Wormann B (2017) Breast cancer: basics, screening, diagnostics and treatment. Med Monatsschr Pharm 40(2):55–64
Wu YH, Chang TH, Huang YF, Huang HD, Chou CY (2014) COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 33(26):3432–3440
Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ et al (2018) A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med 16(1):220
Yang D, Zhang D (2021) miR-1253, a novel tumor suppressor gene in colon cancer, is associated with poor patients prognosis. Clin Exp Med 21(4):563–571
Zheng X, Liu X, Zheng H, Wang H, Hong D (2021) Integrated bioinformatics analysis identified COL11A1 as an immune infiltrates correlated prognosticator in pancreatic adenocarcinoma. Int Immunopharmacol 90:106982
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
XL designed and supervised the study. WM conducted the experiments and drafted the manuscript. YW collected and analyzed the data. FW contributed the methodology and analyzed the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
XL, WM, YW and FW declared that they have no conflict of the interest.
Ethical approval
This study was approved by the Ethics Committee of People’s Hospital of Yangxin County. Animal experiments were approved by the Animal Ethics Committee of People’s Hospital of Yangxin County.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ming, W., Wu, Y., Wang, F. et al. Circ_0000732 promotes breast cancer cell proliferation, metastasis and stemness by mediating COL11A1 through the regulation of miR-1253. Mol. Cell. Toxicol. (2023). https://doi.org/10.1007/s13273-023-00396-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s13273-023-00396-5