Abstract
This paper provides illustrated descriptions of micro-fungi newly found on Pandanaceae in China and Thailand. The fungi are accommodated in 31 families. New taxa described include a new family, seven new genera, 65 new species, 16 previously known species. A new family: Malaysiascaceae (Glomerellales). New genera are Acremoniisimulans (Plectosphaerellaceae), Pandanaceomyces, Pseudoachroiostachy (Nectriaceae), Pseudohyaloseta (Niessliaceae), Pseudoornatispora (Stachybotriaceae) and Yunnanomyces (Sympoventuriaceae). New species are Acremoniisimulans thailandensis, Beltrania krabiensis, Beltraniella pandanicola, B. thailandicus, Canalisporium krabiense, C. thailandensis, Clonostachys krabiensis, Curvularia chonburiensis, C. pandanicola, C. thailandicum, C. xishuangbannaensis, Cylindrocladiella xishuangbannaensis, Dictyochaeta pandanicola, Dictyocheirospora nabanheensis, D. pandanicola, D. xishuangbannaensis, Dictyosporium appendiculatum, Di. guttulatum, Di. hongkongensis, Di. krabiense, Di. pandanicola, Distoseptispora thailandica, D. xishuangbannaensis, Helicoma freycinetiae, Hermatomyces biconisporus, Lasiodiplodia chonburiensis, L. pandanicola, Lasionectria krabiense, Menisporopsis pandanicola, Montagnula krabiensis, Musicillium pandanicola, Neofusicoccum pandanicola, Neohelicomyces pandanicola, Neooccultibambusa thailandensis, Neopestalotiopsis chiangmaiensis, N. pandanicola, N. phangngaensis, Pandanaceomyces krabiensis, Paracylindrocarpon nabanheensis, P. pandanicola, P. xishuangbannaensis, Parasarcopodium hongkongensis, Pestalotiopsis krabiensis, P. pandanicola, Polyplosphaeria nabanheensis, P. pandanicola, P. xishuangbannaensis, Pseudoachroiostachys krabiense, Pseudoberkleasmium pandanicola, Pseudochaetosphaeronema pandanicola, Pseudohyaloseta pandanicola, Pseudoornatispora krabiense, Pseudopithomyces pandanicola, Rostriconidium pandanicola, Sirastachys phangngaensis, Stictis pandanicola, Terriera pandanicola, Thozetella pandanicola, Tubeufia freycinetiae, T. parvispora, T. pandanicola, Vermiculariopsiella hongkongensis, Volutella krabiense, V. thailandensis and Yunnanomyces pandanicola. Previous studies of micro-fungi on Pandanaceae have not included phylogenetic support. Inspiration for this study came from the book Fungi Associated with Pandanaceae by Whitton, McKenzie and Hyde in 2012. Both studies reveal that the micro-fungi on Pandanaceae is particularly rich in hyphomycetes. All data presented herein are based on morphological examination of specimens, coupled with phylogenetic sequence data to better integrate taxa into appropriate taxonomic ranks and infer their evolutionary relationships.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Table of contents
The numbers of taxa in this study associated with Pandanaceae are organized as in the “Outline of Ascomycetes” (Wijayawardene et al. 2018).
Phylum Ascomycota Caval.-Sm.
Class Dothideomycetes sensu O.E. Erikss & Winka
For recent treatments of Dothideomycetes we follow Hyde et al. (2013) with updates by Liu et al. (2017) and Wijayawardene et al. (2018).
Subclass Dothideomycetidae P.M. Kirk et al.
Capnodiales Woron.
Mycosphaerellaceae Lindau
840. Cercospora capsici Heald & F.A. Wolf, Mycologia 3 (1): 15 (1911), new host record
Subclass Pleosporomycetidae C.L. Schoch et al.
Pleosporales Luttrell ex M.E. Barr
Dictyosporiaceae Boonmee & K.D. Hyde
841. Dictyocheirospora nabanheensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 10 (2018), new species
842. Dictyocheirospora pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 10 (2018), new species
843. Dictyocheirospora xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 14 (2018), new species
844. Dictyosporium appendiculatum Tibpromma & K.D. Hyde, in Fungal Diversity 92: 15 (2018), new species
845. Dictyosporium digitatum J.L. Chen, C.H. Hwang & Tzean, Mycological Research 95: 1145 (1991)
846. Dictyosporium guttulatum Tibpromma & K.D. Hyde, in Fungal Diversity 92: 17 (2018), new species
847. Dictyosporium hongkongensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 18 (2018), new species
848. Dictyosporium krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 19 (2018), new species
849. Dictyosporium pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 20 (2018), new species
Didymosphaeriaceae Munk
850. Deniquelata barringtoniae Ariyawansa & K.D. Hyde, Phytotaxa 105 (1): 15 (2013), new host record
851. Montagnula krabiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 23 (2018), new species
852. Pseudopithomyces pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 25 (2018), new species
Hermatomycetaceae Locq.
853. Hermatomyces biconisporus Tibpromma & K.D. Hyde, in Fungal Diversity 92: 28 (2018), new species
Melanommataceae G. Winter (= Pseudodidymellaceae A.Hashim. & Kaz. Tanaka)
854. Byssosphaeria siamensis Boonmee, Q. Tian & K.D. Hyde, Fungal Diversity 74: 283 (2015), new host record
Occultibambusaceae D.Q. Dai & K.D. Hyde
855. Neooccultibambusa thailandensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 31 (2018), new species
Pleosporaceae Nitschke
856. Curvularia chonburiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 33 (2018), new species
857. Curvularia pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 37 (2018), new species
858. Curvularia thailandicum Tibpromma & K.D. Hyde, in Fungal Diversity 92: 38 (2018), new species
859. Curvularia xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 39 (2018), new species
Roussoellaceae J.K. Liu et al.
860. Roussoella solani Crous & M.J. Wingf., Persoonia 36: 341 (2016), new host record
Tetraplosphaeriaceae Kaz. Tanaka & K. Hiray
861. Polyplosphaeria nabanheensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 42 (2018), new species
862. Polyplosphaeria pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 42 (2018), new species
863. Polyplosphaeria xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 44 (2018), new species
Torulaceae Corda
864. Rostriconidium pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 45 (2018), new species
865. Torula chromolaenae J.F. Li, Phook., Mapook & K.D. Hyde, Mycological Progress 16 (4): 454 (2017), new host record
866. Torula ficus Crous, IMA Fungus 6 (1): 192 (2015), new host record
Pleosporales genera incertae sedis
867. Pseudoberkleasmium Tibpromma & K.D. Hyde, in Fungal Diversity 92: 50 (2018), new genus
868. Pseudoberkleasmium pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 51 (2018), new species
869. Pseudochaetosphaeronema pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 53 (2018), new species
Dothideomycetes orders incertae sedis
Botryosphaeriales C.L. Schoch et al.
Botryosphaeriaceae Theiss. & H. Syd.
870. Lasiodiplodia chonburiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 54 (2018), new species
871. Lasiodiplodia hyalina Zh.P. Dou & Y. Zhang, Mycosphere 8 (2): 1016 (2017), new host record
872. Lasiodiplodia pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 58 (2018), new species
873. Lasiodiplodia pseudotheobromae A.J.L. Phillips, A. Alves & Crous, Fungal Diversity 28: 8 (2008), new host record
874. Neofusicoccum pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 60 (2018), new species
Pseudofusicoccumaceae Yao Tan & Crous
875. Pseudofusicoccum adansoniae Pavlic, T.I. Burgess & M.J. Wingf., Mycologia 100 (6): 855 (2008), new host record
Tubeufiales Boonmee & K.D. Hyde
Tubeufiaceae M.E. Barr
876. Helicoma freycinetiae Tibpromma & K.D. Hyde, in Fungal Diversity 92: 68 (2018), new species
877. Neohelicomyces pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 69 (2018), new species
878. Tubeufia freycinetiae Tibpromma & K.D. Hyde, in Fungal Diversity 92: 69 (2018), new species
879. Tubeufia inaequalis Y.Z Lu, J.C. Kang & K.D. Hyde, Fungal Diversity 92 (2018), new host record
880. Tubeufia pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 73 (2018), new species
881. Tubeufia parvispora Tibpromma & K.D. Hyde, in Fungal Diversity 92: 74 (2018), new species
Venturiales Y. Zhang ter et al.
Sympoventuriaceae Y. Zhang ter et al.
882. Yunnanomyces Tibpromma & K.D. Hyde, in Fungal Diversity 92: 75 (2018), new genus
883. Yunnanomyces pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 75 (2018), new species
Class Lecanoromycetes O.E. Erikss. & Winka
Subclass Ostropomycetidae Reeb et al.
Ostropales Nannf.
Stictidaceae Fr.
884. Stictis pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 78 (2018), new species
Class Leotiomycetes O.E. Erikss. & Winka
Rhytismatales M.E. Barr ex Minter
Rhytismataceae Chevall. (= Hypodermataceae Rehm; = Cryptomycetaceae Höhn. nom. inval. fide Jaklitsch et al. 2016)
885. Terriera pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 79 (2018), new species
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Diaporthomycetidae Senan. et al.
Diaporthomycetidae families incertae sedis
Distoseptisporaceae K.D. Hyde & McKenzie
886. Distoseptispora thailandica Tibpromma & K.D. Hyde, in Fungal Diversity 92: 79 (2018), new species
887. Distoseptispora xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 82 (2018), new species
Subclass Hypocreomycetidae O.E. Erikss. & Winka
Glomerellales Chadef. ex Reblová et al.
Glomerellaceae Locq. ex Seifert & W. Gams
888. Colletotrichum pandanicola Tibpromma & K.D. Hyde, Mycokeys 33: 47 (2018)
889. Malaysiascaceae Tibpromma & K.D. Hyde, in Fungal Diversity 92: 88 (2018), new family
890. Malaysiasca phaii Crous & M.J. Wingf., Persoonia 36: 373 (2016), new host record
Plectosphaerellaceae W. Gams et al.
891. Acremoniisimulans Tibpromma & K.D. Hyde, in Fungal Diversity 92: 88 (2018), new genus
892. Acremoniisimulans thailandensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 89 (2018), new species
893. Musicillium pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 91 (2018), new species
Hypocreales Lindau
Bionectriaceae Samuels & Rossman
894. Clonostachys krabiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 93 (2018), new species
895. Lasionectria krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 95 (2018), new species
896. Paracylindrocarpon nabanheensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 102 (2018), new species
897. Paracylindrocarpon pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 103 (2018), new species
898. Paracylindrocarpon xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 104 (2018), new species
Nectriaceae Tul. & C. Tul.
899. Cylindrocladiella xishuangbannaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 105 (2018), new species
900. Pandanaceomyces Tibpromma & K.D. Hyde, in Fungal Diversity 92: 107 (2018), new genus
901. Pandanaceomyces krabiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 107 (2018), new species
902. Pseudoachroiostachys Tibpromma & K.D. Hyde, in Fungal Diversity 92: 107 (2018), new genus
903. Pseudoachroiostachys krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 108 (2018), new species
904. Volutella krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 110 (2018), new species
905. Volutella thailandensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 110 (2018), new species
Niessliaceae Kirschst.
906. Pseudohyaloseta Tibpromma & K.D. Hyde, in Fungal Diversity 92: 113 (2018), new genus
907. Pseudohyaloseta pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 113 (2018), new species
Stachybotryaceae L. Lombard & Crous
908. Parasarcopodium hongkongensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 114 (2018), new species
909. Pseudoornatispora Tibpromma & K.D. Hyde, in Fungal Diversity 92: 115 (2018), new genus
910. Pseudoornatispora krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 115 (2018), new species
911. Sirastachys phangngaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 118 (2018), new species
Microascales Luttr. ex Benny & Kimbr.
Microascaceae Luttr. ex Malloch
912. Parascedosporium putredinis (Corda) Lackner & de Hoog, IMA Fungus 2 (1): 44 (2011), new host record
Subclass Savoryellomycetidae Hongsanan et al.
Savoryellales Boonyuen et al.
Savoryellaceae Jaklitsch & Réblová
913. Canalisporium krabiense Tibpromma & K.D. Hyde, in Fungal Diversity 92: 122 (2018), new species
914. Canalisporium thailandensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 122 (2018), new species
Subclass Sordariomycetidae O.E. Erikss & Winka (= Meliolomycetidae P.M. Kirk & K.D. Hyde)
Chaetosphaeriales Huhndorf et al.
Chaetosphaeriaceae Réblová et al.
915. Dictyochaeta pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 127 (2018), new species
916. Dictyochaeta siamensis J. Yang, K.D. Hyde & J.K. Liu, Mycological Progress 15 (10): 1159 (2016), new host record
917. Menisporopsis pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 128 (2018), new species
918. Thozetella pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 130 (2018), new species
Sordariales Chad. ex D. Hawksw. & O.E. Erikss.
Chaetomiaceae G. Winter
919. Chaetomium globosum Kunze ex Fr., Systema Mycologicum 3: 255 (1829)
Subclass Xylariomycetidae O.E. Erikss & Winka
Amphisphaeriales D. Hawksw. & O.E. Erikss.
Beltraniaceae Nann.
920. Beltrania krabiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 134 (2018), new species
921. Beltraniella pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 134 (2018), new species
922. Beltraniella thailandicus Tibpromma & K.D. Hyde, in Fungal Diversity 92: 136 (2018), new species
Sporocadaceae Corda.
923. Neopestalotiopsis chiangmaiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 139 (2018), new species
924. Neopestalotiopsis pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 141 (2018), new species
925. Neopestalotiopsis phangngaensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 142 (2018), new species
926. Pestalotiopsis krabiensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 143 (2018), new species
927. Pestalotiopsis pandanicola Tibpromma & K.D. Hyde, in Fungal Diversity 92: 145 (2018), new species
Sordariomycetes orders incertae sedis
Vermiculariopsiellales Hern.-Restr. et al.
Vermiculariopsiellaceae Hern.-Restr. et al.
928. Vermiculariopsiella hongkongensis Tibpromma & K.D. Hyde, in Fungal Diversity 92: 146 (2018), new species
Introduction
Pandanaceae is a monocotyledonous family that comprises five genera: Benstonea, Freycinetia, Martellidendron, Pandanus, Sararanga (Wardah and Setyowati 2009; Callmander et al. 2012). Taxonomy of the Pandanaceae was dealt with by St. John and Stone (1962, 1967, 1968, 1969), however, their identification is relatively difficult because of the lack of literature or absence of flowering parts in the field (Nadaf and Zanan 2012; Whitton et al. 2012). The distribution of Pandanaceae is predominantly tropical, but they are also distributed throughout the Pacific, and New Zealand (Nadaf and Zanan 2012; Whitton et al. 2012).
Micro-fungi on Pandanaceae have long been studied and all taxa described from Pandanaceae were listed by McKenzie and Hyde (1996), while Whitton et al. (2012) listed all fungi recorded on Pandanaceae. McKenzie (1991a, b, c, 1995); McKenzie and Kuthubutheen (1993); Hyde (1994), Hyde and Hawksworth (1997b), McKenzie and Hyde (1996); Dulymamode et al. (1998a, b, c, d, e), Whitton et al. (1998, 1999, 2000a, b, 2001a, b, c, d, 2002a, b, 2003), and Thongkantha et al. (2008) provided a series of papers in which many new species of fungi on Pandanaceae were described. More recently, a few papers on micro-fungi from Pandanaceae have been published (Hosagoudar 2012; Crous et al. 2015a; Lombard et al. 2016; Tibpromma et al. 2016a, b, c, 2017a, b, 2018; Hyde et al. 2017, 2018; Zhang et al. 2017) that include both morphological and phylogenetic analysis. Hawksworth (2000) estimated that there may be 1.5 million species of fungi and later Hawksworth and Lücking (2017) revised the estimate upwards and suggested there were 2.2–3.8 million species. It is a huge challenge to find all of the unknown fungi in the world as currently only 120,000 species have been described (Hawksworth and Lücking 2017). Tropical fungi are a major component of biodiversity and play major roles in ecosystem functioning (Hawksworth 2002) and are very significant for the survival of other organisms in tropical forests (Lodge et al. 1996). Pandanaceae has proven to be a good source of new, interesting and often infrequently collected micro-fungi (Whitton et al. 2012). McKenzie et al. (2002) estimated that less than 40% of fungi on a single member of Pandanaceae are ‘unique’, but with time it was understood that many can be found on other members of the Pandanaceae, or on other host families.
