Abstract
Plant-specific SnRK2 (sucrose nonfermenting-1-related protein kinase 2) genes play crucial roles in the coordination of plant growth and development and responses to stress. However, comprehensive studies have not been performed for this gene family in pepper (Capsicum annuum), a very important Solanaceous vegetable worldwide. To fully understand the status of SnRK2s in chili pepper, a total of 9 putative SnRK2 genes (named CaSnRK2.1-2.9) were identified in pepper in the present study. These genes were located on 7 different chromosomes and classified into three subfamilies based on the phylogenetic tree. Their conserved motif compositions and exon-intron structures were systematically analyzed, and the results strongly supported the classification. Furthermore, a total of 81 putative cis-elements were found in the promoter regions, and the cis-elements related to hormone and stress signaling were abundant. Finally, the CaSnRK2 gene expression profiles among different tissues, especially developing fruit tissue, and under various abiotic stresses were investigated to identify tissue-specific or stress-responsive candidates. This study was the first to comprehensively investigate the SnRK2 family in pepper, and the results provide important clues for further functional analyses of fruit development and abiotic stress responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Harsh environments greatly limit plant growth and crop yields. To adapt to changing environments, plants have formed a series of different defense-related metabolic mechanisms. Protein kinases and protein phosphorylation/dephosphorylation play important roles in signal transduction pathways with respect to the recognition and transmission of stress signals to different parts of cells. Members of the SnRK (or SNF1-related protein kinase) family are specific types of serine/threonine protein kinases that exist widely in plants and function significantly in a host of processes, including growth and development, defense against various stresses, and hormone-mediated signaling (Wang et al. 2018; Zhao et al. 2018; Lin et al. 2015; Zhang et al. 2016; Yang et al. 2012; Coello et al. 2012).
In higher plants, the SnRK (sucrose nonfermenting-1-related protein kinase) family consists of three subfamilies (denoted SnRKl, SnRK2 and SnRK3) based on their sequence similarity and C-terminal domain structure characteristics (Hrabak et al. 2003; Kobayashi et al. 2004). Members of the SnRK1 subfamily are most closely related to SNF1 from yeast and AMP-activated protein kinase (AMPK) from mammals (Hardy et al. 1994; Hardie 2007). The SnRK1 subfamily typically consists of relatively few members (only 3 members in Arabidopsis) that both encode relatively large proteins (55–60 kDa) and play a major role in global regulation of carbon metabolism and energy status (Hrabak et al. 2003; Coello et al. 2012). The SnRK2 and SnRK3 subgroups appear to be unique to plants and are more diverse (10 SnRK2 genes and 25 SnRK3 genes have been described in Arabidopsis), and members of these subfamilies are implicated in the response to various abiotic stresses, including heat, cold, drought, salinity, and osmotic stresses (Coello et al. 2012; McLoughlin et al. 2012; Yang et al. 2012; Fujita et al. 2013; Liang et al. 2015; Song et al. 2016; Zhang et al. 2016; Bai et al. 2017; Soma et al. 2017; Tan et al. 2018; Wang et al. 2018).
The first SnRK2 gene, namely, PKABA1, was cloned from wheat and could be induced by dehydration, cold temperature and osmotic stress (Holappa and Walker-Simmons 1995; Gomez-Cadenas et al. 2001). To date, members of the SnRK2 family have been identified and comprehensively analyzed in many different plant species, including the dicotyledons Arabidopsis (Hrabak et al. 2003), apple (Shao et al. 2014), strawberry (Han et al. 2015), pak-choi (Huang et al. 2015), cotton (Liu et al. 2017b), and potato (Bai et al. 2017), and the monocotyledons rice (Kobayashi et al. 2004), maize (Huai et al. 2008), wheat (Zhang et al. 2016) and sugarcane (Li et al. 2017). SnRK2 N-termini contain conserved kinase domains, such as an ATP-binding domain (GXGXXGX) and a serine/threonine activation loop, while the C-terminus contains regulatory domains rich in acidic amino acids (AAs), such as aspartic acid (D) and glutamic acid (E). A phylogenetic analysis revealed that the SnRK2 family could be divided into three subgroups. Members of subgroups I and II are weak or unresponsive to abscisic acid (ABA) (Soma et al. 2017), while those of subgroup III are strongly induced by ABA (Wang et al. 2013).
Cumulative evidence has shown that SnRK2s are core components of the ABA-mediated signaling transduction network involved in the SnRK2-PP2C (a type 2C protein phosphatase) complex structure (Soon et al. 2012; Shen et al. 2017; Zhao et al. 2018). Among the ten AtSnRK2s (named AtSnRK2.1-AtSnRK2.10) identified in the model plant Arabidopsis thaliana, subgroup I members (including AtSnRK2.1, AtSnRK2.4, AtSnRK2.5, AtSnRK2.9, and AtSnRK2.10) are involved in the response to osmotic or salt stress and improve plant resistance to drought (Szymanska et al. 2019; Julkowska et al. 2015; McLoughlin et al. 2012; Kulik et al. 2012). Two members in subgroup II (AtSnRK2.7 and AtSnRK2.8) are responsive to salt stress and could also strengthen plant tolerance to drought (Lee et al. 2015). Subgroup III contains three members (AtSnRK2.2, AtSnRK2.3, and AtSnRK2.6) that are involved in ABA signaling during seed germination/dormancy, root growth (Fujita et al. 2009; Nakashima et al. 2009; Fujii et al. 2011; Zheng et al. 2010), the development and ripening of some nonclimacteric fruit (Han et al. 2015) and the response to drought and chilling stress (Wang et al. 2018; Tan et al. 2018). In rice, all 10 SAPK1 (stress/ABA-activated protein kinase) to SAPK10 members are activated by hyperosmotic stress, and three of them (SAPK8, SAPK9, and SAPK10) are also activated by ABA (Kobayashi et al. 2004; Yu et al. 2012; Lou et al. 2018). The sensitivity of these SnRK2 genes to ABA and various stresses differ between Arabidopsis and rice, indicating that the functions of these core ABA signaling components are diverse. However, research on SnRK2 genes in pepper is very limited.
Pepper (Capsicum spp.) is an important vegetable and spice plant. The Capsicum genus consists of more than 30 species, five of which are domesticated (C. annuum, C. baccatum, C. chinense, C. frutescens, and C. pubescens) (Carrizo Garcia et al. 2016). Given the importance of the SnRK2 gene family in the stress resistance of plants and completion of whole-genome sequencing of the pepper (Kim et al. 2014; Qin et al. 2014), all CaSnRK2s predicted to belong to the SnRK2 family were identified and a comprehensive bioinformatics analysis was carried out in this study. The expression patterns of all CaSnRK2 genes in different tissues, especially the fruit, and under different abiotic stresses were analyzed via transcriptome data and fluorescence quantitative PCR (qRT-PCR). The goal of this study is to fully understand the status of SnRK2s related to growth and abiotic stress in chili pepper (Capsicum annuum L.). This study provides a basis for further characterization of the physiological and biochemical functions of the CaSnRK2s in peppers.
