Abstract
Background
Fruit abscission in an isolated region called abscission zone (AZ) is regulated by several genes including JOINTLESS, MACROCALYX and SEPALLATA, MADS-box genes, in tomato.
Objective
The surviving central pedicels and the abscised lateral pedicels were examined in fruit clusters in order to investigate apple MADS-box genes from fruit pedicels of self-abscising apple ‘Saika’ during early fruit abscission.
Methods
After performing RNA-Seq, transcription profiling was conducted on the MADS-box genes from apple central and lateral pedicels. The JOINTLESS homolog of apple (MdJOINTLESS) was amplified using degenerate primers annealing to a highly conserved domain based on the orthologous genes of various crops, including JOINTLESS gene of tomato. The expression pattern of MdJOINTLESS was investigated in central and lateral pedicles by real-time PCR.
Results
Some homologs were found which similar to JOINTLESS, MACROCALYX and SEPALLATA of tomato MADS-box genes from transcriptome analysis and RACE. Using phylogenetic analyses with the MADS-box gene family, MdJOINTLESS was classified into the SHORT VEGETATIVE PHASE (SVP) clade that included Arabidopsis and other crops. The expression level of MdJOINTLESS in central pedicel was more than twice as high as that of lateral pedicel.
Conclusion
In the current study, we could find apple homologs of JOINTLESS, MACROCALYX, SEPALLATA, which were known to regulate pedicel AZ development in tomato. Furthermore, MdJOINTLESS might contribute to auxin gradation, influencing hierarchical ranking of auxin transport between fruit pedicels of self-abscising apple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fruit abscission typically occurs in a developmentally separated region called the abscission zone (AZ). The AZ, formed at the junction between the fruit and the branch of the plant, is composed of six to eight layers of small, square, and densely packed cells (Sexton and Roberts 1982; Sun et al. 2009). The cells in the AZ are arrested during growth and development and arranged in transverse sections relative to the adjacent vascular cells (Sun et al. 2009). Moreover, the treachery elements in the AZ are less developed and their lignins are less abundant (Patterson 2001). Below the AZ, the protective layers are formed in the proximal region of the peduncle. Abscission is processed by the expanded cells of the AZ and the immature vascular system that does not support the pedicels and fruits.
AZ differentiation is known to be regulated by several genes and transcription factors. In tomato, the LATERAL SUPRRESSOR gene, which encodes a member of VHIID protein family, is required for AZ development in the pedicel (Schumacher et al. 1999). The MADS-box protein, JOINTLESS (J) also plays a crucial role in the development of the AZ in the tomato (Mao et al. 2000) and interacts with the other MADS-box protein, such as MACROCALYX (MC) and SlMBP21, forming protein complexes that regulate AZ formation (Liu et al. 2014; Nakano et al. 2012). SlMBP21 is thought to attach J to MC (AP1 family), as a glue, and was reported that SlMBP21 belongs to SEPALLATA (SEP) family in the quartet model describing floral organ formation (Liu et al. 2014).
In Arabidopsis, the double mutant lacking the transcription factors BLADE-ON-PETIOLE 1 (BOP1) and BOP2 failed to abscise the floral organs, even after senescence and wilting (McKim et al. 2008). BOP1 and BOP2 encode BTB/POZ domain and ankyrin repeat-containing proteins, which redundantly regulate AZ differentiation in floral organs (McKim et al. 2008). The INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptide ligand is known to trigger abscission mechanism by the receptor-like kinases, HAESA (HAE) and HAESA-LIKE2 (HSL2) (Butenko et al. 2003; Jinn et al. 2000; Stenvik et al. 2008), and the signal is transduced into the nucleus by mitogen activating protein kinase cascade (Cho et al. 2008). Shi et al. (2011) reported that KNOTTED-like homeobox (KNOX) genes may regulate transcription of genes related to cell separation. AUXIN RESISTANT 1 (AUX1), LIKE AUX1 (LAX1), and LAX3 were strongly expressed at the site for floral organ abscission and the AXR3 gene, which induced in Aux/IAA transcription factor family, was interfered with auxin signaling at the AZ (Basu et al. 2013), suggesting the involvement of auxin signaling in abscission (Heo et al. 2016).