The objective of this study is to provide a description of recently collected micro-fungi from Pandanaceae, based on both morphology and phylogenetic support to reveal the correct taxonomic placements of these fungi. Thus, information will be compared with previous studies on micro-fungi on Pandanaceae and other hosts. In particular, a comparison will be made with Whitton et al. (2012) who provided a book entitled Fungi associated with Pandanaceae in which 114 genera and 226 species of fungi from Pandanaceae based on morphology are listed. The final outcome will be a firm taxonomy and phylogeny that can provide support to researchers who need to identify fungi for their research work.
Materials and methods
Specimens were collected from Thailand and China, especially from southern Thailand and Yunnan Province of China. Each sample was placed into separate zip-lock bags or envelopes together with collection details (host, place and date) and returned to the laboratory for observation following the methods outlined by Tibpromma et al. (2016a, b, c). Single spore isolation was done following the method described in Chomnunti et al. (2014). Germinated spores were observed with a stereo microscope and transferred to malt extract agar (MEA: 33.6 g/l sterile distilled water, Difco malt extract) or potato dextrose agar (PDA: 39 g/l sterile distilled water, Difco potato dextrose) plates and incubated at room temperature for 4 weeks. Hyphomycetes were immediately isolated because conidia are easy to release under dry conditions. Cultures were subcultured and transferred to MEA or PDA containing sterile toothpicks, pine needles (Phookamsak et al. 2015) or Pandanaceae leaves and incubated at room temperature for 3 months to induce the sexual or asexual morph. Herbarium specimens were deposited in Mae Fah Luang University Herbarium (MFLU), Chiang Rai, Thailand and Kunming Institute of Botany Academia Sinica (HKAS). Living cultures were deposited in the Mae Fah Luang University Culture Collection (MFLUCC) and Kunming Institute of Botany Culture Collection (KMUCC). Facesoffungi numbers (FoF) and Index Fungorum (IF) numbers were obtained as explained in Jayasiri et al. (2015) and Index Fungorum (2018). New taxa were established based on recommendations outlined by Jeewon and Hyde (2016).
Morphological classification
The fruiting structures were examined with a Carl Zeiss GmbH (AxioCam ERC 5 S) or JNOEC JSZ4 (ser. No. 030233) stereo microscope. Fruiting bodies were rehydrated in water, 100% lactic acid or 5% KOH. Sections were cut with a razor blade and studied with a Nikon ECLIPSE 80i or Nikon ECLIPSE Ni compound microscope. Photographs were taken with a Canon 550D or Canon 600D digital camera mounted on the microscope. All microscopic structures were measured using Tarosoft® Image Framework program v.0.9.0.7 and images used for figures were processed with Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems, USA).
DNA extraction, PCR amplification and DNA sequencing
Genomic DNA was extracted from pure fungal cultures by using Biospin Fungal Genomic DNA extraction Kit-BSC14S1 (BioFlux, P.R. China). If cultures were unavailable, DNA was extracted directly from fruiting bodies using a Forensic DNA Kit–D3591-01 (OMEGA bio-tek) following the manufacturer’s instructions. Polymerase chain reaction (PCR) was used to amplify partial gene regions using primers shown in Table 1. The total volume of PCR mixtures for amplifications were 25 μL containing 8.5 μL ddH2O, 12.5 μL 2× Easy Taq PCR Super Mix (mixture of Easy Taq TM DNA Polymerase, dNTPs, and optimized buffer (Beijing Trans Gen Biotech Co., Chaoyang District, Beijing, PR China), 2 μL of DNA template, 1 μL of each forward and reverse primers (10 pM). The quality of PCR products was checked on 1% agarose gel electrophoresis stained with 4S green nucleic acid (Life Science Products and Services, Cat. No: A616694). Purification and sequencing of PCR products were carried out by Sangon Biotech Co., Shanghai, China.
Phylogenetic analysis
LSU or ITS sequence data generated in this study were subjected to BLAST searches in the nucleotide database of GenBank (www http://blast.ncbi.nlm.nih.gov/) to determine their most probable closely related taxa. Sequence data were retrieved from GenBank based on recent publications. Raw forward and reverse sequences were assembled using Geneious Pro.v4.8.5.
Single gene sequence alignments were made with MAFFT v. 7.215 (Katoh and Standley 2016: http://mafft.cbrc.jp/alignment/server/index.html) and edited manually if necessary in BioEdit v. 7.0 (Hall 2004). The sequence datasets were combined using BioEdit v.7.2.5 (Hall 2004). FASTA alignment formats were exchanged to PHYLIP and NEXUS formats by the website (http://sing.ei.uvigo.es/ALTER/).
Phylogenetic analysis of both individual and combined aligned data were based on maximum likelihood (ML), maximum parsimony (MP) and Bayesian analysis (BYPP). The MP analysis was performed using PAUP v. 4.0b10 (Swofford 2002). MP analysis by bootstrap analysis with 1000 replicates using 10 rounds of heuristic search replicates with random addition of sequences and subsequent TBR branch swapping during each bootstrap replicate. Descriptive tree statistics calculated for parsimony were: tree length (TL), consistency index (CI), retention index (RI), relative consistency index (RC) and homoplasy index (HI). A ML analysis was performed via the CIPRES portal (Miller et al. 2010) using RAxML-HPC BlackBox (8.2.4) (Stamatakis 2006, 2014) with the general time reversible model (GTR) with a discrete gamma distribution as the evolutionary model. Posterior probabilities (PP) (Rannala and Yang 1996) were established by Markov chain Monte Carlo sampling (MCMC) in MrBayes v.3.0b4 (Liu et al. 2012). The model of evolution for the Bayesian analysis was determined by using MrModeltest 2.2 (Nylander 2004). Six simultaneous Markov chains were run for at least 1,000,000 generations, or depending on individual settings for the fungal group, and trees were sampled every 100th or 1,000th generation (Cai et al. 2006). The burn-in was set to 0.25, and the run was automatically stopped when the average standard deviation of split frequencies reached below 0.01 (Maharachchikumbura et al. 2015). The best scoring phylogenetic trees were configured and visualized in FigTree 1.4.2 (Rambaut 2014) with bootstrap values (MP, ML and PP) given at the nodes and edited using Microsoft Office PowerPoint 2007 and Adobe Illustrator CS3 (Adobe Systems, USA). MP/ML bootstrap supports (greater than or equal to 60%) and Bayesian posterior probability (greater than or equal to 0.90) are shown below or above each branch. The resulting phylogenetic trees are presented under each relevant description.
Taxonomy
Phylum Ascomycota Caval.-Sm.
The taxa are arranged as in Outline of the Ascomycota – 2017 (Wijayawardene et al. 2018)
Class Dothideomycetes sensu O.E. Erikss & Winka
For the classification of Dothideomycetes we follow Hyde et al. (2013) updates provided by Liu et al. (2017) and Wijayawardene et al. (2018).
Subclass Dothideomycetidae P.M. Kirk et al.
Capnodiales Woron
Mycosphaerellaceae Lindau
Mycosphaerellaceae was erected by Lindau (1897) with the family type Ramularia Unger. Members of Mycosphaerellaceae are pathogens, endophytes, saprobes, epiphytes and fungicolous (Ávila et al. 2005; Crous et al. 2006; Arzanlou et al. 2007; Churchill 2010; Hyde et al. 2013; Chang et al. 2016; de Wit 2016). Morphology of the sexual morph was provided by Schoch et al. (2006) and the asexual morphs are coelomycetes and hyphomycetes (Braun et al. 2003; Aptroot 2006; Schoch et al. 2006; Wijayawardene et al. 2017a). Hawksworth et al. (1995) placed Mycosphaerellaceae in Dothideales, while Kirk et al. (2001) assigned the family to the Mycosphaerellales. Kirk et al. (2008) listed Mycosphaerellaceae in Capnodiales. Wijayawardene et al. (2018) accepted 129 genera. We record Cercospora capsici from Pandanaceae for the first time.
Cercospora Fresen.
Cercospora was erected by Fresenius with C. apii Fresen. as the type species (Fuckel 1863). Cercospora contains numerous important plant pathogenic fungi from a diverse range of hosts (Groenewald et al. 2013a, b) with 3178 epithets are listed in Index Fungorum (2018). The unidentified Cercospora sp. has been reported from Thailand on Pandanus amaryllifolius (Thongkantha et al. 2008).
Cercospora capsici Heald & F.A. Wolf, Mycologia 3 (1): 15 (1911)
Facesoffungi number: FoF04570; Fig. 1
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS and TEF1 partial sequence data. Thirty strains are included in the sequence analysis, which comprise 1884 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Mycosphaerella laricina, M. madeirae and M. marksii are used as outgroup taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 8740.769206 is presented. The matrix had 565 distinct alignment patterns, with 12.46% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.237646, C = 0.247630, G = 0.280191, T = 0.234533; substitution rates AC = 1.626221, AG = 2.901339, AT = 1.381886, CG = 1.148995, CT = 6.553424, GT = 1.000000; gamma distribution shape parameter α = 0.460899. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Saprobic on dead leaves of Pandanus amaryllifolius. Mycelium superficial, rough, branched, septate, light brown to dark brown. Sexual morph Undetermined. Asexual morph Hyphomycetous. Caespituli fasciculate to sporodochial, brown to dark brown, thick-walled, predominantly epiphyllous. Conidiophores 90–215 × 4–7 μm (\( \bar{x} \) = 160 × 5 μm, n = 20), aggregated in dense fascicles, pale brown, cylindrical, up to 6-septate, branched, roughened with scars, straight to curved. Conidiogenous cells 20–65 × 4–7 μm (\( \bar{x} \) = 45 × 6 μm, n = 20), holoblastic, integrated, cylindrical, pale brown, hyaline at the apex, thick-walled. Conidia 85–220 × 4–8 μm (\( \bar{x} \) = 151 × 6 μm, n = 20), cylindrical, base truncate with distinctive scar, apex rounded, solitary, hyaline, straight to slightly curved, guttulate, up to 15-septate, not constricted at septa, thick-walled, without a mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, entire edge, smooth, pulvinate, white–grey in middle, white at margin, velvety.
Material examined: THAILAND, Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on dead leaf of Pandanus amaryllifolius Roxb., 16 December 2017, S. Tibpromma P12 (MFLU 18-0031, HKAS 101799); living culture, MFLUCC 18-0117.
GenBank numbers LSU: MH260285; ITS: MH275053; SSU: MH260331; TEF1: MH412762; RPB2: MH412752; TUB2: MH412741.
Notes: Cercospora sp. was recorded from Thailand on Pandanus amaryllifolius (Thongkantha et al. 2008). We also collected Cercospora from Pandanus amaryllifolius in Thailand, but unfortunately cannot compare the morphology with the species in Thongkantha et al. (2008) as it lacked a description. Cercospora capsici (MFLUCC 18-0117) grouped with C. sojina (CPC 12322) and C. capsici (CBS 118712) (Fig. 2). Cercospora capsici has acicular conidia, 2–12-septate and subacute at the apex (Groenewald et al. 2013a, b), while C. sojina has cylindrical to obclavate or fusiform conidia, with 1–5-septate (Crous et al. 2013). However, when comparing the nucleotide sequences our isolate is almost identifical with Cercospora capsici (CBS 118712 and CPC 12307) with three ITS base pair (0.59%) differences.
Subclass Pleosporomycetidae C.L. Schoch et al.
Pleosporales Luttrell ex M.E. Barr
Dictyosporiaceae Boonmee & K.D. Hyde
Dictyosporiaceae was informally erected by Liu et al. (2015b) and later it was formally introduced by Boonmee et al. (2016) with Dictyosporium Corda. as the type genus based on morphology and multi-gene analysis. Members of Dictyosporiaceae are often saprobes on decaying wood in both terrestrial and freshwater habitats (Boonmee et al. 2016). The sexual morphs of Dictyosporiaceae are characterized by immersed to erumpent or superficial, globose to subglobose, dark brown to black ascomata, bitunicate asci with septate, hyaline, sheathed ascospores; the asexual morphs are cheirosporous hyphomycetes (Boonmee et al. 2016). Illustrations were provided by Tanaka et al. (2015). There are twelve genera in the family (Wijayawardene et al. 2017b, 2018). Asexual morph taxa belonging to Dictyocheirospora and Dictyosporium were found on Pandanaceae. These are described together with an updated tree for the family.
Dictyocheirospora D’souza et al.
Dictyocheirospora was erected with D. rotunda D’souza, Bhat & K.D. Hyde as type species (Boonmee et al. 2016). The genus is characterized by dark sporodochial colonies with aeroaquatic cheiroid dictyospores and members of this genus are saprobes (Boonmee et al. 2016). Index Fungorum lists ten epithets for Dictyocheirospora (Index Fungorum 2018). We introduce three new species from Pandanaceae, a genus was not previously found on Pandanaceae.
Dictyocheirospora nabanheensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554474, Facesoffungi number: FoF04483; Fig. 3
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS partial sequence data. Fifty-five strains are included in the sequence analysis, which comprise 1460 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Murilentithecium clematidis (Lentitheciaceae) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 7300.219123 is presented. The matrix had 474 distinct alignment patterns, with 19.03% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240784, C = 0.235601, G = 0.275234, T = 0.248381; substitution rates AC = 1.951883, AG = 3.307241, AT = 2.649573, CG = 0.576092, CT = 8.294481, GT = 1.000000; gamma distribution shape parameter α = 0.744325. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Nabanhe, the place where the fungus was first discovered.
Holotype: HKAS 101807
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, scattered, dark brown. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 9–20 × 5–7 μm (\( \bar{x} \) = 12 × 5.5 μm, n = 10), holoblastic, cylindrical, wide at the top, sometimes flat at base, dull pale brown, mostly remaining attached to the conidia. Conidia 35–40 × 18–21 μm (\( \bar{x} \) = 38 × 20 μm, n = 20), solitary, oval to ellipsoid, cheiroid, consisting of 40–48 cells, with a basal connecting cell, pale brown to yellow–brown, smooth-walled, guttulate, individual cells discoid, 27–39 × 5–6 μm, arranged in 6 compact rows, with 6–10 cells per row; with 1–2 rounded to cylindrical appendages, 5–16 × 5–6.5 μm, arising from near middle of conidial rows, hyaline.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA, circular, entire edge with white margin and cream to yellow–orange in the central, raised on surface.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 2 August 2016, S. Tibpromma NBH21 (HKAS 101807, holotype); ex-type living culture, KUMCC 16-0152 = MFLUCC 17-0562.
GenBank numbers LSU: MH376712; ITS: MH388340; TEF1: MH388375.
Notes: Dictyocheirospora nabanheensis is introduced as a new species based on morphology and phylogeny. Dictyocheirospora nabanheensis is well-separated in a clade with other members of Dictyocheirospora (Fig. 4) and is distinguished by cylindrical conidiogenous cells with wide top, oval to ellipsoid, cheiroid conidia consisting of 40–48 cells, pale brown to yellow–brown, 6–10 cells in each row and 1–2 rounded to cylindrical appendages. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0152 is 99% identical with the two strains of D. rotunda strain DLUCC 0577 (KY320512) and D. garethjonesii strain KUMCC 15-0396 (KY320510), while the closest matches with the TEF1 sequence were with 98% identity with D. garethjonesii strain DUCC 0848 (MF953166) and 96% identity D. rotunda strain DUCC 0804 (MF953170).