Materials and methods
CaSnRK2 identification and sequence analysis
To identify SnRK2 genes in the pepper genome, a BlastP search was performed against the Sol Genomics Network (https://solgenomics.net/) (Mueller et al. 2005) based on the Capsicum annuum cv. Zunla (v2.0), cv. CM334 (v1.55) and var. glabriusculum (v2.0) genomes using SnRK2 protein sequences from Arabidopsis as queries. The default parameters used a cut-off value of 1e–10. Additionally, the candidate sequences were analyzed via PROSITE (http://prosite.expasy.org/scanprosite) and the SMART program (http://smart.embl-heidelberg.de) to confirm the presence of related domains. The molecular weight (MW) and theoretical isoelectric point (pI)of the CaSnRK2 proteins were computed by ProtParam (https://web.expasy.org/protparam/).
Chromosomal location, gene structure analysis and conserved motif prediction
The chromosomal location information for CaSnRK2s was obtained by a BlastP query against the Pepper Genome Database (PGD, http://peppersequence.genomics.cn/page/species/index.jsp) (cultivar Zunla-1), which was constructed by our group and visualized using MapChart 2.2. The gene structures of all CaSnRK2 genes were drawn by the online software GSDS (http://gsds.cbi.pku.edu.cn/index.php) (Hu et al. 2015). The conserved motifs in the CaSnRK2s were analyzed using the online motif alignment and search tool MEME Suite 5.0.3 (http://meme-suite.org/tools/meme) (Bailey et al. 2015).
Multiple sequence alignment and phylogenetic analysis
The full-length SnRK2 AA sequences of Arabidopsis (Arabidopsis thaliana) (Hrabak et al. 2003), potato (Solanum tuberosum) (Bai et al. 2017), tomato (Solanum lycopersicum), apple (Malus domestica), corn (Zea mays) (Huai et al. 2008), rice (Oryza sativa Japonica) (Kobayashi et al. 2004) and pepper were aligned using ClustalW software. A phylogenetic tree based on the alignment was constructed using the Molecular Evolutionary Genetics Analysis (MEGA) 6.0 software (Tamura et al. 2013) with the NJ method and 1000 bootstrap replicates.
Analysis of cis-elements within the promoters
To analyze the cis-elements within the CaSnRK2 promoters, the 1500 bp (bp) sequence upstream of the initiation codons (ATG) of each CaSnRK2 gene was retrieved from the PGD. The cis-elements were predicted by the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002), including those for phytohormones and the stress responses. The data were processed using the Microsoft Office Excel software, and then the tables and figures were constructed.
Expression pattern analysis based on transcriptome data
To evaluate the expression patterns of the CaSnRK2 genes, previously generated Illumina transcriptome sequencing data were utilized (Liu et al. 2017a; Qin et al. 2014). The transcriptome data included different tissues, such as root, stem, leaf, floral bud and fully blossomed flower tissues as well as tissues from seeds and fruits at different developmental stages. For stress treatments, the 40-day old seedlings were subjected to 200 mM NaCl (salt stress), 400 mM D-mannitol (osmotic stress), 30 mM H2O2 (oxidative stress), 42 ℃ or 10 ℃ (heat or cold stress). Leaf and root tissues from five plants were collected from treated and control plants at 0, 1.5, 3, 6, 12 and 24 h post treatment and mixed as one biological replicate. Only genes with fragments per kilobase million (FPKM) > 1 were collected for further analysis. The expression levels of the CaSnRK2 genes were calculated using the log2 (FPKM). A heatmap was subsequently created using MeV 4.9 (Saeed et al. 2003).
Plant materials, total RNA extraction and qRT-PCR analysis
To validate the transcriptome data downloaded from public databases, we designed qRT-PCR primers using Zunla-1 (Capsicum annuum L.) seedlings. The seedlings at the six-leaf stage were subjected to heat (38 ℃) and NaCl (300 mM) treatments for 3, 6, and 12 h. Leaves harvested at 0 h were regarded as the controls (Wu et al. 2016). Eight plants were included in each group, and their leaves were collected from the same position on the seedlings with three biological replicates.
Total RNA was extracted from developing fruits using the EZNA™ Total RNA Kit I (Omega Biotek, Norcross, GA, USA) and subsequently treated with RNase-free DNase I (Takara, Dalian, China) to remove any genomic DNA contamination. First-strand cDNA was synthesized from the total RNA with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Bio Inc., Dalian, China) using random primers in accordance with the product manual. The TB Green™ Premix Ex Taq™ II kit (Takara Bio Inc., Dalian, China) was used for qRT-PCR analysis on the CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) following the manufacturers’ instructions. The PCR program was initiated for 30 s at 95 °C, followed by 40 cycles of 95 °C for 5 s and 55 °C for 30 s, and the program was completed with a melting curve analysis. CaActin (AY572427 in GenBank) was used as an internal control. The transcription level of each CaSnRK2 gene was calculated using the 2−ΔΔCq method as previously described (Livak and Schmittgen 2001). qRT-PCR was conducted for three technical replicates, and SPSS 18.0 (SPSS Inc., USA) was used for the statistical analysis. The primers used for the qRT-PCR analysis of the CaSnRK2 genes were designed using the Primer Premier 5.0 software.
Results
Pepper genome contains nine SnRK2 genes
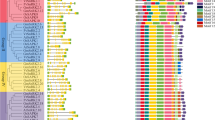
The first Blast search identified 22, 21 and 21 SnRK2 candidates in the Zunla-1, CM334 and chiltepin genomes, respectively. After genes with a percent identity < 50.00 were removed, 9, 8 and 9 suspected SnRK2 genes remained in the Zunla-1, CM334 and chiltepin genomes, respectively. All these protein sequences were downloaded for further study. A total of 9 full-length CaSnRK2 genes were ultimately confirmed in the cultivated pepper Zunla-1 (Capsicum annuum L.) and its wild progenitor chiltepin (Capsicum annuum var. glabriusculum). These genes were named CaSnRK2.1-CaSnRK2.9 according to their genomic positions. The lengths of the CaSnRK2 proteins ranged from 236 AA (CaSnRK2.4) to 331 AA (CaSnRK2.6), and the corresponding open reading frames (ORFs) ranged from 711 to 996 bp (sequences supplied in Supplemental File S1). The MWs ranged from 26.67 to 38.15 kDa, and the theoretical isoelectric point (pI) ranged from 4.46 (CaSnRK2.2) to 7.63 (CaSnRK2.5) (Table 1). All CaSnRK2s have a highly conserved N-terminal catalytic domain that contains an ATP-binding site and a protein kinase activation signature. The C-terminal regions of the CaSnRK2s are quite divergent and contain regions rich in the D/E AA (Fig. 1).