In rice, differentiation of the pedicel AZ is regulated by SHATTERING 4 (SH4) (Li et al. 2006), qSH1 (Konishi et al. 2006), and SHATTERING ABORTION 1 (SHAT1) (Zhou et al. 2012). SH4 and qSH1 both activate AZ formation at the pedicel. SH4 encodes a Myb3 DNA-binding domain-containing protein (Li et al. 2006), whereas qSH1 is a BELL homeobox gene such as REPLUMLESS (or VAAMANA) (Konishi et al. 2006). SHAT1 belongs to the AP2 transcription factor family and is involved in rice grain shattering (Zhou et al. 2012).
In apple, the genes related to polar auxin transport might be involved in abscission induction during the shedding of immature fruit (Dal Cin et al. 2009a, b). Dal Cin et al. (2009a) reported that MdLAX1, MdLAX2, MdPIN1, and MdPIN10 genes may be responsible for the abscission induction and the expression level of auxin hydrogen symporter (AHS) gene was lower in the cortex of abscising fruit (Dal Cin et al. 2009b). Our previous study revealed that IAA3/SHY2 or IAA14/SLR were differentially expressed in the surviving central fruits or the abscised lateral fruits (Heo et al. 2016). The Aux/IAA genes which were related to auxin signal transduction might participate in fruit pedicel abscission by performing comparative analysis using RNA-Seq and validating the expression profiles by qPCR (Heo et al. 2016).
In terms of MADS-box genes, Nakano et al. (2015) reported three J homologs exist in apple ‘Fuji’ genome. MdJa1, 2 and MdJb, respectively, interacted with tomato MC and SlMBP21 (SEP), forming a tetramer that act as transcription factor for regulating pedicel AZ specification or development. MdJb complemented the j mutant phenotype in tomato, inducing AZ development. However, MdJa did not restore the J deficient phenotype, showing incomplete AZ structure in pedicel and the genes that J induced in wild type tomato were not expressed, such as polygalacturonase, cellulase and AZ-specific transcription factors (Nakano et al. 2015).
Fruit trees generally produce excessive fruitlets to the extent that trees can not support the fruit load (Heo et al. 2015). Moreover, most apple cultivars bear five flowers in a flower cluster and remain to grow not having any abscission in their pedicels. Therefore, fruit thinning is an essential practice for producing apples that have commercial quality. Chemical thinning has been widely used with similar substance to plant hormones, such as auxin or ethylene. By spraying chemicals, AZ cells perceive an abscission-stimulating signal and cell adhesion at AZ finally become to be loosened through cell wall remodeling mechanism, activating ethylene-induced enzymes (Nakano et al. 2015; Roberts et al. 2002). However, thinning chemicals have harmful side effects on trees, environment and even fruits, causing resetting, and malformation. In Korea, hand thinning is generally practiced to obtain bigger and sweeter fruits, but it is still labor- and time-intensive work. Therefore, the breeding of self-thinning cultivars or development of new thinning chemical based on abscission mechanism can be an effective alternatives and suit modern apple production for low labor input.
Apple cultivars were classified into three groups as a non-abscising, a June drop, and a self-abscising, according to the fruit abscission patterns from full bloom (FB) to 30 days after full bloom (DAFB) (Heo et al. 2015). Unlike June drop and pre-harvest drop, the self-abscission characteristics shows the only one central fruit remains to grow and the other lateral fruits are abscised in a fruit cluster. Here, we validated the MADS-box genes reported from Nakano et al. (2015) in different apple cultivar, which were expressed in fruit pedicels of self-abscising apple during early fruit abscission. The comparative and phylogenetic analyses were also performed to identify the apple MADS-box (MdMADS) genes which might be involved in the AZ development.
Materials and methods
Plant materials
Ten-year-old, self-abscising ‘Saika’ apple (Malus × domestica Borkh.) trees, grafted on M.9 rootstocks were grown at the National Institute of Horticultural and Herbal Science, Korea. All flowers were artificially pollinated with mixed pollens collected from various cultivars to avoid the self-incompatibility resulting from the same S-genotypes. The central pedicels (CP) and lateral pedicels (LP) in clusters were collected from FB to 20 DAFB. They were immediately frozen in liquid nitrogen and stored at − 80 °C until use.
Microscopic observation
The pedicel tissues including AZ were prepared under a light microscope (Axioskop 2, Carl Zeiss Inc., Oberkochen, Germany) and the specimen fixation, post-fixation treatment, and observation were performed as described by Heo et al. (2016).