Dictyocheirospora pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554475, Facesoffungi number: FoF04484; Fig. 5
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1933
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate, solitary or in small groups, scattered, reddish brown. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 9–15 × 1.5–3 μm (\( \bar{x} \) = 12 × 2.5 μm, n = 10), holoblastic, cylindrical, sometimes flat at base, subhyaline, mostly not remaining attached to the conidia. Conidia 60–75 × 18.5–35.5 μm (\( \bar{x} \) = 65 × 24 μm, n = 30), solitary, oval to ellipsoid, cheiroid, consisting of 95–120 cells, with a basal connecting cell, pale brown, guttulate, individual cells discoid, 53–65 × 6–7.5 μm, arranged in 5–7 compact rows, of which 6 in peripheral positions and one central, each three rows connected to a large basal cell, 13–18 cells per row, smooth-walled, without appendages
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, circular, entire edge, yellow brown, velvety, umbonate on media surface.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on Pandanus sp., 15 July 2015, S. Tibpromma SF15-031 (MFLU 16-1933, holotype; HKAS 96282, isotype); ex-type living culture, MFLUCC 16-0365.
GenBank numbers LSU: MH376713; ITS: MH388341; SSU: MH388309; TEF1: MH388376.
Notes: In the phylogenetic analysis Dictyocheirospora pandanicola clustered with D. vinaya D’souza, Bhat & K.D. Hyde. Figure 4, but the two species differ in morphology. Dictyocheirospora pandanicola has oval to ellipsoid, cheiroid conidia 60–75 × 18.5–35.5 μm with 13–18 cells in each arm, while D. vinaya has cheiroid conidia 58–67 × 15.5–26.5 μm with 9–13 cells in each arm (Boonmee et al. 2016). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0365 is 100% identical D. digitatum strain 30404 (KP714383), while the closest matches with the TEF1 sequence were with 95% identity with D. pseudomusae strain EF9a (AB808492).
Dictyocheirospora xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554476, Facesoffungi number: FoF04485; Fig. 6
Etymology: named after Xishuangbanna, place where fungus was first discovered.
Holotype: HKAS 99628
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark brown, with base attached on surface of host plant. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 3–7 × 3.5–7 μm (\( \bar{x} \) = 5 × 4.5 μm, n = 20), holoblastic, cylindrical, sometimes flat at base, hyaline to pale brown, mostly remaining attached to the conidia. Conidia 35–50 × 17–25 μm (\( \bar{x} \) = 47 × 21 μm, n = 20), solitary, oval to ellipsoid, cheiroid, consisting of 54–65 cells, with a basal connecting cell, pale brown to green–brown, guttulate, 40–48 × 7–9 μm, arranged in 6 compact rows, of which 6 in peripheral positions connected to large basal cell, with individual cells discoid, 6–12 cells per row, without appendages.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA, circular, entire edge with yellowish at the margin and yellow to green in the central, raised on surface media.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on Pandanus sp., 27 April 2017, R. Phookamsak & N.I. de Silva XTBG20 (HKAS 99628, holotype); ex-type living culture, KUMCC 17-0181 = MFLUCC 17-2267.
GenBank numbers LSU: MH376714; ITS: MH388342; SSU: MH388310; TEF1: MH388377; RPB2: MH412728.
Notes: Dictyocheirospora xishuangbannaensis clustered with D. heptaspora (Garov) D’souza, Boonmee & K.D. Hyde in the phylogeny (Fig. 4). There were 22 bp (4.22%) differences in 521 ITS (+5.8S) nucleotides sequences between D. xishuangbannaensis and D. heptaspora. Dictyocheirospora xishuangbannaensis has conidia 35–50 × 17–25 μm, oval to ellipsoid, arranged in 6 compact rows, while D. heptaspora has conidia 50–80 × 20–30 μm, cylindrical with 7 compact rows (Goh et al. 1999). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0181 is 98% identical D. heptasporum strain CBS 396.59 (DQ018090), while the closest matches with the TEF1 sequence were with 96% identity with D. rotunda strain DUCC 0804 (MF953170).
Dictyosporium Corda
Dictyosporium was introduced with the single species Di. elegans Corda. The members of this genus are saprobes in terrestrial or aquatic environments (Hyde et al. 2011). There are 74 epithets are listed in Index Fungorum (2018). Whitton et al. (2012), provided a key to Dictyosporium species and eight species have been described from Pandanaceae. We found five species on Pandanaceae of which only one, Di. digitatum had been previously described. This suggests that Dictyosporium is very common on Pandanaceae.
Dictyosporium appendiculatum Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555327, Facesoffungi number: FoF04486; Fig. 7
Etymology: name refers to its cylindrical appendages.
Holotype: MFLU 18-0019
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark brown, with base attached on surface of host plant. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 2–10 × 1–4 μm (\( \bar{x} \) = 6 × 2.5 μm, n = 10), holoblastic, cylindrical to obovoid, subhyaline. Conidia 30–40 × 12–25 μm (\( \bar{x} \) = 34 × 22 μm, n = 30), solitary, oval to ellipsoid, cheiroid, not complanate, yellow–brown, consisting of 30–40 cells arranged in (4–)5 rows, with a basal connecting cell, not easy to separate rows, guttulate, smooth-walled; 1–3-appendages rounded to cylindrical, 18–25 × 4–6 μm, apical on peripheral rows, hyaline, guttulate.
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, circular, entire edge with white at the margin and cream to yellow–orange in the central, raised on surface media.
Material examined: THAILAND, Nakhon Si Thammarat Province, Khanom District, on Pandanus sp., 22 July 2017, S. Tibpromma SR06 (MFLU 18-0019, holotype; HKAS 100846, isotype); ex-type living culture, MFLUCC 17-2259 = KUMCC 17-0311.
GenBank numbers LSU: MH376715; ITS: MH388343.
Notes: Dictyosporium appendiculatum clusters with Di. thailandicum M.J. D’souza, Bhat & K.D. Hyde (Liu et al. 2015b), but morphological comparison showed Di. appendiculatum is a distinct species. Dictyosporium appendiculatum has conidia 34 × 22 μm, yellow–brown, with 30–40 cells and (4–)5 rows, with 1–3 hyaline appendages, while Di. thailandicum has conidia 30.6 × 19 μm, dark brown to black at maturity, with 28–32 cells and 5 rows, with two hyaline appendages (Liu et al. 2015b). Based on morphology Di. appendiculatum is similar to Di. zhejiangense Wongs., H.K. Wang, K.D. Hyde & F.C. Lin, but the latter has complanate conidia, 25–35 × 17–24 μm and 23–37 cells per conidium (Wongsawas et al. 2009). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 17-2259 is 98% identical D. strelitziae strain CBS 123359 (NR_156216).
Dictyosporium digitatum J.L. Chen, C.H. Hwang and Tzean, Mycological Research 95: 1145 (1991)
Facesoffungi number: FoF04487; Fig. 8
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark brown, with base attached on surface of host plant. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 3–6 × 1–3.5 µm (\( \bar{x} \) = 3.5 × 1.5 μm, n = 20), holoblastic, cylindrical, pale brown to yellow–brown. Conidia 60–70 × 30–40 μm (\( \bar{x} \) = 62.5 × 33.5 μm, n = 20), solitary, oval to ellipsoid, cheiroid, not complanate, consisting of 65–90 cells arranged in 6–8 rows, with a basal connecting cell, not easy to separate, yellow–brown to brown, guttulate, subhyaline at the tip of peripheral rows, smooth-walled.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, entire edge with cream to yellow–orange, raised on surface media, producing yellow–brown pigment in media. Sporulating in culture after 3 months producing conidia similar in shape to those recorded on natural dead leaves.
Material examined: HONG KONG, Lantau Island, Pui O Beach, on Pandanus sp., 20 September 2016, S. Tibpromma HK08 (HKAS 100860); living culture, KUMCC 17-0269 = MFLUCC 17-0635.
GenBank numbers LSU: MH376716; ITS: MH388344; SSU: MH388311; TEF1: MH388378.
Notes: Dictyosporium digitatum was first described from Taiwan (Chen et al. 1991). Morphologically our isolate is identical to Di. digitatum (Chen 1991). Dictyosporium digitatum is common on Pandanaceae and has been found on Pandanus spp. in Australia, Brunei, Hong Kong, Mauritius, Philippines, Seychelles, Taiwan and Thailand (Whitton et al. 2012) which characterised by thick-walled conidia, often flexuous appendages at the apices of each conidial row. Based on a BLASTn search on NCBI GenBank nucleotide database, the closest matches for the ITS sequence of our isolate is Di. digitatum (KT 2660) (identities = 503/503 (100%)).
Dictyosporium guttulatum Tibpromma & K.D. Hyde, sp. nov.
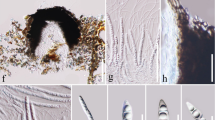
Index Fungorum number: IF555328, Facesoffungi number: FoF04488; Fig. 9
Dictyosporium guttulatum (MFLU 16-1914, holotype). a, b Colonies on dead leaves of Pandanus sp. c–f Conidiogenous cells with conidiophores and conidia. g Germinating conidium. h, i Colony on MEA from above and below. j–m Conidia formed in culture. Scale bars: a = 100 μm, b = 50 μm, c, d, j–m = 20 μm, e–g = 10 μm
Etymology: name refers to the guttulate conidia.
Holotype: MFLU 16-1914
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark, with base attached on surface of host plant. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 4–8.5 × 3–4 μm (\( \bar{x} \) = 6 × 3.5 μm, n = 20), holoblastic, cylindrical, pale brown to green–brown. Conidia 30–40 × 16–23 μm (\( \bar{x} \) = 37 × 20 μm, n = 30), solitary, oval to ellipsoid, cheiroid, not complanate, yellow–brown to brown, consisting of 40–44 cells arranged in (4–)5 rows, with a basal connecting cell, not easy to separate, guttulate, smooth-walled; appendages rounded to cylindrical, 3–11 × 3–5 μm, apical on peripheral rows, hyaline.
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, circular, entire edge with white at the margin and cream to yellow–orange in the centre, raised on surface media. Sporulating in culture after 3 months and producing conidia similar in shape to those recorded on dead leaves.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 15 December 2015, S. Tibpromma KB036 (MFLU 16-1914, holotype; HKAS 96263, isotype); ex-type living culture, MFLUCC 16-0258 = KUMCC 17-0288; Krabi Province, Mueang Krabi District, on Pandanus sp., 16 December 2015, S. Tibpromma KB039 (MFLU 16-1917, paratype).
GenBank numbers LSU: MH376717; ITS: MH388345; SSU: MH388312; TEF1: MH388379.
Notes: Dictyosporium guttulatum clusters with Di. hongkongensis but is well-separated with high bootstrap support (85% in ML, 1 in BYPP, Fig. 4). Dictyosporium guttulatum and Di. hongkongensis are both found on Pandanaceae but differ in morphology (Figs. 9, 10). Dictyosporium guttulatum has rounded to cylindrical appendages, while Di. hongkongensis lacks appendages. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0258 is 96% identical Di. hughesii strain KT1847 (LC014548), while the closest matches with the TEF1 sequence were with 97% identity with Di. digitatum strain yone 280 (AB808488).
Dictyosporium hongkongensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554479, Facesoffungi number: FoF04489; Fig. 10
Etymology: named after Hong Kong, where the fungus was first discovered.
Holotype: HKAS 100864
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark brown, with base attached on surface of host plant. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 4.5–8 × 3–5 μm (\( \bar{x} \) = 6 × 4 μm, n = 20), holoblastic, cylindrical, sometimes flat at base. Conidia 28–41 × 18–26 μm (\( \bar{x} \) = 35.4 × 22 μm, n = 20), solitary, oval to ellipsoid, yellow–brown, cheiroid, not complanate, consisting of 16–30 cells arranged in 4–5 rows, 5–7 cells per row with a basal connecting cell, guttulate, without appendages.
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, circular, rough, entire edge with yellow–brown to dark–brown, yellow–brown in the central, become dark–brown at the margin, raised on surface media.
Material examined: HONG KONG, around Tai Tam Tuk Reservoir Dam, on Pandanus sp., 21 September 2016, S. Tibpromma HK03 (HKAS 100864, holotype); ex-type living culture, KUMCC 17-0268 = MFLUCC 17-0633.
GenBank numbers LSU: MH376718; ITS: MH388346; SSU: MH388313; TEF1: MH388380.
Notes: There were 23 bp (4.22%) differences of the 545 ITS (+5.8S) nucleotides and 17 bp (1.88%) differences in 900 TEF1 nucleotides between Di. hongkongensis and Di. guttulatum. Also, Di. hongkongensis has similar conidia to Di. alatum Emden. Dictyosporium alatum has 5 rows of cells, and 4–6 cells per row with appendages (Whitton et al. 2012), while Di. hongkongensis has 4–5 rows of cell and 5–7 cells per row but lacks appendages. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0268 is 96% identical Di. zhejiangense strain MW-2009a (FJ456893), while the closest matches with the TEF1 sequence were with 97% identity with Di. digitatum strain yone 280 (AB808488).
Dictyosporium krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554480, Facesoffungi number: FoF04490; Fig. 11
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1890
Colonies on leaf sheath of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies black, flat on host surface. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 2–3.5 × 2–3 μm (\( \bar{x} \) = 3 × 2.5 μm, n = 20), holoblastic, integrated, terminal, smooth, thin-walled. Conidia 14–17 × 15–20 μm (\( \bar{x} \) = 16 × 17 μm, n = 40), solitary, yellow green, composed of 4–5 rows, inflated, rows not separating, always in group, each row consisting of 4–6 cell with distinct guttules in each cell; appendages 1–2, cylindrical, conical at apex, hyaline, apical on outside rows.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on dead leaf sheath Pandanus sp., 14 December 2017, S. Tibpromma KB012 (MFLU 16-1890, holotype; HKAS 96240, isotype).
GenBank numbers LSU: MH376719; SSU: MH388314; TEF1: MH388381.
Notes: Dictyosporium krabiense is characterized by 14–17 × 15–20 μm, yellow–green conidia, composed of 4–5 rows, with 1–2 long cylindrical appendages. Based on phylogenetic evidence, Di. krabiense is well-separated from other Dictyosporium species (Fig. 4). Morphologically Di. krabiense has similar conidia to Di. oblongum (Fuckel) S. Hughes and Di. polystichum (Höhn.) Damon but Di. oblongum has conidia 30–50 × 12–30 μm, uniformly medium to dark brown and without appendages (Goh et al. 1999), while Di. polystichum has conidia 26–34 × 23–34 μm, uniformly medium to dark brown and without appendages (Goh et al. 1999). In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of MFLU 16-1890 is 98% identical Di. bulbosum strain yone 221 (AB808487).
Dictyosporium pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554481, Facesoffungi number: FoF04491; Fig. 12
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1886
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia on natural substrate in small groups, dark brown. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 3.5–8 × 3–3.5 μm (\( \bar{x} \) = 5.5 × 3 μm, n = 10), holoblastic, cylindrical sometimes flat at base, hyaline to pale brown. Conidia 30–50 × 15–33 μm (\( \bar{x} \) = 41 × 23 μm, n = 20), solitary, oval to ellipsoid, cheiroid, not complanate, consisting of 34–62 cells arranged in 5–6 rows, each with 6–8 cells, with a basal connecting cell, yellow–brown to brown with age, without appendages or mucilaginous sheath.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB008 (MFLU 16-1886, holotype; HKAS 96236, isotype).
GenBank numbers LSU: MH376720; ITS: MH388347; TEF1: MH388382.