Multiple sequence alignment of the CaSnRK2 amino acid sequences. The CaSnRK2 proteins were aligned using the ClustalX2 program with the default settings. Identical amino acid residues are covered in black, similar residues are indicated in gray, and gaps in the sequences are indicated by dashes. The predicted functional domains are boxed. The ATP-binding domain, serine/threonine protein kinases active-site and the D/E rich domain in C-terminus are marked by red, blue and green box, respectively
CaSnRK2 positions and gene and protein structures
Based on the pepper genome sequence, nine CaSnRK2 genes were mapped onto different chromosomes (Supplemental Fig. 1). CaSnRK2.6 and CaSnRK2.7 were mapped onto chromosome 8, and CaSnRK2.1, CaSnRK2.2, CaSnRK2.3, CaSnRK2.4, CaSnRK2.5 and CaSnRK2.8 were mapped onto chromosomes 1, 2, 4, 5, 6 and 12, respectively. CaSnRK2.9 could not be mapped on any chromosome but on a pseudo-chromosome designated as Chr00.
To understand the structure of the CaSnRK2 genes, we compared the genomic DNA sequences to analyze the exon/intron organization using the online tool Gene Structure Display Server (GSDS) 2.0. The results revealed that pepper SnRK2 genes contained 4 to 8 introns, with four out of nine CaSnRK2 genes containing five introns. The detailed gene structure is presented in Fig. 2a.
The CaSnRK2 gene structures. a The intron/exon distribution in all CaSnRK2 genes. The black lines and red boxes indicate introns and exons, respectively. The blue boxes indicate upstream and downstream. b MEME-based analysis to investigate 10 conserved motifs in the CaSnRK2 proteins. Each colored box represents a conserved region. Details are provided in Supplementary Table 1
To further interpret the structural diversity of the SnRK2 proteins, 10 conserved motifs were predicted using MEME (Fig. 2b). The length of these motifs varied from 7 to 41 AAs; the specific AA sequences of each motif are also provided (Supplementary Table 1). Motifs 1, 2, 4, 6 and 10 were detected in all SnRK2 members. Furthermore, some conserved motifs in specific proteins were identified; for instance, motif 8 was detected only in CaSnRK2.1, CaSnRK2.5 and CaSnRK2.6.
Phylogenetic analysis
To further investigate the evolutionary relationships among the CaSnRK2 proteins and proteins in other plant species, a phylogenetic tree was constructed based on a multiple alignment of 64 selected full-length SnRK2 protein sequences (Supplemental File S2). These sequences included 10 AtSnRK2s from Arabidopsis (Arabidopsis thaliana), 10 OsSAPKs from rice (Oryza sativa Japonica), 10 ZmSnRK2s from corn (Zea mays), 9 MdSnRK2s from apple (Malus domestica), 8 StSnRK2s from potato (Solanum tuberosum), 8 SlSnRK2s from tomato (Solanum lycopersicum) and 9 CaSnRK2s from pepper (Capsicum annuum) (Supplementary Table 2). The phylogenetic tree showed that all SnRK2 proteins could be clustered into three subgroups (I, II and III) corresponding to the Arabidopsis SnRK2 gene classification (Hrabak et al. 2003). Subgroup I consisted of 4 members (CaSnRK2.1, CaSnRK2.5, CaSnRK2.6 and CaSnRK2.9). CaSnRK2.3, CaSnRK2.4 and CaSnRK2.8 belonged to subgroup II, and CaSnRK2.2 and CaSnRK2.7 were included in subgroup III (Fig. 3).
Phylogenetic analysis of SnRK2 members from Arabidopsis (Arabidopsis thaliana), potato (Solanum tuberosum), tomato (Solanum lycopersicum), apple (Malus domestica), corn (Zea mays), rice (Oryza sativa Japonica) and pepper (Capsicum annuum). The phylogenetic tree was constructed by MEGA 6.0 with the neighbor-joining (NJ) method and 1000 bootstrap replications. Values less than 50 were removed. Members in the same clade with a unique color belong to the same subgroup
Analysis of cis-elements in the promoter region of all CaSnRK2s
To better understand the possible functions of the CaSnRK2 genes, the likely cis-elements within the promoter region were identified using PlantCARE online tools (Supplementary Table 3). The cis-elements that responsive to the plant hormones and stresses were focused in this study. In total, 81 cis-acting elements were detected in the promoters of the CaSnRK2 genes, including 9 types of hormone-related elements and 7 stress-related elements (Fig. 4). With respect to the hormone responsive cis-elements, the abscisic acid-responsive elements (ABREs) were found to be the most abundant elements in 7 of the nine CaSnRK2s. Subgroup I CaSnRK2s (including CaSnRK2.1, CaSnRK2.5, CaSnRK2.6 and CaSnRK2.9), which were not induced by ABA, contained several ABREs in their promoter regions. Ethylene-responsive elements (EREs) and methyl jasmonate (MeJA)-responsive elements (the CGTCA and TGACG motifs, respectively) were also found in 5 and 6 CaSnRK2 genes, respectively. Other hormone-related cis-elements, such as salicylic acid-responsive elements (TCA elements), gibberellin-responsive elements (GARE motifs, P-boxes and the TATC-box), and auxin-responsive elements (the TGA-box and TGA elements), were also found in some CaSnRK2 genes.
With respect to stress-responsive cis-elements, heat stress elements (HSEs) and anoxic stress (GC motif)-related elements were detected only in CaSnRK2.2. The wound stress (WUN motif)-responsive element was the most frequent element within the 6 CaSnRK2 promoters. Moreover, one WUN motif was detected in CaSnRK2.1, CaSnRK2.2 CaSnRK2.3, CaSnRK2.5, CaSnRK2.6 and CaSnRK2.7. The CaSnRK2 promoter also contained other stress-responsive elements, such as an MBS (a MYB binding site involved in drought inducibility), ARE (a cis-acting regulatory element essential for anaerobic induction), W-box (a fungal elicitor-responsive element) and TC-rich repeat region (a defense-related and stress-responsive element). These results indicated that CaSnRK2 genes might be involved in transcriptional control of both the hormone and stress responses.
CaSnRK2 expression profiles in different tissues/organs and developing fruits
To understand the specific expression of CaSnRK2 genes in pepper, we analyzed the expression profiles of these genes in the roots, stems, leaves, buds, and flowers (including whole flowers, petals, ovaries and anthers) and developing fruits (which also included the pericarp, placenta and seeds) based on published transcriptome data (Fig. 5 and Supplemental Fig. 2), and their FPKM values are shown in Supplementary Table 4. Some genes, including CaSnRK2.2, CaSnRK2.3, CaSnRK2.4 and CaSnRK2.6, presented high transcript accumulation in all tissues. CaSnRK2.5 and CaSnRK2.8 were expressed at low levels in nearly all tissues, especially in the roots, stems and leaves. Some genes were expressed at relatively low levels in most tissues but at relatively high levels in specific tissues (e.g., CaSnRK2.1 in the roots and CaSnRK2.9 in the roots and buds). The expression patterns of the CaSnRK2 genes varied in the pepper tissues, implying multiple roles in pepper tissue development.