Genomic DNA and RNA extraction
Frozen pedicel tissue samples were pulverized with a mortar and pestle using liquid nitrogen. One gram of frozen sample was suspended in 2.5 mL of extraction buffer consisting of 2% (w/v) cetyltrimethyl ammonium bromide (CTAB), 100 mM Tris–Cl (pH 8.0), 20 mM EDTA, 1.42 M NaCl, 5 mM ascorbic acid, and 2% (w/v) polyvinyl pyrrolidone. Genomic DNA was isolated according to the method of Heo et al. (2016), with minor modifications. Approximately 100 mg of powdered tissue was used to extract total RNA using to the CTAB method (Chang et al. 1993). The extracted RNA was precipitated, and the resulting pellet was resuspended in diethylpyrocarbonate-treated water. RNA quality was assessed by electrophoresis on an 1% agarose gel and quantified using a NanoDrop ND-1000 spectrophotometer (ThermoScientific, Waltham, MA, USA).
Pyrosequencing, trimming, assembly, and annotation of the pedicel transcriptome
As described in our previous research (Heo et al. 2016), the pedicel transcriptome was generated by pyrosequencing on Roche 454 GS-FLX sequencer (Basel, Switzerland). The reads were mapped against Malus×domestica reference sequences from Phytozome (http://www.phytozome.net). The expression values were normalized to reads per kilobase of exon model per million (RPKM) mapped read values and the contigs related to MADS-box genes were screened by annotation informations.
Identification of JOINTLESS homologue, MdJOINTLESS
For isolation of the JOINTLESS homologue, MdJOINTLESS, 1 μg of total RNA was subjected to first-strand cDNA synthesis using T (18) primers and PowerScript reverse transcriptase (Clontech, Mountain View, CA, USA). For second-strand cDNA synthesis, two degenerate primers were designed using the J-CODEHOP program targeting the conserved amino acid sequences MAREKIQIK from the MADS-box domain and RQMRGEDLQG from the K-box region. The sequences of the primers were 5′-ATGGCKAGAGARAARATTMAGATMAAGAA-3′ and 5′-CCTTGRAGHTCYTCHCCYCTCATYTGCCT-3′. Second-strand cDNA synthesis was performed according to the following procedure: initial denaturation at 94 °C for 4 min, 35 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 3 min, and a final extension for 5 min at 72 °C. A single DNA band was detected by agarose gel electrophoresis and was extracted using the QIAquick gel extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The extracted DNA was cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) according to the manufacturer’s instructions, and sequenced. Rapid amplification of cDNA ends (RACE) was performed using the SMARTer RACE cDNA amplification kit (Clontech) with adaptors and specific primers that were based on a partial cDNA sequence. The polymerase chain reaction (PCR) products were cloned and sequenced as described above.
Quantitative real-time reverse-transcription PCR
The cDNA was synthesized from 1 μg of total RNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Real-time PCR reactions were performed with Applied Biosystems 7500 Fast Real-Time PCR System and TaqMan Universal PCR Master Mix (Applied Biosystems). The mixture is composed of a 20 μl reaction solution containing 10 μl 2× master mix, 7 μl ddH2O, 2 μl of template, and 1 μl 20× TaqMan assay mix. The primer and probe sequence for MdJOINTLESS gene are as follows: F: GGGATGCAATTGATGGAAGAGAATG, R: GCCTCCGGCCATCAGA TTT and TaqMan probe: FAM-TCCGCCACTTGCTGTC-TAMRA. Transcript values normalized and calibrated relative to Malus × domestica actin (CN935584) protein as a housekeeping internal standard gene according to the 2−ΔΔCT method (Livak and Schmittgen 2001). The sequences are: F: GCCTCCAGAGTATATGCCAGAGAAT, R: TCTGCTTTTGTTATA TGTTGGTTTTGTGG and TaqMan probe: FAM-CAAGTATGGCACCT CCC-TAMRA. Three biological and technical replicates were used in this experiment.
Phylogenetic analysis
To survey the phylogenetic relationship of MdMADS proteins with those of other plants, the phylogenetic tree was constructed using MEGA X. Dendrograms were generated using neighbor-joining method with default parameters. The MADS-box genes of other crops were assorted with MdMADS genes reported by Velasco et al. (2010).
Results
AZ of self-abscising apple ‘Saika’
As described in Fig. 1, apple cultivars which have the self-abscission characteristics retain their central fruits and abscise lateral fruits within 30 DAFB (Heo et al. 2015). During early fruit abscission, the AZ was developed, and the abscission process occurred in only lateral pedicel of the self-abscising apple cultivar ‘Saika’ (Fig. 1). At 20 DAFB in the LP, the AZ was developed at the junction between the pedicel and the peduncle, with the protective layers forming immediately below the AZ (Fig. 2a). The AZ was composed of six to eight layers of small, square, and densely packed cells (Fig. 2a). The cells of the AZ were transversely arranged relative to vascular system and adjacent cells (Fig. 2b).