Notes: Dictyosporium pandanicola clustered with Di. strelitziae Crous & A.R. Wood in the phylogenetic analysis, but conidia of Di. strelitziae are arranged in (4–)5(–6) rows with 7–11 cells and with globose, apical appendage (Crous et al. 2009), while conidia of Di. pandanicola are arranged in 5–6 rows with 6–8 cells, and lack appendages. Dictyosporium pandanicola is also similar to Di. pandani but the latter has conidia measuring 22–48 × 14–28 μm with 4–5 rows of cells and 29–49 cells per conidium (Whitton et al. 2012). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLU 16-1886 is Di. strelitziae with 99% identity to the strain CBS 123359 (NR_156216), while the closest matches with the TEF1 sequence were with 99% identical Di. bulbosum strain yone 221 (AB808487).
Didymosphaeriaceae Munk
Didymosphaeriaceae was erected by Munk (1953) with the type genus Didymosphaeria Fuckel. and belongs to the order Pleosporales. In previous studies, Didymosphaeriaceae placement was not clear (Barr 1990a; Lumbsch and Huhndorf 2007; Zhang et al. 2012). Wijayawardene et al. (2018) accepted and provided details of 28 genera. We provide an updated tree following Thambugala et al. (2017) and propose a new species of Pseudopithomyces and of Montagnula from Pandanaceae in Thailand, based on morphology and phylogeny analysis (Fig. 13). We also provide a description of Deniquelata barringtoniae, which is newly recorded on Pandanaceae in Thailand.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF1 and ITS sequence data. Related sequences were obtained from Tennakoon et al. (2016). Seventy-two strains are included in the combined sequence analysis, which comprise 3168 characters with gaps. Pleospora herbarum (CBS 191.86) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 15836.943543 is presented. The matrix had 1031 distinct alignment patterns, with 39.64% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239912, C = 0.243956, G = 0.276827, T = 0.239305; substitution rates AC = 1.294307, AG = 1.980610, AT = 1.324440, CG = 0.930745, CT = 7.141409, GT = 1.000000; gamma distribution shape parameter α = 0.184270. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Deniquelata Ariyaw. & K.D. Hyde
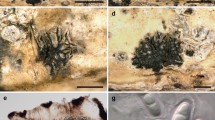
The monotypic genus Deniquelata was erected by Ariyawansa et al. (2013) to accommodate D. barringtoniae Ariyawansa & K.D. Hyde, which was collected from a leaf of Barringtonia asiatica in Chiang Rai, Thailand. We found another collection of Deniquelata barringtoniae on dead leaves of Pandanus sp. from Prachuap Khiri Khan in Thailand.
Deniquelata barringtoniae Ariyawansa and K.D. Hyde, Phytotaxa 105 (1): 15 (2013)
Facesoffungi number: FoF04492; Fig. 14
Deniquelata barringtoniae (MFLU 16-0556). a Appearance of ascomata on host. b Section of ascoma. c Section of peridium. d Pseudoparaphyses. e–g Asci and ascospores at different stages of maturity. h Ascospore mounted in India ink. i–m Ascospores. n Germinating ascospore. o, p Colony on MEA from above and below. Scale bars: a = 500 μm, b, c = 50 μm, d, i–n = 5 μm, e–g = 20 μm, h = 10 μm
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 120–220 × 90–130 µm (\( \bar{x} \) = 159.5 × 109 µm, n = 5), scattered to gregarious, immersed, conspicuous on host surface, dark brown, dull, solitary, uniloculate, globose to subglobose, without papilla and ostiole. Peridium 22–30 µm wide, composed of several layers of thick-walled, yellow–brown cells of textura angularis. Hamathecium comprising numerous 2–3 µm wide, filiform, filamentous, unbranched, guttulate, septate pseudoparaphyses. Asci 60–130 × 10–25 μm (\( \bar{x} \) = 88.5 × 17 μm, n = 20), (6–)8-spored, bitunicate, cylindrical to clavate, with a short furcate pedicel, apically rounded. Ascospores 10–20 × 5–10 µm (\( \bar{x} \) = 14 × 8 µm, n = 40), overlapping 2–3-seriate, muriform, oblong to narrowly oblong, straight or somewhat curved, pale brown to yellow–brown, with 3 transverse septa and 1−2 longitudinal septa in the middle cells, constricted at septa, verruculose, guttulate, with mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, white–grey on the surface, circular, with entire edge, raised, not yellow–white in reverse, with smooth margin.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-035 (MFLU 16-0556); living culture, MFLUCC 16-0271 = KUMCC 16-0161.
GenBank numbers LSU: MH260291; ITS: MH275059; SSU: MH260333; TEF1: MH412766; RPB2: MH412753.
Notes: In the molecular analysis our isolate clustered with Deniquelata barringtoniae (CBS 109027) with high bootstrap support (100% in ML, 1 in BYPP, Fig. 13). The morphology of our isolate was similar to that of D. barringtoniae described by Ariyawansa et al. (2013), although we note that the ascospores have a mucilaginous sheath. This is the first report of Deniquelata from Pandanaceae.
Montagnula Berl.
Montagnula was erected by Berlese (1896) with M. infernalis (Niessl) Berl. as type species. The genus is characterized by globose or sphaerical, immersed ascomata with a clypeus, claviform asci, fusoid or ellipsoid ascospores with transverse septa. The members species can be saprobes growing on dead plants, especially dead wood and bark, sometimes on dead leaves (Ariyawansa et al. 2014). Presently, there are 38 epithets are listed in Index Fungorum (2018). We provide an updated tree and introduce a new species from Pandanaceae in Thailand based on morphology and phylogenetic analysis (Fig. 13).
Montagnula krabiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554482, Facesoffungi number: FoF04493; Fig. 15
Montagnula krabiensis (MFLU 16-1891, holotype). a Appearance of ascomata on host. b Section of ascoma. c Section of peridium. d Pseudoparaphyses. e–h Asci and ascospores at different stages of maturity. i-k Ascospores. l Germinating ascospore. m, n Colony on MEA from above and below. Scale bars: a = 200 μm, b = 50 μm, c, i–l = 10 μm, d = 5 μm, e–h = 20 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1891
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 140–160 × 150–170 µm (\( \bar{x} \) = 152 × 160 µm, n = 5), immersed, under clypeus, inconspicuous on host surface, solitary or scattered, as small dark brown dots, solitary, uniloculate, globose, without papilla and ostiole. Peridium 12–26 µm wide, comprising several layers, composed of dark brown to black cells of textura angularis. Hamathecium comprising 2–4 µm, numerous filamentous, unbranched, guttulate, septate pseudoparaphyses. Asci 70–125 × 15–20 μm (\( \bar{x} \) = 94 × 16 μm, n = 10), 8-spored, bitunicate, cylindrical to clavate, long-pedicellate, furcate at base, apically rounded. Ascospores 25–32 × 6–7 µm (\( \bar{x} \) = 29 × 6.5 µm, n = 20), 1–2-seriate, fusiform, 1-septate, septum median, widest at the centre and tapering towards the narrow ends, constricted at the septa, yellow–brown to brown with age, guttulate, with mucilaginous sheath, thick-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, reddish brown to brown, circular, with entire edge, raised, dark brown in reverse, with smooth margin.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB013 (MFLU 16-1891, holotype; HKAS 96241, isotype); ex-type living culture, MFLUCC 16-0250 = KUMCC 16-0138.
GenBank numbers LSU: MH260303; ITS: MH275070; SSU: MH260343; TEF1: MH412776.
Notes: Montagnula krabiensis has cylindrical to clavate, long-pedicellate asci, and fusiform, narrow 1-septate, yellow brown to brown ascospores, with a rough mucilaginous sheath. Based on phylogenic analysis, M. krabiensis clusters with M. appendiculata (Aptroot) Wanas., E.B.G. Jones & K.D. Hyde (0.90 in BYPP, Fig. 13). However, M. appendiculata has reddish brown, broadly fusiform ascospores, with 2–5 greenish oil droplets and two polar hyaline appendages (Aptroot 2004). This is the first report of Montagnula species from Pandanaceae. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0250 is M. scabiosae with 93% identity to the strain MFLUCC 14-0954 (NR_155378).
Pseudopithomyces Ariyaw. & K.D. Hyde
Pseudopithomyces was erected by Ariyawansa et al. (2015b) to accommodate P. chartarum (Berk. & M.A. Curtis) J.F. Li, Ariyawansa & K.D. Hyde. The members can be found as saprobes (Ariyawansa et al. 2015b). Nine epithets are listed in Index Fungorum (2018). We introduce a new species of Pseudopithomyces based on morphology and phylogeny from Pandanaceae. Pseudopithomyces has never been reported from Pandanaceae.
Pseudopithomyces pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554483, Facesoffungi number: FoF04494; Fig. 16
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0029
Saprobic on dead leaves of Pandanus amaryllifolius. Colonies effuse, dark brown to black. Mycelium mostly superficial or partly immersed on the substrate, composed of septate, branched, smooth, thin-walled, hyaline hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores micro- to macronematous, mononematous, hyaline. Conidiogenous cells holoblastic, terminal, hyaline, cylindrical. Conidia 10–25 × 7–15 μm (\( \bar{x} \) = 19 × 11 μm, n = 20), muriform with 2–3 transverse septa and 1–2 longitudinal septa, verruculose to echinulate, amygdaliform or ovoid, yellow to brown, often carrying part of broken conidiogenous cell at base.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, grey on the surface, with dense, circular, with entire edge, raised, dark brown in reverse, with smooth margin.
Material examined: THAILAND, Chiang Rai Province, Mueang District, Mae Fah Luang University, on Pandanus amaryllifolius Roxb., 15 December 2017, S. Tibpromma P10 (MFLU 18-0029, holotype); ex-type living culture, MFLUCC 18-0116.
GenBank numbers LSU: MH376738; ITS: MH388364; SSU: MH388329; TEF1: MH388399; RPB2: MH412734; TUB2: MH412724.
Notes: Pseudopithomyces pandanicola clusters with P. kunmingnensis Karun. & K.D. Hyde and P. chartarum (Berk. & M.A. Curtis) Jin F. Li, Ariyaw. & K.D. Hyde. However, P. kunmingnensis has globose or subglobose conidiogenous cells with verruculose to echinulate and light brown to brown conidia (Hyde et al. 2017), while P. chartarum has broadly ellipsoid, mid brown to dark brown conidia (Ariyawansa et al. 2015a, b). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 18-0116 is P. chartarum with 100% identity to the strain OA28 (JQ406588), while the closest matches with the RPB2 sequence were with 99% identical P. chartarum strain UTHSC 04-678 (LK936414).
Hermatomycetaceae Locq.
Hermatomycetaceae was proposed by Locquin (1984) and formalised by Hashimoto et al. (2017). The type genus, Hermatomyces was placed within Ascomycota as ‘incertae sedis’, in previous studies (Wijayawardene et al. 2012), while Doilom et al. (2017) and Tibpromma et al. (2016b) suggested it belongs in Lophiotremataceae. Five species of Hermatomyces have been previously found on Pandanaceae (Tibpromma et al. 2016b, 2017b; Hyde et al. 2017). In addition, we describe a new taxon belonging to Hermatomyces which was collected from Pandanaceae in China.
Hermatomyces Speg.
Hermatomyces was introduced as a hyphomycetous genus by Spegazzini (1911) with H. tucumanensis Speg. as the type species. The characteristic features of Hermatomyces are sporodochial conidiomata with one to two types of conidia (lenticular and cylindrical) (Chang 1995). Sexual morphs have not been found (Hashimoto et al. 2017). There are 23 epithets for Hermatomyces listed in Index Fungorum (2018). We provide an updated phylogenetic tree (Fig. 17) for this genus. Hermatomyces tucumanensis has been found on leaves of Pandanus sp., P. furcatus, P. monticola and P. tectorius in Hong Kong (Whitton et al. 2012). Tibpromma et al. (2017b) described four species of Hermatomyces on Pandanus spp. from Thailand, while Hyde et al. (2017) described H. nabanheensis on Pandanus sp. from China.
Phylogram generated from maximum likelihood analysis based on combined LSU, TEF1, RPB2, TUB2 and ITS sequence data. Related sequences were obtained from Tibpromma et al. (2017b) and Koukol et al. (2018). Forty-six strains are included in the combined sequence analysis, which comprise 3835 characters with gaps. Verruculina enalia (BCC 18401) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 11360.152489 is presented. The matrix had 793 distinct alignment patterns, with 39.31% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.241274, C = 0.263021, G = 0.260518, T = 0.235186; substitution rates AC = 1.445186, AG = 4.215831, AT = 1.256927, CG = 0.850126, CT = 11.957308, GT = 1.000000; gamma distribution shape parameter α = 0.166181. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red bold while black bold are previous sequences from Hermatomyces species from Pandanaceae
Hermatomyces biconisporus Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554484, Facesoffungi number: FoF04495; Fig. 18
Etymology: biconisporus refers to two types of conidia.
Holotype: HKAS 99630
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural blackish brown, velvety, shiny, in small groups, glistening, conidia readily liberated when disturbed. Mycelium superficial, composed of a network of branched, septate, hyaline to pale brown, thick-walled hyphae 1.7–3.4 μm wide. Conidiophores 2.5–4 × 1.5–2.5 μm, micronematous, straight or flexuous, short, hyaline to pale brown, aseptate, smooth, unbranched, arising from prostrate hyphae at the centre of colony. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, cylindrical, hyaline to subhyaline. Conidia dimorphic, thick-walled, smooth: lenticular conidia 28–34 × 15–25 µm (\( \bar{x} \) = 31 × 18 µm, n = 30), numerous, central cells dark brown to black, peripheral cells subhyaline to pale brown, slightly constricted at septa, smooth, in lateral view obovoid, guttulate; cylindrical conidia 32–39 × 14.5–26 μm in broadest part of lower cells, (\( \bar{x} \) = 36 × 19 μm, n = 20), with 1–2 forked columns of 3–4 cells arising from a common basal cell, each column of rectangular to globose cells, constricted at septa, subhyaline, upper part of terminal cells dark brown, granulate, smooth.
Culture characteristics: Colonies on PDA at room temperature reaching 9 cm in 4 weeks, circular, yellow–white to grey mycelium with white margin, smooth surface, velvety and raised, white to yellow–brown from below.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on dead leaves of Pandanus sp., 12 November 2016, T. Aluthwaththa XTBG22 (HKAS 99630, holotype); living cultures, KUMCC 17-0183, MFLUCC 17-2267).
GenBank numbers LSU: MH260296; ITS: MH275063; SSU: MH260338; TEF1: MH412771; RPB2: MH412755.
Notes: Hermatomyces sphaericus was introduced by Hughes (1953) with a single conidium type (Hughes 1953). Koukol et al. (2018) synonymized several species under H. sphaericus (H. chromolaenae Jin F. Li, Mapook & K.D. Hyde, H. saikhuensis Tibpromma, Bhat & K.D. Hyde and H. tectonae Bhat & K.D. Hyde) based on morphological and molecular comparisions. Our isolate of H. biconisporus (KUMCC 17-0183) has two conidial types which are similar to H. tectonae, but in the phylogenetic analysis, the sequences of our isolate clustered with the H. sphaericus clade with a single conidium type. Therefore, we do not agree with Koukol et al. (2018) who synonymized H. saikhuensis and H. tectonae under H. sphaericus. There are important differences in base pairs (bp) even though they clustered together in the phylogenetic analysis. Although the lenticular conidia are similar, we believe that H. sphaericus is a species complex comprising several species.
In a BLASTn search in the NCBI GenBank, the closest match to the ITS sequence of H. biconisporus KUMCC 17-0183 is H. sphaericus strain PMA:116081 (LS398283) with 99% similarity, while the closest match to the TEF1 sequence is H. sphaericus strain PRC 4100 (LS398429) with 99% similarity. The closest match to the RPB2 sequence is H. tectonae strain KH 409 (LC194456) with 99% similarity and the closest match to the RPB2 sequence is H. tectonae strain KH 409 (LC194332) with 99% similarity.