Expression abundance of CaSnRK2sin different tissues and at different developmental stages. The tissues include roots, stems, leaves, buds and flowers as well as whole fruits at different developmental stages. Dev1–5 indicate 0–1 cm, 1–3 cm, 3–4 cm, 4–5 cm, and mature green fruits, respectively. Dev6 indicates the breaker stage fruit. Dev7–9 indicate 3, 5, and 7 days after breaking. The bar in the upper left corner indicates the log2-corrected FPKM values, and the different colors indicate different expression levels. Red indicates relatively high expression, and green indicates relatively low expression. ZL1 indicates pepper (Capsicum annuum) cultivar Zunla-1
To further explore the functions of the CaSnRK2s during fruit development and ripening, we investigated their expression characteristics. CaSnRK2.2 and CaSnRK2.6 were both strongly expressed throughout the fruit development and ripening process. CaSnRK2.1, CaSnRK2.3 and CaSnRK2.7 were expressed at moderate levels, while CaSnRK2.5, CaSnRK2.8 and CaSnRK2.9 were expressed at low levels at all stages. During the fruit development and expansion stage (from Dev1 to Dev5), CaSnRK2.2, CaSnRK2.4, CaSnRK2.6 and CaSnRK2.7 expression continued to increase and peaked at the Dev3 stage, followed by a decreasing expression pattern. During the fruit color change process until ripening (from Dev5 to Dev9, the fruit size remained unchanged), the expression of most of the CaSnRK2s except CaSnRK2.5, CaSnRK2.8 and CaSnRK2.9 was strongly induced and peaked at the Dev7 stage, followed by a sharp decrease.
Expression patterns of CaSnRK2s in response to abiotic stress
Research has increasingly shown that SnRK2 genes are highly important for tolerance to various abiotic stresses. To further explore the biological functions of the CaSnRK2s under abiotic stresses (cold, oxidative, heat, drought, and salinity), available transcriptome data (Liu et al. 2017a) were gathered to construct heatmaps (Fig. 6), and the FPKM values of these genes are shown in Supplementary Table 5.
In the leaf tissue, two genes (CaSnRK2.5 and CaSnRK2.9) with nonexistent or extremely low expression (FPKM < 1) were removed from the subsequent analysis. Overall, the other seven members in the leaf tissue responded to at least one treatment (Fig. 6a). Among these members, CaSnRK2.1, CaSnRK2.3 and CaSnRK2.8 were significantly induced by four treatments. CaSnRK2.8 showed immediate transcript accumulation that peaked at 3 h in response to those treatments. CaSnRK2.1 responded after 6 or 12 h. CaSnRK2.7 appeared to be upregulated by H2O2, heat and mannitol. CaSnRK2.4 and CaSnRK2.6 expression was neither upregulated nor downregulated significantly under any of these stresses despite stress exposure varying from 0 to 24 h. This finding implied that these genes might not have any function in leaf tissue. Moreover, CaSnRK2.2 expression was downregulated at the early stage (from 0 to 3 h) in response to these stresses but not to a very high degree (fold change (FC), treatment FPKM/control FPKM < 2).
In the root tissue (Fig. 6b), the expression of CaSnRKs was time-dependent under different stresses, demonstrating that the responses of the plants to stress involved a complex process. Most CaSnRKs, including CaSnRK2.1, CaSnRK2.2, CaSnRK2.3, CaSnRK2.4, CaSnRK2.5 and CaSnRK2.7, were inducible at 1.5 h in the early stage, while CaSnRK2.8 expression was downregulated. CaSnRK2.5 exhibited a significantly high FC (> 10-fold) of upregulated expression under cold, mannitol and NaCl stresses, especially during the 6–24 h time period. With respect to different stresses, CaSnRK2.1 was significantly induced under cold stress, CaSnRK2.7 was induced under heat stress, and CaSnRK2.3 was induced under mannitol and NaCl stress. Some genes demonstrated interesting and unique expression profiles. For example, CaSnRK2.2 was induced only at 24 h under cold/heat stress (log2 FC > 1), while CaSnRK2.4 was repressed only at 3 or 6 h under mannitol/NaCl stress (log2 FC < -1), suggesting their potential functions in response to these stresses. In addition, the expression of one gene, CaSnRK2.6, did not vary in response to any of the tested stresses (|log2 FC| < 1).
To further validate the transcriptome data, qRT-PCR was employed to assess the expression patterns of these genes under heat and salt stress (Fig. 7). The primers used are listed in Supplementary Table 6. Apart from two genes (CaSnRK2.1 and CaSnRK2.9), the other members were all inducible by the two stresses. The expression profiles of most genes were basically consistent with the published data. CaSnRK2.2, CaSnRK2.3 and CaSnRK2.8 were all significantly upregulated in response to heat stress (p < 0.01), while the expression levels of CaSnRK2.6 and CaSnRK2.7 did not change significantly under the two stresses.
The expression profiles of CaSnRK2s under heat (a) and salt (b) stress viaqRT-PCR. Total RNA was extracted from leaves of six-leaf stage pepper seedlings. For heat and salt stress, seedlings were suffered to 38 ℃ and 300 mM NaCl for 0, 3, 6, and 12 h. Eight plants were included in each group, and their leaves were collected from the same position on the seedlings with three biological replicates. The characters on top of the error bars represent the significant difference (α < 0.05) compared to the control
Discussion
Plant perception of adversity and growth regulation are achieved through the signal transduction mechanism in vivo, and the key step of signal transduction is the phosphorylation and dephosphorylation by protein kinase and protein phosphatase. SnRK2 is a serine/threonine protein kinase unique to plants that has been cloned in Arabidopsis, rice, maize, tobacco and many other plants. SnRK2 responds to environmental signals, regulates the expression of related genes and protein activity, participates in a variety of signal transduction pathways in plants, and plays an important role in plant growth, development and stress resistance (Kulik et al. 2011; Rosenberger and Chen 2018). However, this gene family is poorly understood in pepper (Capsicum annuum). Fortunately, whole-genome sequencing data are available for pepper (Qin et al. 2014; Kim et al. 2014) and provide a great opportunity to explore this gene family at the genome level comprehensively.