The abscission zone (AZ) developed at the pedicel of the lateral fruits in the self-abscising apple ‘Saika’. The AZs located between the pedicel and peduncle were manually dissected from longitudinal sections (a). Scanning electron micrograph of the pedicel AZ (b). The square cell layers are seen at the lower right corner. These cells were transversely arranged relative to the adjacent cells. Scale bar: 100 μm in a; 50 μm in b. Each parts of the fruit cluster is designated by arrowheads
Hierarchical clustering analysis
In the previous research (Heo et al. 2016), the self-abscising cultivar ‘Saika’ was used for RNA-Seq and differentially expressed genes analysis between the surviving central pedicels and the abscised lateral pedicels. Total 65,876 contigs were assembled and mapped to apple reference transcriptome. Among the assembled transcripts, MADS-box genes were screened and assigned according to hierarchical clustering (Fig. 3). Twenty-one contigs containing MdMADS genes were found to be expressed in fruit pedicels, while other MADS-box genes exhibit no expression by RNA-Seq. MdMADS genes were divided into three groups, with six genes each appearing specifically in CP and LP. The gene that expressed in both CP and LP generally belonged to AG family. In this study, apple MC homolog (MdMC) was MDP0000269921 and SlMBP21 (MdSEP) homolog was MDP0000326390 (Fig. 3). MdMC gene was differentially expressed in LP, but MdSEP gene was similarly expressed in both pedicels.
Isolation and sequence analysis of J homolog from apple fruit pedicels
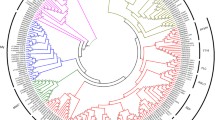
The full-length protein sequences of MADS-box proteins of Arabidopsis, field mustard, sweet potato, bean, and tomato were aligned using ClustalW2 for designing degenerate primers to discover J-like proteins in apple (Fig. 4a). The reverse transcription (RT)-PCR produced a single cDNA band at the expected size of 350 bp, from which full-length cDNA was reconstructed using 3′-RACE. The final product, named MdJOINTLESS (MdJ, GenBank accession No. ABD66219.2), is a MADS-box gene isolated from the pedicel tissue of young fruit. MdJ showed the highest sequence similarity to members of the SHORT VEGETATIVE PHASE (SVP) family of MADS-box genes (Fig. 4b). The MdJ gene encodes a putative 224 amino acid protein that is 77% and 78% identical to J (tomato) and IbMADS3 (sweet potato), respectively (Fig. 3b). The tree was divided into some distinct clades according to the representing MdMADS genes with those of other crops, such as Arabidopsis and tomato. The orthologs of SVP gene are known to be repressors of floral transition during the vegetative phase (Hartmann et al. 2000), and after the floral transition, they are expressed in the floral meristem (Gregis et al. 2013).
Multiple alignment (a) and phylogenetic analysis (b) of the deduced amino acid sequence of MdJOINTLESS with other MADS-box protein homologs. a Comparison of MdJOINTLESS with SVP from Brassica campestris (DQ922944), IbMADS3 from Ipomoea batatas (AB054255), SVP from Arabidopsis thaliana (NP_179840), SVP from Pisum sativum (AY830919), and JOINTLESS from Solanum lycopersicum (AF275345). Dark boxes indicate amino acid identity or similarity between all six sequences, whereas grey boxes indicate amino acid identity or similarity among any four sequences. b An unrooted tree showing the phylogenetic relationships between MdJOINTLESS and MADS-box proteins from apple (Malus × domestica Borkh.) and other plants, including: StMADS16 (AF008651) from Solanum tuberosum; MdMADS1 (AAC25922), MdMADS2 (AAC83170), MdMADS3 (AAD51422), MdMADS4 (AAD51423), MdMADS5 (CAA04321), MdMADS6 (CAA04322), MdMADS7 (CAA04323), MdMADS8 (CAA04919), MdMADS10 (CAA04324), MdMADS11 (CAA04325), MdMADS12 (CAC86183), MdMADS13 (CAC80856), MdMADS14 (CAC80857), MdMADS15 (CAC80858), MdMADS16 (AB370212), MdTM6 (AB081093), MdPI (CAC28021), MdAGAMOUS (AAQ03090), MdAP1 (AAL61543), and MdSOC1 (DQ887181). The phylogenetic tree was generated in MEGA X using the neighbour-joining method with the P-distance amino acid substitution model and 1000 bootstrap replicates
Analysis of expression pattern of MdJ genes in pedicels during early fruit abscission
The DNA gel blot revealed a single hybridization band with EcoRV and two bands with HindIII (data not shown). These data suggest that apple genome has J homologs. Although ‘Saika’ is a triploid apple, it seems to be common that low-copy-number of MdJ is in diploid apple cultivars. Quantification of expression levels of MdJ gene was performed through quantitative real-time PCR analysis in CP and LP of self-abscising apple (Fig. 5). Based on normalization of gene expression to that of MdActin, MdJ was expressed higher in CP than in lateral fruit at FB. MdJ was detectable in both pedicels at 10 DAFB and its expression levels were lower than those at FB. However, the relative expression of MdJ was higher in CP than in LP (Fig. 5). In LP, the expression of MdJ showed similar level from FB to 10 DAFB, while, in CP, the expression level at 10 DAFB was the half of that at FB. According to the quantitative real-time polymerase chain reaction (RT-qPCR) result, MdJ might not be involved in formation of AZ in apple. The highest expression level of MdJ was found actually in CPs, although its expression should have up-regulated in LPs.