There is evidence that our strain is a new species based on the recommendations of Jeewon and Hyde (2016). Thus, we introduce our collection as a new species based on morphological differences and difference in nucleotide base pairs. Hermatomyces chromolaenae, H. saikhuensis and H. tectonae clustered with the H. sphaericus clade, but these taxa lack TUB2 gene sequence data which is needed when introducing new species of Hermatomyces. We also maintain these names as distinct species until further research proves otherwise.
Melanommataceae G. Winter (= Pseudodidymellaceae A. Hashim. & Kaz. Tanaka)
Melanommataceae was established by Winter (1885a, b) with Melanomma Nitschke ex Fuckel as type genus. The family is characterized by globose or depressed perithecial ascomata, bitunicate and fissitunicate asci and pigmented and phragmosporous ascospores (Sivanesan 1984; Barr 1990a; Zhang et al. 2012; Hyde et al. 2013). A key to genera of Melanommataceae is provided in Tian et al. (2015). There are twenty-four genera in the family (Wijayawardene et al. 2017b, 2018). We collected Byssosphaeria siamensis on decaying dead leaves of Pandanus sp.; it has superficial ascomata, surrounded by brown to dark brown setae.
Byssosphaeria Cooke
Byssosphaeria was erected by Cooke and Plowright (1879) with B. keithii (Berk. & Broome) Cooke as type species. There are 25 epithets are listed in Index Fungorum (2018) which are commonly are found as saprobes (Lumbsch and Huhndorf 2010; Wijayawardene et al. 2017a, b). Tian et al. (2015) provided an update to Byssosphaeria based on morphology and phylogeny with new species and records have been introduced by Hyde et al. (2018). We found Byssosphaeria siamensis on Pandanaceae in Thailand. This species was originally described from decaying wood in Thailand.
Byssosphaeria siamensis Boonmee, Q. Tian and K.D. Hyde, Fungal Diversity 74: 283 (2015)
Facesoffungi number: FoF04499; Fig. 19
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF1 and RPB2 sequence data. Related sequences were obtained from Tian et al. (2015). Twenty-eight strains are included in the combined sequence analysis, which comprise 4229 characters with gaps. Trematosphaeria pertusa (CBS 122371) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 4630.742815 is presented. The matrix had 366 distinct alignment patterns, with 20.38% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.247303, C = 0.233841, G = 0.315115, T = 0.203741; substitution rates AC = 0.959786, AG = 2.362319, AT = 1.107025, CG = 1.419953, CT = 9.165549, GT = 1.000000; gamma distribution shape parameter α = 0.321701. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 290–405 × 335–465 µm (\( \bar{x} \) = 352 × 378.5 µm, n = 5), superficial, with flat base, conspicuous at the surface, globose to subglobose, uni-loculate, black, hairy, ostiole at central, with pore-like opening, surrounded by orange to yellow disc. Peridium 50–65 µm wide, outer layer comprising 4–6 layers of flattened, brown cells, arranged in a textura angularis, inner layer comprising 3–5 layers, hyaline cells, arranged in textura angularis to textura prismatica. Hamathecium comprising numerous, dense, 1–2.5 µm wide, filiform, filamentous, branched, guttulate, septate pseudoparaphyses. Asci 85–170 × 9–16 μm (\( \bar{x} \) = 129 × 14 μm, n = 20), 8-spored, bitunicate, fissitunicate, clavate, long-pedicellate with knob-like pedicel, apically rounded. Ascospores 30–40 × 5.5–7.5 µm (\( \bar{x} \) = 34 × 7 µm, n = 20), overlapping 1–2-seriate, fusiform, conical at each end, hyaline to pale brown with age, 1-septate, constricted at the septum, smooth-walled, guttulate, with mucilaginous sheath. Asexual morph Undetermined.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Pandanus sp., 20 December 2017, N. Chaiwan P13 (MFLU 18-0032 = HKAS 101800).
GenBank numbers LSU: MH376706; ITS: MH388334; SSU: MH388303; TEF1: MH388370; TUB2: 18-0032.
Notes: Our new strain of Byssosphaeria siamensis has the same characteristics as that reported on decaying wood in Thailand (MFLUCC 10-0099) (Tian et al. 2015). They both have superficial, globose to subglobose ascomata, with orange to yellow disc around the pore, clavate asci with long and knob-like pedicel and fusiform ascospores with conical ends and a mucilaginous sheath. Phylogenetic analyses using combined LSU, SSU, TEF1 and RPB2 sequence data demonstrate that our strain is B. siamensis (Fig. 20). The ascospores of this fungus failed to germinate and grow in culture and we extracted DNA directly from fruiting bodies. This is the first record of B. siamensis on Pandanaceae.
Occultibambusaceae D.Q. Dai & K.D. Hyde
Occultibambusaceae was erected by Dai et al. (2017) with Occultibambusa as the type genus. Members of the family Occultibambusaceae are usually found on monocotyledons hosts, but can also be found on hardwood trees (Dai et al. 2017). The characteristic features of asci and ascospores of Occultibambusaceae are similar to Bambusicola (Bambusicolaceae), Lophiostoma (Lophiostomataceae) and Massarina (Massarinaceae) (Zhang et al. 2009; Dai et al. 2012, 2015). There are four genera in the family (Wijayawardene et al. 2018). Neooccultibambusa pandanicola (asexual morph), which belongs in Occultibambusaceae, has been reported on Pandanaceae (Hyde et al. 2018). We introduce a new species of Neooccultibambusa.
Neooccultibambusa Doilom & K.D. Hyde
Neooccultibambusa was erected by Doilom et al. (2017) to accommodate N. chiangraiensis Doilom & K.D. Hyde, which is provided with both sexual and asexual morphs. Neooccultibambusa shares similar morphology with Occultibambusa (Doilom et al. 2017). There are three species for Neooccultibambusa are listed in Index Fungorum (2018).
Neooccultibambusa thailandensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554488, Facesoffungi number: FoF04500; Fig. 21
Phylogram generated from maximum likelihood analysis based on combined LSU, RPB2, SSU and ITS sequence data. Related sequences were obtained from Zhang et al. (2016). Fifty-five strains are included in the combined sequence analysis, which comprise 3605 characters with gaps. Melanomma pulvispyrius (CBS 124080) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 4630.742815 is presented. The matrix had 366 distinct alignment patterns, with 20.38% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.247303, C = 0.233841, G = 0.315115, T = 0.203741; substitution rates AC = 0.959786, AG = 2.362319, AT = 1.107025, CG = 1.419953, CT = 9.165549, GT = 1.000000; gamma distribution shape parameter α = 0.463512. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 18-0017
Saprobic on dead leaf of Pandanus sp. Sexual morph Ascomata 65–80 × 44–61 µm (\( \bar{x} \) = 72 × 51 µm, n = 5), superficial, globose to subglobose, flat at the base, solitary, papillate, ostiole central, black, smooth-walled. Peridium 5.6–14 µm wide, composed several layers of dark brown to black cells of textura angularis. Hamathecium comprising 0.7–1.7 µm wide, aseptate pseudoparaphyses. Asci 34–51 × 5–8 µm (= 45 × 6 µm, n = 20), (6–)8-spored, bitunicate, fissitunicate, cylindrical to cylindric-clavate, short-pedicellate, apically rounded with an ocular chamber. Ascospores 6–11 × 2–3.5 µm (\( \bar{x} \) = 9 × 2.6 µm, n = 40), overlapping uni- to bi-seriate, yellow–brown, fusiform, 1-septate, constricted at the septum, conical at each end, guttulate, smooth-walled, without appendages or mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 24 h. Colonies on MEA circular, convex, aerial in the center, edge entire, rough, dark brown to black mycelium.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on dead leaf of Pandanus sp., 30 July 2015, S. Tibpromma & K.D Hyde SF15-044 (MFLU 18-0017, holotype; HKAS 100843, isotype); ex-type living culture, MFLUCC 16-0274 = KUMCC 17-0309.
GenBank numbers LSU: MH260308; ITS: MH275074; SSU: MH260348; TEF1: MH412780; RPB2: MH412758.
Notes: Based on phylogeny this species group with Neooccultibambusa (Fig. 22). The previous species was reported with an asexual morph and we compare morphological characteristics of N. thailandensis (sexual morph) is distinct. Neooccultibambusa thailandensis and differs from N. chiangraiensis Doilom & K.D. Hyde which has 1–3 transverse septate ascospores surrounded by a mucilaginous sheath, while N. thailandensis has 1-septate ascospores and without a mucilaginous sheath. Lacking asexual morph is also a factor to distinguish two species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0274 is Neokalmusia didymospora with 97% identity to the strain MFLUCC 11-0613 (KP091433), while the closest matches with the TEF1 sequence were with 95% identical Occultibambusa maolanensis strain GZCC 16-0116 (KY814757).
Neooccultibambusa thailandensis (MFLU 18-0017, holotype). a Colonies on dead leaves of Pandanus sp. b Section of ascoma. c Peridium. d Hamathecium. e–g Asci. h-j Ascospores. k Germinating ascospore. l, m Colony on PDA from above and below. Scale bars a = 100 µm, b = 10 µm, c, f, g = 5 µm, d, f, h–k = 2 µm, e = 20 µm
Pleosporaceae Nitschke
Pleosporaceae was erected by Nitschke (1869) with Pleospora Rabenh. ex Ces. & De Not. as the type genus, based on structure of ascomata and pseudoparaphyses and was assigned to the order Sphaeriales. The placement of Pleosporaceae was not clear (Theissen and Sydow 1917) and later Lumbsch and Huhndorf (2010), Zhang et al. (2012), Hyde et al. (2013), Ariyawansa et al. (2015a) and Thambugala et al. (2017) used multi-gene phylogenetic analyses to establish the placement of this family. Pathogens or saprobes on wood, dead herbaceous stems or leaves can be found in Pleosporaceae (Hyde et al. 2013). Curvularia (Pleosporaceae) comprises species associated with plants (saprobes and pathogens), living organisms including humans, fresh-water and soils worldwide (Sivanesan 1987; Manamgoda et al. 2011, 2012a, b; da Cunha et al. 2013; de Aldana et al. 2013; Rangaswamy et al. 2013; Verma et al. 2013; Hyde et al. 2014). Seven species of Curvularia are known from Pandanaceae (Whitton et al. 2012). We introduce four new species of saprobic Curvularia from Pandanaceae based on both morphology and phylogeny.
Curvularia Boedijn
Curvularia was erected by Boedijn (1933) with C. lunata (Wakker) Boedijn as the type species and characterised by long, dark coloured, polytretic conidiophores, and conidia that are usually curved, often versicoloured and multi-septate (Ellis 1971, 1976). More than 150 epithets are listed in Index Fungorum (2018). Whitton et al. (2012) reported seven species of Curvularia as known from Pandanaceae. An updated phylogenetic tree for Curvularia is presented (Fig. 23).
Phylogram generated from maximum likelihood analysis based on combined ITS, GPDH and TEF1 sequence data of Curvularia. Related sequences were obtained from Marin-Felix et al. (2017) and Hyde et al. (2017). Eighty-three strains are included in the combined sequence analysis, which comprise 2221 characters with gaps. Bipolaris bamagaensis (BRIP 15934) and Bipolaris panici-miliacei (CBS199.29) are used as the outgroup taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 15729.11775 is presented. The matrix had 897 distinct alignment patterns, with 23.55% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.232486, C = 0.300291, G = 0.241124, T = 0.226099; substitution rates AC = 0.833112, AG = 2.362707, AT = 1.030919, CG = 1.030374, CT = 5.190668, GT = 1.000000; gamma distribution shape parameter α = 0.737704. Bootstrap support values for ML equal to or greater than 70% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Curvularia chonburiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554492, Facesoffungi number: FoF04501; Fig. 24
Etymology: named after Chonburi Province, where the fungus was first discovered.
Holotype: MFLU 16-1876
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium composed of septate, branched, smooth, subhyaline hyphae. Conidiophores 70–200 × 4–7 µm (\( \bar{x} \) = 116 × 6 µm, n = 10), unbranched, septate, flexuous, geniculate, pale brown. Conidiogenous cells polytretic, sympodial, integrated, smooth, swollen, brown. Conidia 17–28 × 7–15 µm (\( \bar{x} \) = 23 × 11 µm, n = 40), ovoid to obclavate, narrowing towards rounded ends, (2–)3-distoseptate, with a dark brown band at septa, with end cells hyaline to pale brown, with 2 middle cells dark brown, third cell from base often larger, with a scar (hilum) at base.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 1 week at room temperature, beginning white and become black with age, circular, entire edge with curled, raised on media surface, smooth, velvety.
Material examined: THAILAND, Chonburi Province, Bang Lamung District, on dead leaf of Pandanus sp., 18 July 2016, W. Jaidee PTY01 (MFLU 16-1876, holotype); HKAS 96270, isotype; ex-type living culture, MFLUCC 16-0375 = KUMCC 17-0298.
GenBank numbers LSU: MH260287; ITS: MH275055; GPDH: MH412747.
Notes: Based on multi-gene analysis (ITS, GPDH and TEF1), our collection clustered with Curvularia hominis K.C. Cunha, Madrid, Gené & Cano with 97% in ML, 0.99 in BYPP (Fig. 23). Curvularia hominis is characterized by versicoloured conidia that are 18–30 × 7–14 μm, slightly curved, and 4–5-distoseptate (Madrid et al. 2014). Curvularia chonburiensis has larger conidia (17–28 × 7–15 µm) that are ovoid to obclavate, and 2–3-distoseptate. Thus, based on both morphological and molecular characteristics we introduce the new species, C. chonburiensis. In a BLASTn search on NCBI GenBank, the closest matches of GPDH sequence of MFLUCC 16-0375 is C.lunata with 95% identity (X58718), while the closest matches with the TEF1 sequence were with 99% identical C. hominis strain MFLUCC 12-0178 (KM196591) and ITS sequence were with 100% identical C. platzii strain BRIP27703 (MH414906).
Curvularia pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554491, Facesoffungi number: FoF04502; Fig. 25
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0009
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium composed of septate, branched, smooth, subhyaline hyphae. Conidiophores 100–180 × 1.5–5 µm (\( \bar{x} \) = 140 × 3 µm, n = 10), arising in groups, unbranched, septate, flexuous, nodose, pale brown. Conidiogenous cells polytretic, sympodial, integrated, smooth, swollen, brown. Conidia 9–17 × 4–8 µm (\( \bar{x} \) = 12 × 6 µm, n = 10), ovoid to obclavate, narrowing towards rounded ends, 3(–4)-distoseptate, with middle septum appearing as a thick black band, pale brown at end cells, dark brown in 2 middle cells, slightly curved, with a scar (hilum) at base.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 1 week at room temperature, yellow–brown to brown, circular, entire edge with raised on media surface, smooth.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on dead leaf of Pandanus sp., 8 December 2014, S. Tibpromma & K.D Hyde SF14-042 (MFLU 18-0009, holotype; HKAS 100833, isotype); ex-type living cultures, MFLUCC 15-0746 = KUMCC 16-0156.
GenBank numbers LSU: MH260288; ITS: MH275056; TEF1: MH412763; ACT: MH412737; GPDH: MH412748.
Notes: Curvularia pandanicola has similar conidia to C. eragrostidis (Henn.) J.A. Mey. However, the conidia of C. eragrostidis are 20 × 12 µm, ellipsoidal or barrel shaped, and 3-distoseptate (Su et al. 2015). Multi-gene phylogenetic analysis (Fig. 23) showed that C. pandanicola is distinct from C. eragrostidis and other Curvularia. In a BLASTn search on NCBI GenBank, the closest matches of GPDH sequence of MFLUCC 15-0746 is C.lunata with 94% identity (X58718), while the closest matches with the TEF1 sequence were with 99% identical Curvularia sp. strain 5854 (KT012577) and ITS sequence were with 100% identical C. eragrostidis strain CBS 189.48 (HG778986).