Identification and characterization of SnRK2 genes in pepper
Thus far, comprehensive studies have been conducted on genome-wide identification of SnRK2 genes, and researchers have identified these genes in a variety of plants, such as 10 members in Arabidopsis thaliana (Hrabak et al. 2003), Oryza sativa (Kobayashi et al. 2004) and Zea mays (Huai et al. 2008), nine members in Malus domestica (Shao et al. 2014) and Fragaria ananassa (Han et al. 2015), 15 members in Brassica rapa ssp. chinensis (Huang et al. 2015), 20 members in Gossypium hirsutum (Liu et al. 2017b), etc. In the current study, a genome-wide identification revealed nine CaSnRK2 genes from the ‘CM334’ and ‘Zunla-1’ databases of pepper genome named CaSnRK2.1-CaSnRK2.9. Multiple sequence alignment results demonstrated that the N-terminus of this family contained a relatively conservative protein kinase catalytic domain while the C-terminus is relatively specific and consists of two parts, an activated domain responding to abiotic stress and an ABA AA regulatory domain (D/E). The result is similar to the discoveries for Malus prunifolia (Shao et al. 2014) and Solanum tuberosum (Bai et al. 2017). The SnRK2 gene number in pepper is very similar to those in the Arabidopsis, rice, apple and strawberry genomes but less similar to those in Brassica rapa and cotton. Brassica rapa has more SnRK2s than pepper due to the salicoid duplication and Brassica-specific whole genome duplication (WGD) events (Huang et al. 2015). Thus, gene duplication likely contributed more to pepper SnRK2 expansion than to genome size (pepper genome is 3.48 Gb).
Through the phylogenetic analysis of 64 SnRK2 genes from pepper, tomato, potato, apple, Arabidopsis, rice and maize, we conducted a comprehensive phylogenetic tree. Consistent with previous reports, the SnRK2 genes were divided into three groups (Hrabak et al. 2003; Bai et al. 2017), with each group including two to four pepper SnRK2 genes. The gene structure analysis revealed that CaSnRK2s contained four to eight introns. Similar characteristics have been reported in potato (Bai et al. 2017), Arabidopsis (Hrabak et al. 2003) and maize (Huai et al. 2008), suggesting their evolutionary conservation. The conserved motif identification showed an irregular distribution of 10 motifs in CaSnRK2s. Among them, motifs 1, 2, 4, 6 and 10 were in all SnRK2 members, motif 8 was only present in subgroup I, and motif 7 was unique to groups II and III. The results implied that they might contribute to the functional specificity of different groups, which requires deeper research.
Potential functions of CaSnRK2 genes in growth and development
The tissue-specific expression of SnRK2 genes provides insights into their potential function and has been investigated in many plants. In Arabidopsis, SnRK2.10 was predominantly expressed in the vascular tissue while SnRK2.4 was expressed throughout the roots. Both genes are involved in root growth and architecture (McLoughlin et al. 2012). SnRK2.8/SRK2C was expressed mainly in the roots, whereas SnRK2.7/SRK2F was expressed primarily in guard cells and vascular tissues of the roots, leaves and apical meristems (Mizoguchi et al. 2010). TaSnRK2.3 in wheat was strongly expressed in booting spindles but less so in the seedling roots, leaves and emerging spikes (Tian et al. 2013). The rice SAPK9 gene was most highly expressed in the leaves, and its expression was upregulated in response to drought stress and ABA treatment (Dey et al. 2016). In potato, StSnRK2.1, StSnRK2.2, StSnRK2.5 and StSnRK2.6 expression was highest in the roots while StSnRK2.3, StSnRK 2.7 and StSnRK 2.8 expression was significantly higher in the leaves and stems (Bai et al. 2017). In tomato, SlSnRK2.1 was expressed only in the roots while all the other SlSnRK2s were expressed in all vegetative organs (Sun et al. 2011). Here, we assessed expression data in different tissues and organs obtained from public databases. Most CaSnRKs exhibited moderate expression in different tissues and organs, although some genes showed very high or very low expression in certain tissues. For example, CaSnRK2.1 was expressed at relatively low levels in most tissues but at an especially high level in the roots, and CaSnRK2.9 was also expressed in the roots and buds, implying vital roles for these genes in the regulation of root and flower growth.
Fruit development and ripening is a complex process that involves a series of complex physiological and biochemical reactions, such as cell division, expansion, and aging. The phytohormone ABA is an important regulator of fruit ripening, and it is likely involved in the regulation of fleshy fruit development and ripening in both climacteric and nonclimacteric fruits (Hou et al. 2018; Liao et al. 2018; Sun et al. 2017; Forlani et al. 2019). SnRK2s act as the core components of the ABA-mediated signaling network, and their roles in fruit development and ripening have attracted considerable attention from researchers. In the nonclimacteric fruit strawberry, FaSnRK2.6, FaSnRK2.3 and FaSnRK2.5 expression declined throughout fruit growth and development. FaSnRK2.6 was proven to be a negative regulator of strawberry fruit development and ripening (Han et al. 2015). In tomato fruits, which are climacteric, SlSnRK2.2, SlSnRK2.3, SlSnRK2.4, and SlSnRK2C were expressed at high levels at all fruit maturity stages while SlSnRK2.1 and SlSnRK2.5 were expressed at low levels. The researchers speculated that SlSnRk2.3 and SlSnRK2.4 might play key roles in the regulation of fruit development and ripening at all maturity stages and that SlSnRK2.3 might play a more important role during the breaker stage (Sun et al. 2011). In cucumber fruits, which are also nonclimacteric, the absolute CsSnRK2.1 expression level was much greater than that of CsSnRK2.2 during all fruit developmental stages (Wang et al. 2012). To understand the function of CaSnRK in pepper fruit development, we analyzed their expression levels in nine developmental stages from fruit development to ripening. In our study, CaSnRK2.2 and CaSnRK2.6 were both strongly expressed throughout the fruit development and ripening process. Especially during the ripening process when the fruit turns color, the expression of most of the CaSnRK2s except SnRK2.5, SnRK2.8 and SnRK2.9 was strongly induced and peaked at the breaker stage (Dev7). These results demonstrated that variations in CaSnRK2 expression were associated with the pepper fruit development and ripening process. We can also presume that ABA signal transduction may be involved in regulation of fruit development and ripening at the transcriptional level.
Possible functions of CaSnRK2 members under abiotic stress
When plants are subjected to abiotic stresses, such as drought, high temperature, low temperature or salinity, the signals related to these stresses will be transformed into resistance factors through a series of signal transduction pathways. These resistance factors will activate the transcription factors related to adversity and combine with the cis-acting regulatory elements of corresponding genes to start the expression of corresponding genes and respond to adversity stress. Zhang et al. (2016) found a large number of cis-acting elements, such as ABRE, LTREs, ACGTATERD1 and DRE, in response to different hormones and abiotic stresses in the study of the SnRK2 gene family of wheat (Zhang et al. 2016). Similar results were also verified in potato (Bai et al. 2017) and cotton (Liu et al. 2017b). In this study, the results showed that almost all genes in the SnRK2 gene family of pepper contain ABRE cis-acting elements except CaSnRK2.4 and CaSnRK2.8. CaSnRK2.2 contains TGA, GARE, ERE, TCA, P-boxes, TATC-box, etc. and a total of 21 cis-acting elements, while other genes contain 3 to 11 elements. All genes in the SnRK2 gene family may be involved in the drought resistance stress response mechanism and wound or fungal elicitor-responses in plants, and CaSnRK2.2 is more comprehensively involved in stress and hormone responses.