Relative expression levels of MdJOINTLESS gene between central pedicel (CP) and lateral pedicel (LP) of self-abscising apple ‘Saika’ from full bloom (FB) to 10 days after full bloom (DAFB). Data were normalized using housekeeping MdActin gene. Vertical bars are the standard errors of the means. Statistical analysis was performed using Student’s T test (***p < 0.001, NS, not significant). The primers and probe sequences are listed in “Materials and methods”
Phylogenetic analysis with MdMADS and MADS-box genes from other crops
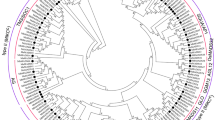
MADS-box genes contribute to reproductive development by regulating flowering timing and floral organogenesis (Theissen and Saedler 2001; Weigel 1995). However, several genes are also involved in vegetative development, including the regulation of the AZ (Liu et al. 2014; Mao et al. 2000; Nakano et al. 2012). The MADS-box genes in pedicel transcriptome from ‘Saika’ were compared to the MdMADS genes reported from apple genome sequencing project (Velasco et al. 2010) (Fig. 6).
Phylogenetic tree of apple MADS-box genes, using the neighbour-joining method. The MADS-box gene or transcripts which expressed in central pedicel, lateral pedicel, and both pedicels marked with triangle, star, and square shape, respectively. The MADS-box genes, except apple transcripts, have the abbreviation of scientific name in front of their gene name: At, Arabidopsis thaliana; Sl, Solanum lycopersicum; Md, Malus × domestica. The abbreviation for MADS-box clades are as follows: SEP SEPALLATA, AP1 APETALA1, AGL AGAMOUS-LIKE GENE, SOC1 SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, AG AGAMOUS, FLC FLOWERING LOCUS C, DAM DORMANCY ASSOCIATED MADS-BOX, AP3 APETALA3, PI PISTILLATA, SVP SHORT VEGETATIVE PHASE, AP2 APETALA2
The SlMBP21, MC-like genes that are involved in pedicel AZ formation in tomato were discovered in pedicel transcriptome. Considering only MADS-box genes that were specific to the central or lateral fruitlets, except those that were expressed at the same time, the central fruitlets had a high expression of genes belonging to AG, AP2 groups and the MdMADS genes which belong to AP1, SEP, AP3, and AG groups were highly expressed in lateral fruitlets. AP1 family contained MC gene of tomato and the similar contigs to SlMBP21 gene were included in SEPALLATA family. The SEPALLATA-like genes were significantly upregulated in pedicel as follows: MDP0000326390, MDP0000366022, MDP0000370413, MDP0000300752, and MDP0000326906. The following MC homologs were also discovered: MDP0000269921, MDP0000013331, MDP0000289836 and MDP0000132738.
Discussion
Abscission is progressed by four steps: (1) differentiation of abscission zone, (2) excess of the threshold to abscission-promoting signals during the pre-abscission stage, (3) activation of abscission, and (4) suturing abscission zone in the proximal region (Nakano et al. 2013; Patterson 2001). In step 1, the differentiation of AZ is initiated by transcription factors and other regulators. In tomato, J (Mao et al. 2000) forms protein complexes to develop the AZ by interacting with other MADS-box genes, MC (Nakano et al. 2012) and SEP (Liu et al. 2014). The important point is that the AZ was formed in step 1, but abscission process was not advanced yet.