Curvularia thailandicum Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554493, Facesoffungi number: FoF04503; Fig. 26
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 18-0010
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 63–95 × 2.5–4 µm (\( \bar{x} \) = 78 × 3 µm, n = 10), arising in groups, unbranched, septate, flexuous, geniculate, pale brown to brown, guttulate, thick-walled. Conidiogenous cells polytretic, sympodial, integrated, smooth, swollen, brown. Conidia 8–15 × 5–8 µm (\( \bar{x} \) = 10.5 × 6 µm, n = 40), ovoid to obclavate, tapering towards rounded ends, subhyaline to yellow–brown or brown with age, basal cell subhyaline, smooth-walled, 3(–4)-distoseptate, with dark brown septa, guttulate, with a scar (hilum) at base.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 1 week at room temperature, white in the beginning and brown to black with age, circular, entire edge with raised on media surface, smooth, velvety.
Material examined: THAILAND, Phang Nga Province, Mueang Phang Nga District, on fallen dead and decaying leaves of Pandanus sp., 6 December 2014, S. Tibpromma SF14-046 (MFLU 18-0010, holotype; HKAS 100835, isotype); ex-type living culture, MFLUCC 15-0747 = KUMCC 17-0302.
GenBank numbers LSU: MH260289; ITS: MH275057; TEF1: MH412764; ACT: MH412738; GPDH: MH412749.
Notes: The verruculose, curved, 3–4(–5)-distoseptate conidia with middle cells unequally enlarged, reniform, pale brown to brown, apical and basal cells paler than middle cells being subhyaline to pale brown are characteristic features of Curvularia soli, while our new species showed ovoid to obclavate, subhyaline to yellow–brown or brown, 3–4-distoseptate conidia with basal cell always subhyaline. Curvularia thailandicum is introduced here as a new saprobic fungus on Pandanus sp. In a BLASTn search on NCBI GenBank, the closest matches of GPDH sequence of MFLUCC 15-0746 is C. lunata with 96% identity (X58718), while the closest matches with the TEF1 sequence were with 99% identical C. soli strain CBS 222.96 (KY905698) and ITS sequence were with 99% identical C. geniculata strain JUF0011 (MH371471).
Curvularia xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554494, Facesoffungi number: FoF04504; Fig. 27
Etymology: named after Xishuangbanna, where the fungus was first discovered.
Holotype: HKAS 99632
Saprobic on dead leaf of Pandanus amaryllifollus. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 100–400 × 6–10 µm (\( \bar{x} \) = 197 × 7 µm, n = 10), arising in groups, branched, septate, flexuous, geniculate, yellow–brown to brown, guttulate, thick-walled. Conidiogenous cell polytretic, sympodial, integrated, smooth, swollen, brown. Conidia 32–46 × 13–20 µm (\( \bar{x} \) = 38 × 16 µm, n = 30), ovoid to obclavate, tapering towards rounded ends, yellow–green to dark brown, 3–5-distoseptate with thick dark brown septa, guttulate, with a dark scar (hilum) at base.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 1 week at room temperature, white in the beginning and brown to black with age, circular, entire edge with raised on media surface, smooth, velvety.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus amaryllifollus Roxb., 15 November 2016, T. Aluthwaththa XTBG24 (HKAS 99632, holotype); ex-type living culture, KUMCC 17-0185 = MFLUCC 17-2271.
GenBank numbers LSU: MH260290; ITS: MH275058; TEF1: MH412765; ACT: MH412739; GPDH: MH412750.
Notes: Curvularia xishuangbannaensis is characterized by ovoid to obclavate conidia that are yellow-green to dark brown, 3–5-distoseptate, thick dark brown septa and rough. In the phylogenetic analysis, this species clustered with C. thailandicum, which has also been collected from Pandanaceae. Curvularia thailandicum differs from C. xishuangbannaensis by having subhyaline to yellow–brown or brown, 3(–4)-distoseptate conidia with basal cell always subhyaline. In a comparison of the 528 ITS (+5.8S) nucleotides of these two strains reveals 18 (3.40%) nucleotide differences with C. thailandicum and C. xishuangbannaensis. In a BLASTn search on NCBI GenBank, the closest matches of GPDH sequence of KUMCC 17-0185 is C. senegalensis with 98% identity to the strain CBS 431.75 (LT715832), while the closest matches with the TEF1 sequence were with 100% identical C. soli strain CBS 222.96 (KY905698) and ITS sequence were with 99% identical C. affinis strain CMRP2493 (MF154614).
Roussoellaceae J.K. Liu et al.
Roussoellaceae was erected by Liu et al. (2014) with Roussoella Sacc. as the type genus, based on both morphology and phylogenetic analyses. Members of Roussoellaceae are known from bamboo and palms (Hyde et al. 2013). Jaklitsch and Voglmayr (2016) treated Roussoellaceae as a synonym of Thyridariaceae, but Tibpromma et al. (2017b) recommended to retain Roussoellaceae based on phylogenetic analysis and also the two families have distinct morphological characteristics. Six genera are listed in Wijayawardene et al. (2018). We describe Roussoella solani on Pandanaceae from Thailand based on both morphology and phylogeny (Fig. 22).
Roussoella Sacc.
Roussoella was introduced with R. nitidula Sacc. & Paol. 1888 as type species. The species are commonly found as saprobes (Liu et al. 2014). There are 42 epithets are listed in Index Fungorum (2018). We describe another species from Pandanaceae, the first report of Roussoella from Pandanaceae.
Roussoella solani Crous & M.J. Wingf., Persoonia 36: 341 (2016)
Facesoffungi number: FoF04505; Fig. 28
Roussoella solani (MFLU 18-0007). a Conidiomata on host substrate. b Section through conidioma. c Section of pycnidial wall. d–h Conidiogenous cells and conidia. i–l Conidia. m Germinating conidium. n, o Colony on MEA from above and below. Scale bars: a = 500 μm, b, c = 200 μm, d–h = 50 μm, i–m = 2 μm
Saprobic on dead root of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata pycnidial, 160–170 × 77–105 μm (\( \bar{x} \) = 166 × 94 μm, n = 5), immersed to erumpent, under clypeus, visible as black, ovoid or obpyriform, solitary, scattered or gregarious, ostiolate, with an ostiole. Pycnidial wall 13–23 μm, composed of thin layers of cells of textura angularis, hyaline to pale brown. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–6 × 3–4 μm (\( \bar{x} \) = 5 × 3.5 μm, n = 10), enteroblastic, phialidic, integrated, ovoid to obpyriform, hyaline to pale brown, smooth. Conidia 2–5 × 1.5–4 μm (\( \bar{x} \) = 4 × 2.5 μm, n = 40), oval to ellipsoidal, initially hyaline, becoming pale brown, aseptate with one or two guttules, thick-walled, smooth-walled.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, brown on the surface, with dense, circular, with entire edge, convex with floosy and velvety, dark brown in reverse, with smooth margin.
Material examination: THAILAND, Phang Nga Province, Mueang Phang Nga District, on dead root of Pandanus sp., 6 December 2014, S. Tibpromma & K.D. Hyde SF14-004 (MFLU 18-0007; HKAS 100827); living culture, MFLUCC 15-0098.
GenBank numbers MFLUCC 15-0098 LSU: MH260309; ITS: MH275075; SSU: MH260349. MFLUCC 18-0007 LSU: MH260310; ITS: MH275076; SSU: MH260350.
Notes: In molecular phylogenetic analysis our isolates clustered with Roussoella solani Crous & M.J. Wingf (Fig. 22). We compared our isolates with R. solani (CPC 26331) and both have similar conidiogenous cells and conidia. Roussoella solani (CPC 26331) has hyaline, ampulliform to doliiform, 4–6 × 3–4 μm conidiogenous cells and (4–)4.5–5(–7) × 2(–3) μm, aseptate, pale brown, subcylindrical conidia (Crous et al. 2016a), while our isolate has 4–6 × 3–4 μm, hyaline to pale brown, ovoid to obpyriform conidiogenous cells and 2–5 × 1.5–4 μm, oval to ellipsoidal, aseptate conidia, initially hyaline, becoming pale brown. This is the first record of Roussoella on Pandanus sp.
Tetraplosphaeriaceae Kaz. Tanaka & K. Hiray
Tetraplosphaeriaceae was introduced with Tetraplosphaeria as the type genus (Tanaka et al. 2009). The morphology of Tetraplosphaeriaceae are Massarina-like sexual morph with hyaline 1(–3)-septate ascospores and/or Tetraploa-like asexual morph with several setose appendages (Tanaka et al. 2009). There are five genera accepted in Wijayawardene et al. (2018). We describe three new species of Polyplosphaeria on Pandanaceae from China.
Polyplosphaeria Kaz. Tanaka & K. Hiray
Polyplosphaeria is typified by P. fusca Kaz. Tanaka & K. Hirayama. The members are saprobes and characteristics including globose ascomata surrounded by numerous brown hyphae and Piricauda-like conidia (e.g. P. cochinensis and P. longispora) (Tanaka et al. 2009). Two epithets are listed in Index Fungorum (2018). This is the first report of Polyplosphaeria from Pandanaceae, and we provide an updated synopsis of Polyplosphaeria species (Table 2).
Polyplosphaeria nabanheensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554496, Facesoffungi number: FoF04506; Fig. 29
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS, TUB2 and TEF1 partial sequence data. Thirty-eight strains are included in the sequence analysis, which comprise 4198 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Muritestudina chiangraiensis (Testudinaceae) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 16498.013303 is presented. The matrix had 940 distinct alignment patterns, with 17.89% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.243803, C = 0.246513, G = 0.272484, T = 0.237200; substitution rates AC = 2.655152, AG = 4.461936, AT = 2.147661, CG = 1.746218, CT = 11.186368, GT = 1.000000; gamma distribution shape parameter α = 0.751821. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Nabanhe, where the fungus was first discovered.
Holotype: HKAS 96219
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, scattered or gregarious, blackish, dull, easy to remove. Mycelium superficial. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells monoblastic, forming directly on creeping hyphae holoblastic, determinate, with guttules, hyaline. Conidia 110–150 × 75–88 μm (\( \bar{x} \) = 143 × 82 μm, n = 10), oval to ellipsoid, made up of cells, 2–3 celled, dark brown, verrucose and dark at base, with setose appendages on surface, rough-walled; appendages unbranched, smooth, brown to black, of two types, long appendages 60–125 × 4–6 μm (\( \bar{x} \) = 86 × 5 μm, n = 10), 3–9-septate, arising from apical part of conidia; short appendages 35–40 × 3.5–6 μm (\( \bar{x} \) = 38 × 4.5 μm, n = 10), wide at the base, 0–2-septate and arising eccentrically from conidial base.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA, circular, undulate with dark brown mycelium. Mycelium superficial, velvety, flossy.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on fallen dead and decaying leaves of Pandanus sp., 2 August 2016, S. Tibpromma NBH19 (HKAS 96219, holotype); ex-type living culture, KUMCC 16-0151.
GenBank numbers LSU: MH260312; ITS: MH275078; SSU: MH260352; TUB2: MH412745.
Notes: In the phylogenetic analysis, Polyplosphaeria nabanheensis is in a distinct lineage, and basal to Polyplosphaeria (Fig. 30). Polyplosphaeria nabanheensis has oval to ellipsoid conidia while other Polyplosphaeria spp. are with globose, subglobose, obovoid, pyriform or ellipsoidal conidia (Tanaka et al. 2009). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0151 is Quadricrura meridionalis with 94% identity to the accession number NR_119401, while the closest matches with the TUB2 sequence were with 91% identical P. fusca strain KT 2124 (AB524853).
Polyplosphaeria pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554497, Facesoffungi number: FoF04507; Fig. 31
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 99627
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate scattered or gregarious, blackish, dull. Mycelium partly superficial. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells monoblastic, forming directly on creeping hyphae, holoblastic, integrated, terminal, determinate, incomplete globose connected to base of conidia, with guttules, hyaline. Conidia 35–70 × 40–75 μm (\( \bar{x} \) = 41.4 × 44.6 μm, n = 20), globose to subglobose, solitary, brown to dark brown, verrucose at base, with setose appendages on surface; appendages of two forms, unbranched, smooth, brown at base and almost hyaline at apex, long appendages 60–105 × 3–5 μm (\( \bar{x} \) = 81 × 4 μm, n = 10), wide at the apex, 4–6-septate, arising from apical part of conidia; short appendages 20–35 × 3.5–5 μm (\( \bar{x} \) = 26.4 × 4 μm, n = 10), wide at the base, 0–2-septate, arising eccentrically from conidial base.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA, circular, undulate with dark brown in the middle and grey at the margin, smooth and raised on surface media. Mycelium superficial, velvety, flossy.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus sp., 28 April 2017, R. Phookamsak & N.I. de Silva XTBG19 (HKAS 99627, holotype); ex-type living culture, KUMCC 17-0180 = MFLUCC 17-2266.
GenBank numbers LSU: MH260313; ITS: MH275079; SSU: MH260353.
Notes: Polyplosphaeria pandanicola shares common features with other Polyplosphaeria spp., but it differs by size of conidia (Table 2). According to our phylogenetic analysis, based on combined multi-gene sequence data, P. pandanicola formed a distinct lineage with 60% in ML bootstrap support (Fig. 30) compared with other known species in Polyplosphaeria. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0180 is P. fusca with 90% identity to the strain KT 1640 (AB524790).
Polyplosphaeria xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554498, Facesoffungi number: FoF04508; Fig. 32
Etymology: named after Xishuangbanna, where the fungus was first discovered.
Holotype: HKAS 101810
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate scattered or gregarious, blackish, shining. Mycelium superficial. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells monoblastic, forming directly on creeping hyphae, holoblastic, integrated, terminal, determinate, incomplete globose connected to base of conidia, with guttules, hyaline. Conidia 53–115 × 45–105 μm (\( \bar{x} \) = 76 × 69 μm, n = 10), subglobose, solitary, black, verrucose at base, with setose appendages on surface; appendages of two forms, unbranched, smooth, brown at base and almost hyaline at apex, long appendages 75–132 × 4–7 μm (\( \bar{x} \) = 105 × 5 μm, n = 20), wide at the apex, 4–7-septate, arising from apical part of conidia; short appendages 20–40 × 2–4.5 μm (\( \bar{x} \) = 31 × 4 μm, n = 20), wide at the base, 1–3-septate, arising eccentrically from conidial base.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA, circular, undulate with dark-grey, smooth and raised on surface media. Mycelium superficial, velvety, flossy.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus sp., 28 April 2017, R. Phookamsak & N.I. de Silva XTBG27 (HKAS 101810, holotype); ex-type living culture, KUMCC 17-0187.
GenBank numbers LSU: MH260314; ITS: MH275080; SSU: MH260354.
Notes: Polyplosphaeria xishuangbannaensis was collected from a dead leaf of Pandanus sp. Phylogenetic analysis placed our collection in Tetraplosphaeriaceae and grouped within Polyplosphaeria (Fig. 30). Polyplosphaeria xishuangbannaensis is distinct by black, subglobose conidia, while other Polyplosphaeria spp. have brown, dark brown or grey conidia (Table 2). Therefore, we refer our new isolates to a new species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0187 is Triplosphaeria sp. with 98% identity to the accession number KY315599.
Torulaceae Corda
Torulaceae was erected by Corda (1829) with Torula Pers. as type genus. Crous et al. (2015b) provided molecular data for Torulaceae. There are four genera in the family (Wijayawardene et al. 2018). We describe two known species of Torula and a new species of Rostriconidium collected on Pandanaceae from Thailand and China.
Rostriconidium Z.L. Luo et al.