Previous investigations have shown that SnRK2 genes are involved in various types of abiotic stresses and play an important role in regulation via ABA-dependent or ABA-independent signaling pathways (Yoshida et al. 2002; Tan et al. 2018; McLoughlin et al. 2012; Kobayashi et al. 2004; Ding et al. 2015; Soma et al. 2017). AtSnRK2.4 and AtSnRK2.10 are rapidly and transiently activated in Arabidopsis roots after exposure to salt (McLoughlin et al. 2012; Szymanska et al. 2019). In rice, all 10 SnRK2 members could be induced and activated by NaCl and the gene expression levels were significantly increased (Kobayashi et al. 2004). The expression profiling revealed that SAPK1 and SAPK2 expression was strongly induced by drought, NaCl, and polyethylene glycol (PEG) treatment but not by ABA treatment (Lou et al. 2018). Overexpression of SAPK4 or SAPK6 (OSRK1) could increase salt tolerance in rice (Diedhiou et al. 2008; Nam et al. 2012), while SAPK9 positively regulated ABA-mediated drought stress signaling pathways (Dey et al. 2016). In wheat (Triticum aestivum), TaSnRK2 expression was induced by water deficit and salt and cold stress, and in Arabidopsis, TaSnRK2.4 overexpression enhanced salt tolerance (Zhang et al. 2016). In potato (Solanum tuberosum), StSnRK2.4 expression was greater in the shoot tissues than root tissues and could be induced by NaCl, ABA and PEG, suggesting that StSnRK2.4 might be sensitive to stress signals and function in osmotic stress responses in potato shoot tissues (Bai et al. 2017). In cotton (Gossypium hirsutum), five GhSnRK2 genes (GhSnRK2.3/2.7/2.8/2.9/2.10) were notably upregulated under salt and PEG treatment (Liu et al. 2017b). In our study, seven SnRK2 members responded to at least one treatment in the leaves, with CaSnRK2.1, CaSnRK2.3 and CaSnRK2.8 significantly induced by four treatments: cold, H2O2, mannitol and NaCl. Roots seem to be more sensitive to stress because the level of gene-induced expression is significantly greater in those tissues than in other tissues. Most CaSnRKs, including CaSnRK2.1, CaSnRK2.2, CaSnRK2.3, CaSnRK2.4, CaSnRK2.5 and CaSnRK2.7, were inducible at the early stage. CaSnRK2.5 exhibited the greatest increase in expression (FC > 10-fold) under cold, mannitol and NaCl stresses. According to the phylogenetic analysis, CaSnRK2.7 is a very close homologue of Arabidopsis OST1/SnRK2.6 kinase, which is the main regulator of stomatal movements in response to stress (Liang et al. 2015). Therefore, CaSnRK2.7 should play a role similar to that of SnRK2.6 in leaves. Subsets of CaSnRK2 genes that exhibit different expression profiles in response to abiotic stress have been identified, and they indicate that CaSnRK2 plays a role in abiotic stress signaling in hot peppers.
In summary, pepper SnRK2 genes were analyzed at the genome-wide level in this study and nine CaSnRK2 genes were identified. Analyses of the gene structure, conserved motifs, cis-elements and expression patterns in different tissues and in response to different abiotic stresses revealed conserved and diverse characteristics of the members of the SnRK2 gene family. Among them, CaSnRK2.2 and CaSnRK2.6 may be involved in pepper fruit development and ripening process, while CaSnRK2.1, CaSnRK2.3, CaSnRK2.5 and CaSnRK2.7 may play important roles in abiotic stresses. This study lays a foundation for an improved understanding of the function of CaSnRK2s during pepper growth and development and response to stress.
References
Bai J, Mao J, Yang H, Khan A, Fan A, Liu S, Zhang J, Wang D, Gao H, Zhang J (2017) Sucrose non-ferment 1 related protein kinase 2 (SnRK2) genes could mediate the stress responses in potato (Solanum tuberosum L.). BMC Genet 18(1):41. doi:https://doi.org/10.1186/s12863-017-0506-6
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43(W1):W39–W49. doi:https://doi.org/10.1093/nar/gkv416
Carrizo Garcia C, Barfuss MH, Sehr EM, Barboza GE, Samuel R, Moscone EA, Ehrendorfer F (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot 118(1):35–51. doi:https://doi.org/10.1093/aob/mcw079
Coello P, Hirano E, Hey SJ, Muttucumaru N, Martinez-Barajas E, Parry MA, Halford NG (2012) Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J Exp Bot 63(2):913–924. doi:https://doi.org/10.1093/jxb/err320
Dey A, Samanta MK, Gayen S, Maiti MK (2016) The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol 16(1):158. doi:https://doi.org/10.1186/s12870-016-0845-x
Diedhiou CJ, Popova OV, Dietz KJ, Golldack D (2008) The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol 8:49. doi:https://doi.org/10.1186/1471-2229-8-49
Ding S, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought Tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27(11):3228–3244. doi:https://doi.org/10.1105/tpc.15.00321
Forlani S, Masiero S, Mizzotti C (2019) Fruit ripening: the role of hormones, cell wall modifications and their intersection with pathogens. J Exp Bot. doi:https://doi.org/10.1093/jxb/erz112
Fujii H, Verslues PE, Zhu JK (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci U S A 108(4):1717–1722. doi:https://doi.org/10.1073/pnas.1018367108
Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50(12):2123–2132. doi:https://doi.org/10.1093/pcp/pcp147
Fujita Y, Yoshida T, Yamaguchi-Shinozaki K (2013) Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant 147(1):15–27. doi:https://doi.org/10.1111/j.1399-3054.2012.01635.x
Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho TH (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13(3):667–679
Han Y, Dang R, Li J, Jiang J, Zhang N, Jia M, Wei L, Li Z, Li B, Jia W (2015) Sucrose nonfermenting1-related protein kinase2.6, an ortholog of open stomata1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol 167(3):915–930. doi:https://doi.org/10.1104/pp.114.251314
Hardie DG (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8(10):774–785. doi:https://doi.org/10.1038/nrm2249
Hardy TA, Huang D, Roach PJ (1994) Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem 269(45):27907–27913
Holappa LD, Walker-Simmons MK (1995) The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold Temperature, and osmotic stress. Plant Physiol 108(3):1203–1210
Hou BZ, Li CL, Han YY, Shen YY (2018) Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biol 18(1):162. doi:https://doi.org/10.1186/s12870-018-1377-3
Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132(2):666–680. doi:https://doi.org/10.1104/pp.102.011999
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. doi:https://doi.org/10.1093/bioinformatics/btu817
Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G (2008) Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep 27(12):1861–1868. doi:https://doi.org/10.1007/s00299-008-0608-8
Huang Z, Tang J, Duan W, Wang Z, Song X, Hou X (2015) Molecular evolution, characterization, and expression analysis of SnRK2 gene family in Pak-choi (Brassica rapa ssp. chinensis). Front Plant Sci 6:879. doi:https://doi.org/10.3389/fpls.2015.00879
Julkowska MM, McLoughlin F, Galvan-Ampudia CS, Rankenberg JM, Kawa D, Klimecka M, Haring MA, Munnik T, Kooijman EE, Testerink C (2015) Identification and functional characterization of the Arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant Cell Environ 38(3):614–624. doi:https://doi.org/10.1111/pce.12421
Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, Jung K, Lee GW, Oh SK, Bae C, Kim SB, Lee HY, Kim SY, Kim MS, Kang BC, Jo YD, Yang HB, Jeong HJ, Kang WH, Kwon JK, Shin C, Lim JY, Park JH, Huh JH, Kim JS, Kim BD, Cohen O, Paran I, Suh MC, Lee SB, Kim YK, Shin Y, Noh SJ, Park J, Seo YS, Kwon SY, Kim HA, Park JM, Kim HJ, Choi SB, Bosland PW, Reeves G, Jo SH, Lee BW, Cho HT, Choi HS, Lee MS, Yu Y, Do Choi Y, Park BS, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai HS, Ahn HK, Yeam I, Giovannoni JJ, Rose JK, Sorensen I, Lee SJ, Kim RW, Choi IY, Choi BS, Lim JS, Lee YH, Choi D (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46(3):270–278. doi:https://doi.org/10.1038/ng.2877
Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16(5):1163–1177. doi:https://doi.org/10.1105/tpc.019943
Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases—key regulators of plant response to abiotic stresses. OMICS 15(12):859–872. doi:https://doi.org/10.1089/omi.2011.0091
Kulik A, Anielska-Mazur A, Bucholc M, Koen E, Szymanska K, Zmienko A, Krzywinska E, Wawer I, McLoughlin F, Ruszkowski D, Figlerowicz M, Testerink C, Sklodowska A, Wendehenne D, Dobrowolska G (2012) SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol 160(2):868–883. doi:https://doi.org/10.1104/pp.112.194472
Lee HJ, Park YJ, Seo PJ, Kim JH, Sim HJ, Kim SG, Park CM (2015) Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 27(12):3425–3438. doi:https://doi.org/10.1105/tpc.15.00371
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li C, Nong Q, Xie J, Wang Z, Liang Q, Solanki MK, Malviya MK, Liu X, Li Y, Htun R, Wei J, Li Y (2017) Molecular characterization and co-expression analysis of the SnRK2 gene family in sugarcane (Saccharum officinarum L.). Sci Rep 7(1):17659. doi:https://doi.org/10.1038/s41598-017-16152-4
Liang S, Lu K, Wu Z, Jiang SC, Yu YT, Bi C, Xin Q, Wang XF, Zhang DP (2015) A link between magnesium-chelatase H subunit and sucrose nonfermenting 1 (SNF1)-related protein kinase SnRK2.6/OST1 in Arabidopsis guard cell signalling in response to abscisic acid. J Exp Bot 66(20):6355–6369. doi:https://doi.org/10.1093/jxb/erv341
Liao X, Li M, Liu B, Yan M, Yu X, Zi H, Liu R, Yamamuro C (2018) Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc Natl Acad Sci USA 115(49):E11542–E11550. doi:https://doi.org/10.1073/pnas.1812575115
Lin Q, Wu F, Sheng P, Zhang Z, Zhang X, Guo X, Wang J, Cheng Z, Wang J, Wang H, Wan J (2015) The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat Commun 6:7981. doi:https://doi.org/10.1038/ncomms8981
Liu F, Yu H, Deng Y, Zheng J, Liu M, Ou L, Yang B, Dai X, Ma Y, Feng S, He S, Li X, Zhang Z, Chen W, Zhou S, Chen R, Liu M, Yang S, Wei R, Li H, Li F, Ouyang B, Zou X (2017a) PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol Plant 10(8):1129–1132. doi:https://doi.org/10.1016/j.molp.2017.03.005
Liu Z, Ge X, Yang Z, Zhang C, Zhao G, Chen E, Liu J, Zhang X, Li F (2017b) Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet 18(1):54. doi:https://doi.org/10.1186/s12863-017-0517-3
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. doi:https://doi.org/10.1006/meth.2001.1262
Lou D, Wang H, Yu D (2018) The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol 18(1):203. doi:https://doi.org/10.1186/s12870-018-1408-0
McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Lauriere C, Munnik T, Haring MA, Testerink C (2012) The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J 72(3):436–449. doi:https://doi.org/10.1111/j.1365-313X.2012.05089.x
Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2010) Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol 51(5):842–847. doi:https://doi.org/10.1093/pcp/pcq041
Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD (2005) The SOL genomics network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138(3):1310–1317. doi:https://doi.org/10.1104/pp.105.060707
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50(7):1345–1363. doi:https://doi.org/10.1093/pcp/pcp083
Nam MH, Huh SM, Kim KM, Park WJ, Seo JB, Cho K, Kim DY, Kim BG, Yoon IS (2012) Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci 10:25. doi:https://doi.org/10.1186/1477-5956-10-25
Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu Z, Mao L, Wu H, Ling-Hu C, Zhou H, Lin H, Gonzalez-Morales S, Trejo-Saavedra DL, Tian H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Bi S, Yang X, Li W, Cai H, Luo X, Montes-Hernandez S, Leyva-Gonzalez MA, Xiong Z, He X, Bai L, Tan S, Tang X, Liu D, Liu J, Zhang S, Chen M, Zhang L, Zhang L, Zhang Y, Liao W, Zhang Y, Wang M, Lv X, Wen B, Liu H, Luan H, Zhang Y, Yang S, Wang X, Xu J, Li X, Li S, Wang J, Palloix A, Bosland PW, Li Y, Krogh A, Rivera-Bustamante RF, Herrera-Estrella L, Yin Y, Yu J, Hu K, Zhang Z (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci USA 111 (14):5135–5140. doi:https://doi.org/10.1073/pnas.1400975111
Rosenberger CL, Chen J (2018) To grow or not to grow: TOR and SnRK2 coordinate growth and stress response in Arabidopsis. Mol Cell 69(1):3–4. doi:https://doi.org/10.1016/j.molcel.2017.12.013
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34(2):374–378. doi:https://doi.org/10.2144/03342mt01
Shao Y, Qin Y, Zou Y, Ma F (2014) Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 552(1):87–97. doi:https://doi.org/10.1016/j.gene.2014.09.017