In step 2, The AUX/IAA genes which related to auxin signal transduction may act as the trigger to initiate abscission process. The AXR3 gene, a member of AUX/IAA transcription factor family, delayed shedding of floral organs by interfering with auxin signaling at the AZ (Basu et al. 2013). Meir et al. (2006) showed that expression levels of Mj-Aux/IAA1 and Mj-Aux/IAA2 were repressed by removal of IAA sources in the AZ of Mirabilis jalapa. Microarray analysis of the transcriptome in the AZ of tomato flowers showed that AUX/IAA genes were down-regulated early after the depletion of auxin and that early modified expression of these genes might mediate auxin regulation of ethylene sensitivity in the AZ (Meir et al. 2010).
In step 3, cell wall degrading and modifying enzymes that expressed by ethylene were activated in the separation layers of AZ (Belfield et al. 2005; Cho and Cosgrove 2000). The combined function of enzymes, such as cellulase, polygalacturonase, expansin, xyloglucan endohydrolase and endotransglycosylase, could modify cell wall and finally detach organ.
In step 4, late phase of abscission, the genes belong to lignin biosynthesis pathway were significantly up-regulated by ethylene exclusively in the AZ. The role of lignin deposition may be associated with the generation of protective layers at the tissues remaining in the plant, such as receptacle or peduncle, during the last step of abscission process (Van Nocker 2009) and lignification could facilitate cell wall separation mechanically (Merelo et al. 2017). On the contrary, repression of lignin biosynthesis may result in grain shattering in rice (Yoon et al. 2017).
In tomato, AZ exists in fruit pedicel and J has an important role for AZ development. This gene belongs to the SVP clade in MADS-box gene family which are generally expressed in vegetative organs and are involved in floral transition and bud dormancy. The SVP genes differ in function compared with other MADS-box genes that are involved in floral organ determination. Based on phylogenetic analyses with MADS-box genes derived from pedicel transcriptome of self-abscising apple, many MADS-box genes were also expressed in both central and lateral pedicels which did not belong to the SVP clade. Some genes were estimated to be similar with SEP (SlMBP21) and MC gene in tomato and most MADS-box genes belonged to AGAMOUS-LIKE (AGL) gene clade. To identify the J homolog that played a similar role in apple, fruit pedicels were used from self-abscising apples during early abscission. Based on multiple sequence alignment and phylogenetic analysis, MdJ was identified as J homolog in apple, while we could find only one J homolog using RACE method. According to our DNA blot analyses (data not shown), ‘Saika’ has three J homologs in its genome. In addition, three SVP homologs were reported to exist in ‘Golden Delicious’ genome (Velasco et al. 2010), and three homologs, MdJa1, MdJa2 and MdJb, were also found in ‘Fuji’ pedicels (Nakano et al. 2015). These J homologs and our MdJ sequence had the same length and were highly homologous to each other. Even our MdJ sequence was 100% identical to MdJa2. Ectopic expression of the MdJa restored J-deficient tomato mutant and MdJa physically interacted with MC and SlMBP21 of tomato (Nakano et al. 2015). The amino acid sequences of tomato J, apple MdJ and Arabidopsis SVP showed significant similarity each other. The SVP orthologs of other crops repressed the development of floral organs in the meristem and the floral differentiation during bud dormancy (Li et al. 2009; Yamane et al. 2011; Wu et al. 2012). The SVP gene might suppress cell differentiation in pedicels AZs. Apple pedicel AZs showed a group of small cells that were arranged transversely to adjacent cells and arrested in an undifferentiated state (Fig. 2). In tomato, these undifferentiated cells were not discovered in j mutant pedicels (Mao et al. 2000; Nakano et al. 2015). These results suggest that MdJ may play a fundamental role in repressing cell differentiation in pedicel AZ and prohibiting transition to reproductive phase.
The expression pattern of MdJ was investigated in central and lateral pedicles by real-time PCR. At FB, the expression of MdJ in central pedicel was more than twice as high as that of lateral pedicels. After 10 days from FB, the expression was still higher in CP than LP, but it did not show much difference between central and lateral pedicels. The expression of MdJ was higher in CF from FB than that from 10 DAFB and this reason is seemed that MdJ may be involved not only in the formation of AZ but also in other function. MdJ has three homologs in apple genome, which are likely to be functionally diversified. Ectopic overexpression of MdJa showed a strong apical dominance phenotype in transgenic ‘Royal Gala’ apple, constraining lateral shoot outgrowth during the first year (Wu et al. 2017). At the second year, the bud break was significantly delayed and leaf senescence was not generated in transgenic lines (Wu et al. 2017). According to these results, MdJ affects apical dominance of shoot in meristem being expressed within pedicels of fruit clusters in self-abscising apple.