Rostriconidium was erected by Su et al. (2018) as a monotypic genus for R. aquaticum Z.L. Luo, K.D. Hyde & H.Y. Su, from decaying wood in a river in China. Rostriconidium is characterized by long conidiophores with monotretic or polytretic conidiogenous cells. The conidia are rostrate with a sheath at the apex, multi-septate with a subhyaline apex and a distinctive, dark basal hilum with a pale coloured cell immediately above. We describe a new species of Rostriconidium. This is the first report of Rostriconidium on Pandanaceae.
Rostriconidium pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554490, Facesoffungi number: FoF04571; Fig. 33
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, RPB2 and TEF1 partial sequence data. Fifty strains are included in the sequence analysis, which comprise 3015 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Ohleria modesta (Ohleriaceae) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 13481.727487 is presented. The matrix had 946 distinct alignment patterns, with 26.17% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.242018, C = 0.268530, G = 0.274306, T = 0.215146; substitution rates AC = 2.017469, AG = 4.885145, AT = 2.212961, CG = 1.112797, CT = 12.027062, GT = 1.000000; gamma distribution shape parameter α = 0.614798. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 99620
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate consisting of long, dark, flexuous conidiophores. Mycelium immersed in the substrate, composed of septate, branched, brown hyphae. Conidiophores 360–485 × 11–13 μm (\( \bar{x} \) = 423 × 12.5 μm, n = 10), macronematous, mononematous, scattered, dark brown to black, rough-walled, septate, unbranched, straight or slightly flexuous. Conidiogenous cells 15–30 × 8.5–12 μm (\( \bar{x} \) = 21.6 × 10.6 μm, n = 20), polyblastic, sympodial, integrated, terminal, later becoming intercalary, cylindrical, dark brown, smooth, with black conidiogenous scars. Conidia 55–110 × 18–26 μm (\( \bar{x} \) = 69.5 × 21 μm, n = 20), solitary, pale brown to dark brown, rostrate, 4–7-septate, with dark bands at the septa, slightly constricted at septa, with truncate, thick scar at base, narrower at base, flattened and paler towards the apex, short apex when young, long rostrate apex when mature, with hyaline thin sheath at the apex, granulate, smooth-walled.
Culture characteristics: Conidia germinating on PDA within 24 h. and germ tubes produced at the apex. Colonies on PDA reaching 9 cm diam., in 2 weeks at room temperature, circular, undulate with white to cream, raised on surface media with flossy with velvety. Mycelium superficial, branched, septate, hyaline to pale yellow, smooth.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus sp., 30 April 2017, R. Phookamsak & N.I. de Silva XTBG12 (HKAS 99620, holotype); ex-type living culture, KUMCC 17-0176 = MFLUCC 17-2262; Yunnan Province, Xishuangbanna, on fallen dead of Pandanus sp., 28 April 2017, R. Phookamsak XTBG28 (HKAS 99626, paratype).
GenBank numbers LSU: MH260318; ITS: MH275084; SSU: MH260358; TEF1: MH412781; RPB2: MH412759.
Notes: The conidia of Rostriconidium aquaticum Z.L. Luo, K.D. Hyde & H.Y. Su are larger conidia (134–180 × 22–26 um) and have more septa (8–9) (Su et al. 2018) than those of R. pandanicola. Maximum likelihood of combined sequence data indicated that Rostriconidium pandanicola and R. aquaticum form a well-supported clade in Torulaceae (Fig. 34). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0176 is R. aquaticum with 95% identity to the strain MFLUCC 16-1113 (MG208164), while the closest matches with the TEF1 sequence were with 98% identical R. aquaticum strain KUMCC 15-0297 (MG207995) and RPB2 sequence were with 96% identical R. aquaticum strain KUMCC 15-0297 (MG207975).
Rostriconidium pandanicola (HKAS 99620, holotype). a Colonies on dead leaf of Pandanus sp. b, c Conidiophores, conidiogenous cells and conidia. d, e Conidiogenous cells and conidia. f, g Conidia. h Germinating conidium. i, j Colonies on PDA from above and below. Scale bars: b, c = 100 μm, d–h = 10 μm
Torula Pers.
Torula was erected by Persoon (1795) to accommodate T. monilis Pers. The genus is characterized by dark or subhyaline moniliform conidia (Persoon 1795). Crous et al. (2015b) accepted Torula in Torulaceae (Pleosporales). More than 500 epithets are listed in Index Fungorum (2018). We collected T. chromolaenae and T. ficus from Pandanaceae.
Torula chromolaenae J.F. Li, Phook., Mapook & K.D. Hyde, Mycological Progress 16 (4): 454 (2017)
Facesoffungi number: FoF04572; Fig. 35
Saprobic on dead leaf of Pandanus tectorius. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate effuse, black, powdery. Conidiophores up 40 μm long, micronematous, reduced to conidiogenous cells, with hyaline to pale brown supporting cell. Mycelium immersed or superficial, composed of 2–4 μm, hyaline, branched, septate, hyphae. Conidiogenous cells 10–35 × 3–5 µm (\( \bar{x} \) = 17 × 4 µm, n = 10), macronematous, solitary, cylindrical, hyaline to pale brown, verruculose at apex, mono- to polyblastic. Conidia 13–30 × 6–10 µm (\( \bar{x} \) = 23 × 8 µm, n = 50), phragmosporous, in short branched chains, 1 − 3-septate, with dark bands at septa, acrogenous, pale brown when young, brown when mature, pale brown at apex, constricted at septa, verrucose, fragmenting into segments.
Culture characteristics: Colonies on PDA attaining 9 cm diam., within 2 weeks at room temperature, brown to yellow–brown, irregular with undulate edge raised on media surface, smooth, velvety.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus tectorius Parkinson ex Du Roi., 15 October 2016, T. Aluthwaththa XTBG06 (HKAS 99614); living culture, KUMCC 17-0174.
GenBank numbers LSU: MH260321; ITS: MH275087; TEF1: MH412784; RPB2: MH412760.
Notes: Torula chromolaenae was described from Chromolaena odorata (Asteraceae) in Thailand (Li et al. 2017). In pair-wise comparison of DNA sequences of RPB2 and TEF1 regions of Torula chromolaenae (KUMCC 17-0174) and T. chromolaenae (KMUCC 16-0035) (type stain), they are almost identical (Fig. 34). Morphologically our isolate is identical to Torula chromolaenae.
Torula ficus Crous, IMA Fungus 6 (1): 192 (2015)
Facesoffungi number: FoF 04609; Fig. 36
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate effuse, black, powdery. Mycelium immersed or superficial, composed of 1–2.5 μm, hyaline, branched, septate hyphae. Conidiophores up to 25 μm, micronematous, reduced to conidiogenous cells, with hyaline to pale brown supporting cell. Conidiogenous cells 3–5 × 2–3 µm (\( \bar{x} \) = 4 × 2.5 µm, n = 10), solitary on mycelium, cylindrical, hyaline to pale brown, verruculose at apex, mono- to polyblastic. Conidia 10–23 × 4–7 µm (\( \bar{x} \) = 15 × 5 µm, n = 20), phragmosporous, in long branched chains, 1–3-septate, with dark bands at the septa, acrogenous, pale brown when young, brown when mature, pale brown at apex, constricted at septa, verrucose, fragmenting into segments.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 2 weeks at room temperature, circular with entire edge, raised on media surface, white at the middle with velvety, pale brown at the margin with flat mycelium.
Material examined: THAILAND, Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on dead leaf of Pandanus sp., 16 December 2017, S. Tibpromma P06 (MFLU 18-0025, HKAS 101794); living culture, MFLUCC 18-0112.
GenBank numbers LSU: MH260322; ITS: MH275088; TEF1: MH412785.
Notes: Torula ficus was originally collected from Ficus in Europe (Crous et al. 2015b) and later found on submerged decaying wood of Chromolaena odorata in Thailand (Li et al. 2017). We provide sequence data (Fig. 34) for an isolate collected from Pandanus sp. (Pandanaceae) in Thailand. The morphology our isolate is identical with T. ficus.
Pleosporales genera incertae sedis
Pseudoberkleasmium Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF555331, FacesofFungi number: FoF04509
Etymology: name refers to its characteristic features of Berkleasmium.
Type species: Pseudoberkleasmium pandanicola Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves and wood in terrestrial habitats. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, in small groups, blackish to brown, velvety, glistening, with conidia readily liberated when disturbed. Mycelium immersed in the substrate, composed of branched, septate, smooth, hyaline to subhyaline hyphae. Conidiophores micronematous, mononematous, fasciculate, septate, hyaline or subhyaline. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, globose, with or without guttules, hyaline to subhyaline. Conidia acrogenous, solitary, broadly ellipsoidal to obovoid, flattened, one-cell thick, muriform, brown olivaceous green, with or without guttules.
Notes: Pseudoberkleasmium is established with Ps. pandanicola as the type species based on morphology and phylogenetic analyses. In a BLASTn search on NCBI GenBank, the closest matches for LSU sequence of KUMCC 17-0178 was Hermatomyces chiangmaiensis strain MFLUCC 16-2819 (KY559394) with 96% similarity, while the closest matches for the ITS sequence of KUMCC 17-0178 was H. krabiensis strain MFLUCC 16-0249 (KX525750) with 92% similarity. Multi-gene (LSU, SSU, TEF1 and ITS genes) analyses of Pseudoberkleasmium indicated that it separates from the family of Hermatomycetaceae with 62% in ML, 0.99 in BYPP support (Fig. 37) and the morphology totally differs from Hermatomyces by having lenticular and cylindrical conidia (one to two types of conidia) (Chang 1995).
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and TEF1 partial sequence data. Sixty-six strains are included in the sequence analysis, which comprise 3307 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Lophiostoma arundinis (CBS 621.86) and Lophiostoma crenatum (CBS 629.86) were used as outgroups taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 26522.759094 is presented. The matrix had 1375 distinct alignment patterns, with 27.73% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.241919, C = 0.250696, G = 0 0.273797, T = 0.233589; substitution rates AC = 1.515500, AG = 2.699099, AT = 1.528593, CG = 1.081356, CT = 7.828217, GT = 1.000000; gamma distribution shape parameter α = 0.209065. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences is in red
The genus has similar morphology to Bahugada K.A. Reddy & V. Rao and Berkleasmium Zobel. Pseudoberkleasmium has funnel/globose shaped conidiogenous cells and brown to olivaceous green conidia, usually with a globose attached conidiogenous cell, while Bahugada has sympodial and denticulate conidiogenous cells and dark brown conidia (Reddy and Rao 1984). Berkleasmium is polyphyletic, as many species clustered in different clades and even in different families in the phylogenetic analysis (Fig. 37). Moreover, the type species of the genus (Berkleasmium concinnum) has been moved to Tubeufiaceae (Tubeufiales) (Tanney and Miller 2017; Lu et al. 2018). Berkleasmium concinnum has obovoid conidia with a scar (hilum) at base and cylindrical conidiogenous cells with a dark apex (Ellis 1971; Bussaban et al. 2001; Seifert et al. 2011; Tanney and Miller 2017). This genus needs to be revised based on both morphology and phylogenetic analysis. Therefore, based on different nature of both morphological and phylogenetic evidence from B. concinnum, we introduce a new genus here to accommodate our collection.
Pseudoberkleasmium pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555332, Facesoffungi number: FoF04510; Fig. 38
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 99622
Saprobic on dead leaf sheath of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, in small groups, blackish brown, velvety, glistening, with conidia readily liberated when disturbed. Mycelium immersed in the substrate, composed of branched, septate, smooth, hyaline hyphae. Conidiophores micronematous, mononematous, fasciculate, hyaline, smooth. Conidiogenous cells 5–11 × 9–12 μm (\( \bar{x} \) = 8.5 × 10 μm, n = 20), holoblastic, monoblastic, integrated, terminal, determinate, subglobose, connected at the base of conidia, with guttules, hyaline. Conidia 26–31 × 15–19 μm (\( \bar{x} \) = 29 × 17 μm, n = 30), acrogenous, solitary, broadly ellipsoidal to obovoid, flattened, one-cell thick, muriform, smooth, brown to olivaceous green, guttulate, usually with conidiogenous cell attached.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA, circular, undulate with dark grey mycelium and raised on surface media. Mycelium superficial, velvety.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaf sheath of Pandanus sp., 28 April 2017, R. Phookamsak & N.I. de Silva XTBG14 (HKAS 99622, holotype); ex-type living culture, KUMCC 17-0178 = MFLUCC 17-2264.
GenBank numbers LSU: MH260304; ITS: MH275071; SSU: MH260344.
Notes: Pseudoberkleasmium pandanicola is the type species of the genus Pseudoberkleasmium, which can be distinguished morphologically. In the phylogenetic tree the strain formed a separate branch from other species based on combined LSU, SSU, ITS and TEF1 data, thus it is considered as a new genus and species (Fig. 37).
Pseudochaetosphaeronema Punith.
Pseudochaetosphaeronema was erected by Punithalingam (1979) with P. larense (Borelli and R. Zamora) Punith. as the type species. Pseudochaetosphaeronema was placed in Macrodiplodiopsidaceae by Crous et al. (2015b), while Wijayawardene et al. (2017a) placed in Pleosporales genera incertae sedis. Members can found as saprobic in terrestrial or aquatic habitats but some also can be human pathogens (Zhang et al. 2012; Ahmed et al. 2015a). Morphological characteristics of asexual morph of this genus are black obpyriform pycnidia with a long neck, hyaline and phialidic conidiophores and unicellular subspherical to ellipsoidal conidia (Zhang et al. 2016). The sexual morph is undetermined (Ahmed et al. 2014). Two epithets are listed in Index Fungorum (2018). We introduce a new species in Pseudochaetosphaeronema based on molecular and morphological characteristics; this is the fully described and illustrated study using morphological and phylogenetic evidences for this genus.
Pseudochaetosphaeronema pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554501, Facesoffungi number: FoF04511; Fig. 39
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and TEF1 sequence data. Related sequences were obtained from Zhang et al. (2016). Twenty-nine strains are included in the combined sequence analysis, which comprise 4502 characters with gaps. Dothidea insculpta (CBS 189.58) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the PP. The best scoring RAxML tree with a final likelihood value of − 16777.220849 is presented. The matrix had 1151 distinct alignment patterns, with 37.92% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.244562, C = 0.237859, G = 0.273035, T = 0.244544; substitution rates AC = 0.994384, AG = 2.031393, AT = 1.306799, CG = 0.988463, CT = 5.904665, GT = 1.000000; gamma distribution shape parameter α = 0.463512. Bootstrap support values for ML equal to or greater than 70% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0016
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 100–110 × 156–167 µm (\( \bar{x} \) = 107 × 161 µm, n = 5), scattered to gregarious, superficial, flat on the base, conspicuous on host surface, dark brown, shiny, solitary, uniloculate, globose to subglobose, without papilla and ostiole. Pycnidial wall 11–18 µm, composed of several layers of thick-walled, dark brown cells of textura prismatica. Conidiogenous cells 8–17 × 1–3 μm (\( \bar{x} \) = 11 × 2 μm, n = 20), monophialidic, cylindrical, thick-walled, smooth, each with a small collarette at the tip. Conidia 2–5 × 1.5–3 µm (\( \bar{x} \) = 3 × 2 µm, n = 30), subglobose to oval, aseptate, hyaline to subhyaline, guttulate without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, grey on the surface, with dense, circular, with entire edge, raised, velvety, brown in reverse, with smooth margin. Sporulating in MEA after 3 months.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-040 (MFLU 18-0016, holotype; HKAS 100842, isotype); ex-type living culture, MFLUCC 16-0272 = KUMCC 17-0308.
GenBank numbers LSU: MH260316; ITS: MH275082; SSU: MH260356.