Shen X, Guo X, Zhao D, Zhang Q, Jiang Y, Wang Y, Peng X, Wei Y, Zhai Z, Zhao W, Li T (2017) Cloning and expression profiling of the PacSnRK2 and PacPP2C gene families during fruit development, ABA treatment, and dehydration stress in sweet cherry. Plant Physiol Biochem 119:275–285. doi:https://doi.org/10.1016/j.plaphy.2017.08.025
Soma F, Mogami J, Yoshida T, Abekura M, Takahashi F, Kidokoro S, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2017) ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat Plants 3:16204. doi:https://doi.org/10.1038/nplants.2016.204
Song X, Yu X, Hori C, Demura T, Ohtani M, Zhuge Q (2016) Heterologous overexpression of poplar SnRK2 genes enhanced salt stress tolerance in Arabidopsis thaliana. Front Plant Sci 7:612. doi:https://doi.org/10.3389/fpls.2016.00612
Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, Brunzelle JS, Zhang H, Yang H, Jiang H, Li J, Yong EL, Cutler S, Zhu JK, Griffin PR, Melcher K, Xu HE (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335(6064):85–88. doi:https://doi.org/10.1126/science.1215106
Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P (2011) Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot 62(15):5659–5669. doi:https://doi.org/10.1093/jxb/err252
Sun Y, Ji K, Liang B, Du Y, Jiang L, Wang J, Kai W, Zhang Y, Zhai X, Chen P, Wang H, Leng P (2017) Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. Plant J 91(4):574–589. doi:https://doi.org/10.1111/tpj.13588
Szymanska KP, Polkowska-Kowalczyk L, Lichocka M, Maszkowska J, Dobrowolska G (2019) SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int J Mol Sci. doi:https://doi.org/10.3390/ijms20010143
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:https://doi.org/10.1093/molbev/mst197
Tan W, Zhang D, Zhou H, Zheng T, Yin Y, Lin H (2018) Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet 14(4):e1007336. doi:https://doi.org/10.1371/journal.pgen.1007336
Tian S, Mao X, Zhang H, Chen S, Zhai C, Yang S, Jing R (2013) Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J Exp Bot 64(7):2063–2080. doi:https://doi.org/10.1093/jxb/ert072
Wang Y, Wu Y, Duan C, Chen P, Li Q, Dai S, Sun L, Ji K, Sun Y, Xu W, Wang C, Luo H, Wang Y, Leng P (2012) The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J Plant Physiol 169(18):1874–1882. doi:https://doi.org/10.1016/j.jplph.2012.07.017
Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110(27):11205–11210. doi:https://doi.org/10.1073/pnas.1308974110
Wang X, Wang L, Wang Y, Liu H, Hu D, Zhang N, Zhang S, Cao H, Cao Q, Zhang Z, Tang S, Song D, Wang C (2018) Arabidopsis PCaP2 plays an important eole in chilling tolerance and ABA response by activating CBF- and SnRK2-mediated transcriptional regulatory network. Front Plant Sci 9:215. doi:https://doi.org/10.3389/fpls.2018.00215
Wu Z, Cheng J, Cui J, Xu X, Liang G, Luo X, Chen X, Tang X, Hu K, Qin C (2016) Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front Plant Sci 7:574. doi:https://doi.org/10.3389/fpls.2016.00574
Yang L, Ji W, Gao P, Li Y, Cai H, Bai X, Chen Q, Zhu Y (2012) GsAPK, an ABA-activated and calcium-independent SnRK2-type kinase from G. soja, mediates the regulation of plant tolerance to salinity and ABA stress. PLoS One 7(3):e33838. doi:https://doi.org/10.1371/journal.pone.0033838
Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43(12):1473–1483
Yu LJ, Luo YF, Liao B, Xie LJ, Chen L, Xiao S, Li JT, Hu SN, Shu WS (2012) Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol 195(1):97–112. doi:https://doi.org/10.1111/j.1469-8137.2012.04154.x
Zhang H, Li W, Mao X, Jing R, Jia H (2016) Differential activation of the wheat SnRK2 family by abiotic stresses. Front Plant Sci 7:420. doi:https://doi.org/10.3389/fpls.2016.00420
Zhao Y, Zhang Z, Gao J, Wang P, Hu T, Wang Z, Hou YJ, Wan Y, Liu W, Xie S, Lu T, Xue L, Liu Y, Macho AP, Tao WA, Bressan RA, Zhu JK (2018) Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep 23(11):3340–3351 e3345. doi:https://doi.org/10.1016/j.celrep.2018.05.044
Zheng Z, Xu X, Crosley RA, Greenwalt SA, Sun Y, Blakeslee B, Wang L, Ni W, Sopko MS, Yao C, Yau K, Burton S, Zhuang M, McCaskill DG, Gachotte D, Thompson M, Greene TW (2010) The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol 153(1):99–113. doi:https://doi.org/10.1104/pp.109.150789
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31672162 and 31701921), the Natural Science Foundation of Guangdong Providence (2018A030313020), the Guangzhou Science and Technology Program (201704020019) and the Zunyi City Science and Technology Plan Program of China (201612). We thank AJE (American Journal Experts) for its English editing during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Data curation, JC and CQ; Formal analysis, JC and XX; Funding acquisition, ZW and KH; Methodology, JC; Project administration, ZW and KH; Resources, XX; Software, FH and CQ; Supervision, KH; Visualization, FH; Writing-original draft, ZW; Writing-review and editing, CQ and KH. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13258_2020_968_MOESM2_ESM.docx
Supplementary File S2. The 64 full-length SnRK2 protein sequences used to construct the phylogenetic tree. These sequences included those of 10 AtSnRK2s from Arabidopsis thaliana, 10 OsSAPKs from Oryza sativa Japonica, 10 ZmSnRK2s from Zea mays, 9 MdSnRK2s from Malus domestica, 8 StSnRK2s from Solanum tuberosum, 8 tomato (Solanum lycopersicum) and 9 CaSnRK2s from Capsicum annuum (docx 22 kb)
13258_2020_968_MOESM9_ESM.pdf
Supplementary Figure 1. Chromosomal locations of the 9 predicted CaSnRK2 genes. The chromosome number is marked on each chromosome. The red/green arrows next to the genes indicate the direction of their transcription (+/-, respectively) (DOC 229 kb)
13258_2020_968_MOESM10_ESM.pdf
Supplementary Figure 2. Expression abundance of CaSnRK2s in flowers and fruits, including the pericarp, placenta and seeds. P10 indicates the petal, O10 indicates the ovary with stigma, and STA10 indicates the stamen. FST0 indicates fruit at 3 days after pollination (DAP), and FST1 indicates fruit at 7 DAP (DOC 48 kb)
Rights and permissions
About this article
Cite this article
Wu, Z., Cheng, J., Hu, F. et al. The SnRK2 family in pepper (Capsicum annuum L.): genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genom 42, 1117–1130 (2020). https://doi.org/10.1007/s13258-020-00968-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-020-00968-y