Bangerth (2000) explained that early abscission of young fruits results from correlative dominance between fruit pedicels by competing polar basipetal IAA transport (Fig. 7). This dominance may be due not only to earlier pollination and fertilization but also to the position of pedicels in one cluster (Celton et al. 2014). In fruit cluster, central fruit (central position) exerts a dominance over lateral fruits enclosed with central pedicel. The auxin gradient may be formed between central pedicel and lateral pedicels, and affects gene expression related to auxin signal transduction. According to our previous research, the competition of auxin transport between CP and LP has resulted in determining the survival of pedicels elevating expression of IAA3/SHY2 gene in the dominant CP and IAA14/SLR in the inferior LPs according to hierarchical ranking (Heo et al. 2016). IAA3/SHY2 homolog was differentially expressed in CP and IAA14/SLR homolog was strongly up-regulated in LP from self-abscising apple (Heo et al. 2016). The function of Aux/IAA transcription factor genes in pedicel AZ of apple is not yet clear, while these genes have been well investigated to play a key role in development of lateral root. IAA3/SHY2 was reported to participate in auxin-mediated meristem differentiation (Koren et al. 2013) or lateral root emergence (Lavenus et al. 2013), which mediate auxin signal transduction. The mechanism of lateral root emergence allows IAA14/SLR signaling cascade to trigger IDA-HAE-HSL2 signaling cascade in Arabidopsis (Okushima et al. 2007). Chrysanthemum gene CmANR1, homologous to MADS-box gene AtANR1, plays a key role in the development of lateral root by accumulating auxin directly in lateral root (Sun et al. 2018). The MADS-box gene, AGL21 regulates auxin accumulation in lateral root primordia and lateral root by enhancing local auxin biosynthesis and stimulates lateral root initiation and growth in Arabidopsis (Yu et al. 2014).
Apple MADS-box genes, expressed in pedicel AZ, MdJ, MdMC, and MdSEP homologs reported from Nakano et al. (2015) in different apple cultivar, were validated in this study. To confirm the function of MdMADS in the apple, transgenic apples are necessary with antisense suppression and sense complementation of MdJ, MdMC, and MdSEP genes to wild type and j mutant tomato. MdJ has the potential to be involved in formation of the AZ and auxin transport in step 1 of abscission process. The self-abscission might result in hierarchical ranking of auxin transport between pedicels according to the order of flowering and location of pedicels. The phase transition to vegetative growth occurred in accordance with the ranks based on the expression degree of MdJ between fruit pedicels. MdJ is thought to induce rapid growth of the shoot as well as vegetative phase transition. It seems that MdJ not only contributes to auxin gradation but also influences auxin signaling and transport in fruit pedicel of self-abscising apple. In future studies, the interaction between AUX/IAA TFs and MADS-box genes, including MdJ, needs to be investigated. Furthermore, additional experiments are needed to determine if the MdMADS proteins which are homologous with J, MC, and SlMBP21 are physically interact with each other, resulting in forming heterodimer.
References
Bangerth F (2000) Abscission and thinning of young fruit and their regulation by plant hormones and bioregulators. Plant Growth Regul 31:43–59
Basu MM, González-Carranza ZH, Azam-Ali S, Tang S, Shahid AA, Roberts JA (2013) The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol 162:96–106
Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56:817–823
Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15:2296–2307
Celton JM, Dheilly E, Guillou MC, Simonneau F, Juchaux M, Costes E, Laurens F, Renou JP (2014) Additional amphivasal bundles in pedicel pith exacerbate central fruit dominance and induce self-thinning of lateral fruitlets in apple. Plant Physiol 164:1930–1951
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Cho HT, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:9783–9788
Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 105:15629–15634
Dal Cin V, Barbaro E, Danesin M, Murayama H, Velasco R, Ramina A (2009a) Fruitlet abscission: a cDNA-AFLP approach to study genes differentially expressed during shedding of immature fruits reveals the involvement of a putative auxin hydrogen symporter in apple (Malus × domestica L. Borkh.). Gene 442:26–36
Dal Cin V, Velasco R, Ramina A (2009b) Dominance induction of fruitlet shedding in Malus × domestica (L. Borkh.): molecular changes associated with polar auxin transport. BMC Plant Biol 9:139
Gregis V, Andrés F, Sessa A, Guerra RF, Simonini S, Mateos JL, Torti S, Zambelli F, Prazzoli GM, Bjerkan KN, Grini PE, Pavesi G, Colombo L, Coupland G, Kater MM (2013) Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol 14:R56
Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21:351–360
Heo S, Hwang JH, Kim D, Yu DJ, Lee HJ (2015) Classification of apple genetic resources using early fruit abscission pattern. J Am Pomol Soc 69:102–108
Heo S, Hwang JH, Jun JH, Lee HJ (2016) Abscission-related genes revealed by RNA-Seq analysis using self-abscising apple (Malus × domestica). J Hortic Sci Biotechnol 91:271–278
Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14:108–117
Konishi S, Izawa T, Lin SY, Ebana K, Fujuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396
Koren D, Resnick N, Gati EM, Belausov E, Weininger S, Kapulnik Y, Koltai H (2013) Strigolactone signalling in the endodermis is sufficient to restore root responses and involves SHORT HYPOCOTYL 2 (SHY2) activity. New Phytol 198:866–874
Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18:450–458
Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Li Z, Reighard GL, Abbott AG, Bielenberg DG (2009) Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60:3521–3530
Liu D, Wang D, Qin Z, Zhang D, Yin L, Wu L, Colasanti J, Li A, Mao L (2014) The SEPALLATA MADS-box protein SlMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J 77:284–296
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406:910–913
McKim SM, Stenvik GE, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW (2008) The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135:1537–1546
Meir S, Hunter DA, Chen JC, Halaly V, Reid MS (2006) Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa. Plant Physiol 141:1604–1616
Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KS, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154:1929–1956
Merelo P, Agustí J, Arbona V, Costa ML, Estornell LH, Gómez-Cadenas A, Coimbra S, Gómez MD, Pérez-Amador MA, Domingo C, Talón M, Tadeo FR (2017) Cell wall remodeling in abscission zone cells during ethylene-promoted fruit abscission in citrus. Front Plant Sci 8:126
Nakano T, Kimbara J, Fujisawa M, Kitagawa M, Ihashi N, Maeda H, Kasumi T, Ito Y (2012) MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol 158:439–450
Nakano T, Fujisawa M, Shima Y, Ito Y (2013) Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol 9:13–40
Nakano T, Kato H, Shima Y, Ito Y (2015) Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol 56:1097–1106
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130
Patterson SE (2001) Cutting loose: abscission and dehiscence in Arabidopsis. Plant Physiol 126:494–500
Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53:131–158
Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96:290–295
Sexton R, Roberts JA (1982) Cell biology of abscission. Annu Rev Plant Physiol 33:133–162
Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23:2553–2567
Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20:1805–1817
Sun L, Bukovac MJ, Forsline PL, van Nocker S (2009) Natural variation in fruit abscission-related traits in apple (Malus). Euphytica 165:55–67
Sun CH, Yu JQ, Duan X, Wang JH, Zhang QY, Gu KD, Hu DG, Zheng CS (2018) The MADS transcription factor CmANR1 positively modulates root system development by directly regulating CmPIN2 in chrysanthemum. Hortic Res 5:52
Theissen G, Saedler H (2001) Floral quartets. Nature 409:469–471
Van Nocker S (2009) Development of the abscission zone. Stewart Posthar Rev. 5:1–6
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Weigel D (1995) The genetics of flower development: from floral induction to ovule morphogenesis. Annu Rev Genet 29:19–39
Wu RM, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E (2012) Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J Exp Bot 63:797–807
Wu R, Tomes S, Karunairetnam S, Tustin SD, Hellens RP, Allan AC, Macknight RC, Varkonyi-Gasic E (2017) SVP-like MADS box genes control dormancy and budbreak in apple. Front Plant Sci 8:477
Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R (2011) Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J Exp Bot 62:3481–3488
Yoon J, Cho LH, Antt HW, Koh HJ, An G (2017) KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol 174:312–325
Yu LH, Miao ZQ, Qi GF, Wu J, Cai XT, Mao JL, Xiang CB (2014) MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol Plant 7:1653–1669
Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, Han B (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24:1034–1048
Acknowledgements
This work was supported by the education, research and student guidance grant funded by Jeju National University in 2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Seong Heo and Yong Suk Chung declare that they have no conflict of interest.
Research involving human and animal participants
This article does not contain any studies with human subjects or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Heo, S., Chung, Y.S. Validation of MADS-box genes from apple fruit pedicels during early fruit abscission by transcriptome analysis and real-time PCR. Genes Genom 41, 1241–1251 (2019). https://doi.org/10.1007/s13258-019-00852-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00852-4