Notes: In the phylogenetic analysis, Pseudochaetosphaeronema pandanicola clustered with P. martinelli S.A. Ahmed, Desbois, Miossec, Atoche, Bonifaz, & de Hoog. but was well-separated with high bootstrap support of 99% in ML and 1 in BYPP (Fig. 40). Pseudochaetosphaeronema martinelli is a human pathogen and hyphae are present in culture (Ahmed et al. 2015a). According to Ahmed et al. (2015a) some Pseudochaetosphaeronema species can produce coelomycetous fruit bodies after extended incubation. Our taxon produced conidia in culture. Based on molecular phylogeny, P. pandanicola is described here as a new species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0272 is P. martinelli with 96% identity to the strain JP105B-2X (MG649295).
Pseudochaetosphaeronema pandanicola (MFLU 18-0016, holotype). a Appearance of ascomata on host substrate. b Section of conidioma. c Section of pycnidial wall. d, e Conidiogenous cell producing conidia. f Conidia. g Germinating conidium. h, i Colony on MEA from above and below. j, k Mycelium formed in culture. l Conidia formed in culture. Scale bars: a = 200 μm, b = 50 μm, c, d–g, j = 5 μm, k, l = 20 μm
Dothideomycetes orders incertae sedis
Botryosphaeriales C.L. Schoch et al.
Botryosphaeriaceae Theiss. & H. Syd.
Botryosphaeriaceae was erected by Theissen and Sydow (1918) with Botryosphaeria Ces. & De Not. as type genus. Members of this family are saprobes, endophytes and opportunistic pathogens and many of them cause disease on ecologically and economically important plants in forestry and agriculture (Schoch et al. 2006; Slippers and Wingfield 2007; Mehl et al. 2014). They are widespread in all climatic regions worldwide, but apparently absent from polar regions (Hyde et al. 2013, 2014; Phillips et al. 2013; Slippers et al. 2013; Dissanayake et al. 2016). Phillips et al. (2013) accepted 17 genera based on morphology and multi-gene analysis, while Wijayawardene et al. (2018) accepted and provided details of 28 genera. This family has been well circumscribed by several authors (Crous et al. 2006; Phillips et al. 2008, 2013; Slippers et al. 2013; Hyde et al. 2014; Dissanayake et al. 2017). New isolates and specimens collected on Pandanaceae in Thailand and China are described.
Lasiodiplodia Ellis & Everh.
Lasiodiplodia was formally erected by Ellis & Everh. (Clendenin 1896), and typified by L. theobromae (Phillips et al. 2013). Members of this genus, which can cause cankers, die-back, fruit or root rot, branch blight or discoloration on a wide range of woody hosts, are mostly distributed in tropical and subtropical regions (Punithalingam 1980; Ismail et al. 2012; Phillips et al. 2013). There are 52 epithets for Lasiodiplodia are listed in Index Fungorum (2018). Lasiodiplodia abnormis Traverso & Spessa and L. theobromae (Pat.) Griffon & Maubl. are known from Pandanaceae (Whitton et al. 2012).
Lasiodiplodia chonburiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554472, Facesoffungi number: FoF04564; Fig. 41
Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2 and TEF1 sequence data. Related sequences were obtained from Dissanayake et al. (2016). Sixty-eight strains are included in the combined sequence analysis, which comprise 1384 characters with gaps. Diplodia mutila (CBS112553) and Diplodia seriata (CBS 112555) are used as the outgroup taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 5260.567452 is presented. The matrix had 439 distinct alignment patterns, with 19.02% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.209713, C = 0.304662, G = 0.257279, T = 0.257279; substitution rates AC = 1.066215, AG = 3.169642, AT = 1.471415, CG = 0.952504, CT = 4.325945, GT = 1.000000; gamma distribution shape parameter α = 0.141773. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Chonburi Province, where the fungus was first discovered.
Holotype: MFLU 16-1877
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 210–250 × 270–300 µm (\( \bar{x} \) = 236 × 287 µm, n = 5), scattered to gregarious, immersed, conspicuous on host surface, dark brown, solitary, uniloculate, globose to subglobose, ostiole without papilla. Pycnidial wall 20–51 µm, composed of several layers of thick-walled, hyaline to dark brown cells of textura angularis. Conidiogenous cells 9–13 × 3–5 μm (\( \bar{x} \) = 11 × 4 μm, n = 20), annellidic, cylindrical, thick-walled, smooth. Conidia 15–30 × 10–15 µm (\( \bar{x} \) = 23 × 12 µm, n = 30), subglobose to oval, aseptate, hyaline to subhyaline with age, guttulate, without longitudinal striations and mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, white at first, becoming black with age, circular, with entire edge, raised, velvety, black in reverse, with smooth margin.
Material examined: THAILAND, Chonburi Province, Bang Lamung District, on dead leaf of Pandanus sp., 18 July 2016, W. Jaidee PTY02 (MFLU 16-1877, holotype; HKAS 96271, isotype); ex-type living culture, MFLUCC 16-0376 = KUMCC 17-0299.
GenBank numbers LSU: MH260299; ITS: MH275066; TEF1: MH412773; TUB2: MH412742.
Notes: Lasiodiplodia chonburiensis clusters with L. caatinguensis I.B.L. Cout., F.C. Freire, C.S. Lima & J.E. Cardoso in phylogenetic analyses (86% in ML, 0.99 in BYPP, Fig. 42). Lasiodiplodia caatinguensis has ovoid to ellipsoids conidia 18.15 × 11.78 μm (Coutinho et al. 2017), while L. chonburiensis has subglobose to oval conidia 23 × 12 µm. Therefore, L. chonburiensis is introduced as a new species based on morphology and phylogeny. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0376 is Lasiodiplodia sp. with 96% identity to the strain CMW40968 (KP872323), while the closest matches with the TEF1 sequence were with 100% identical L. theobromae strain CBS190.73 (EF622048) and closest matches with the TUB2 sequence were with 99% identical L. theobromae strain L10 (KR260829).
Lasiodiplodia chonburiensis (MFLU 16-1877, holotype). a Appearance of conidiomata on host substrate. b Section of conidioma. c Section of pycnidial wall. d, e Conidiogenous cell producing conidia. f–h Conidia. i Germinating conidium. Scale bars: a = 200 μm, b = 100 μm, c = 20 μm, d, e = 5 μm, f–i = 10 μm
Lasiodiplodia hyalina Zh.P. Dou & Y. Zhang, Mycosphere 8 (2): 1016 (2017)
Facesoffungi number: FoF04565; Fig. 43
Lasiodiplodia hyalina (MFLU 18-0023). a Appearance of conidiomata on host substrate. b Section of conidioma. c Section through ostiole. d Pycnidial wall. e–g Conidiogenous cell producing conidia. h–j Conidia. k Conidium with dark longitudinal striations. l Germinating conidium. Scale bars: b = 50 μm, c = 20 μm, d = 10 μm, e–l = 5 μm
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 197–244 × 134–156 µm (\( \bar{x} \) = 222.6 × 148.5 µm, n = 5), scattered to gregarious, immersed, conspicuous on host surface, dark brown, solitary, uniloculate, obpyriform, ostiole central, without papilla, always sporulating on host surface. Pycnidial wall 22–55 µm, composed of several layers of thick-walled, pale brown to dark brown cells of textura angularis. Conidiogenous cells 5–10 × 4–7 μm (\( \bar{x} \) = 7 × 5.5 μm, n = 10), holoblastic, phialidic, ovoid, thick-walled, smooth. Conidia 20–30 × 10–15 µm (\( \bar{x} \) = 23 × 12 µm, n = 30), oval, becoming 1-septate with age, with a dark band at septum, not constricted at septum, initially hyaline to yellow–brown or brown with age, rounded at both ends, guttulate, with longitudinal surface striations from apex to base, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, white at first, becoming black with age, circular, with entire edge, raised, velvety, black in reverse, with smooth margin.
Material examined: THAILAND, Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on dead leaf of Pandanus sp., 16 December 2017, S. Tibpromma P04 (MFLU 18-0023 = HKAS 101792); living culture, MFLUCC 18-0110.
GenBank numbers LSU: MH260300; ITS: MH275067; SSU: MH260341; TUB2: MH412743.
Notes: Our isolate similar with Lasiodiplodia hyalina Z.P. Dou & Ying Zhang which found from cankered stems of Acacia confuse and unidentified woody plant (Dou et al. 2017). Lasiodiplodia hyalina has cylindrical to ampulliform conidiogenous cells and ellipsoid to ovoid, 24 × 13.6 μm conidia (Dou et al. 2017) and similar to our isolate. Lasiodiplodia hyalina (MFLUCC 18-0110) is supported by our molecular data (Fig. 42) and this is the first record of L. hyalina on dead leaves of Pandanus sp.
Lasiodiplodia pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554473, Facesoffungi number: FoF04566; Fig. 44
Lasiodiplodia pandanicola (MFLU 18-0011, holotype). a Appearance of conidiomata on host substrate. b Section of conidioma. c Section of pycnidial wall. d Conidiogenous cell producing conidia. e–g Conidia. h Conidium with longitudinal striations. i Germinating conidium. j, k Colony on MEA from above and below. l, m Conidia formed in culture. Scale bars: a = 200 μm, b = 100 μm, c, d, l, m = 20 μm, e–i = 5 μm
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0011
Saprobic on dead root of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 185–210 × 187–240 μm (\( \bar{x} \) = 198 × 211 μm, n = 5), immersed to erumpent through host, visible as black, subglobose to ovoid, solitary, scattered or gregarious, ostiole. Pycnidial wall 20–42 μm, composed of several layers of textura angularis, dark brown. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 4–6.5 × 5–7 μm (\( \bar{x} \) = 5 × 6 μm, n = 5), phialidic, integrated, cylindrical or cylindric-clavate or irregular swollen cell, hyaline, smooth. Conidia 14–38 × 9–22 μm (\( \bar{x} \) = 27 × 14 μm, n = 30), ellipsoidal to obovate, initially hyaline and aseptate, later becoming brown and 1-septate, thick and rough-walled, guttulate, with longitudinal striations.
Culture characteristics Conidia germinating on MEA within 12 h. Colonies on MEA, initially white–grey and later become black, with dense mycelium, circular, with entire edge, raised, velvety, black in reverse, with smooth margin. Sporulating in culture after 2 months, producing conidia similar in shape to those recorded on natural dead leaves.
Material examined: THAILAND, Phatthalung Province, Mueang Phatthalung District, on dead leaves of Pandanus sp., 14 June 2015, B. Thongbai SF15-009 (MFLU 18-0011, holotype); ex-type living culture, MFLUCC 16-0265 = KUMCC 16-0158.
GenBank numbers LSU: MH260301; ITS: MH275068; TEF1: MH412774; TUB2: MH412744.
Notes: Phylogenetic analyses indicated Lasiodiplodia pandanicola is well-separated with other Lasiodiplodia spp. (95% in ML, 1 in BYPP, Fig. 42). Lasiodiplodia parva A.J.L. Phillips, A. Alves & Crous share similar conidia to L. pandanicola. However, L. parva has conidia 16–23.5 × 10.5–13 μm, ovoid (Alves et al. 2008), while L. pandanicola has conidia 14–38 × 9–22 μm and ellipsoidal to obovate. Therefore, we propose L. pandanicola as a new species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0265 is L. theobromae with 100% identity to the strain Lt-A2 (KX270362), while the closest matches with the TEF1 sequence were with 100% identical L. theobromae strain IRNHM-KB64-2 (KU737511) and closest matches with the TUB2 sequence were with 95% identical Botryosphaeria quercuum strain CBS177.89 (DQ026404).
Lasiodiplodia pseudotheobromae A.J.L. Phillips, A. Alves & Crous
Facesoffungi number: FoF04567; Fig. 45
Lasiodiplodia pseudotheobromae (MFLU 18-0027). a, b Appearance of conidiomata on host substrate. c Section of conidioma. d Section of pycnidial wall. e–g Conidiogenous cell producing conidia. h–j Conidia. k Germinating conidium. l, m Colony on MEA from above and below. Scale bars: c = 50 μm, d–k = 5 μm
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 165–240 × 140–215 µm (\( \bar{x} \) = 193.6 × 180 µm, n = 5), scattered to gregarious, immersed, conspicuous on host surface, dark brown, solitary, uniloculate, obpyriform, ostiole in the central, without papilla, always sporulate on host surface. Pycnidial wall 10–35 µm, composed of several layers, thick-walled, pale brown to dark brown cells of textura angularis. Conidiogenous cell 4.5–8 × 2.5–8 μm (\( \bar{x} \) = 6 × 5 μm, n = 10), holoblastic, cylindrical, flat at the base, thick-walled, smooth. Conidia 20–30 × 10–15 µm (\( \bar{x} \) = 24 × 12.4 µm, n = 30), oval to ellipsoid, 1-septate with age, dark band at the septum, not constricted at septum, initially hyaline becoming yellow–brown or brown with age, rounded at apex, guttulate, longitudinal striations from apex to base, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, white at first and becoming black with age, circular, with entire edge, raised, velvety, black in reverse, with smooth margin.
Material examined: THAILAND, Chiang Rai Province, Muang District, Mae Fah Luang university, on dead leaf of Pandanus sp., 15 December 2017, S. Tibpromma P08 (MFLU 18-0027, HKAS 101796); living culture, MFLUCC 18-0114.
GenBank numbers LSU: MH376724; ITS: MH388351; SSU: MH388318; TEF1: MH388386; TUB2: MH412719.
Notes: In the phylogeny our collection clusters between Lasiodiplodia pseudotheobromae (CBS 447.62) and L. pseudotheobromae (CBS 116459), first reported on grapevine in Brazil as a trunk pathogen (Correia et al. 2013). Conidial dimensions of our collection are similar to the values given by Alves et al. (2008). This is the first record of L. pseudotheobromae on dead leaves of Pandanus sp.
Neofusicoccum Crous et al.
Neofusicoccum was erected by Crous et al. (2006) with N. parvum (Pennycook & Samuels) Crous, Slippers & A.J.L. Phillips, as the type species. Morphologically the genus is Fusicoccum-like, but it forms a Dichomera-like synanamorph with brown, globose to pyriform conidia (Crous et al. 2006). There are 44 epithets for Neofusicoccum are listed in Index Fungorum (2018). Neofusicoccum has never been reported on Pandanaceae.
Neofusicoccum pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554752, Facesoffungi number: FoF04568; Fig. 46
Neofusicoccum pandanicola (HKAS 99631, holotype). a Conidiomata on dead leaves of Pandanus sp. b Cross section of the conidioma. c Conidiomatal wall. d, e Conidia attached to conidiogenous cell. f–h Conidia. i Germinating conidium. j, k Colony on MEA from above and below. Scale bars b = 50 μm, c, i = 20 μm, d, e, h = 10 μm, f, g = 5 μm
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 99631
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 180–250 × 280–295 μm (\( \bar{x} \) = 225 × 289 μm, n = 5), immersed in the host, solitary or gregarious, brown to dark brown, globose to subglobose, uniloculate, ostiolate. Pycnidial wall 33–81 μm, thick-walled, composed of dark brown to pale brown cells of textura angularis. Conidiophores micronematous, reduced to conidiogenous cell. Conidiogenous cells 10–17 × 4–8 μm (\( \bar{x} \) = 15 × 5.4 μm, n = 10), monoblastic, holoblastic, integrated, hyaline, smooth. Conidia 15–26 × 8–12 μm (\( \bar{x} \) = 22 × 10 μm, n = 20), ovoid to ellipsoid, thin-walled, rounded at apex, hyaline, aseptate, rough, without a mucilaginous sheath.
Culture characteristics: Colonies on PDA covering the entire plate after 7 days at room temperature, mycelium velvety and moderately fluffy with an irregular margin, surface initially white and later becoming dark from the middle of the colony and dark in reverse.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus sp., 12 November 2016, T. Aluthwaththa XTBG23 (HKAS 99631 holotype); ex-type living culture, KUMCC 17-0184 = MFLUCC 17-2270.
GenBank numbers LSU: MH260306; ITS: MH275072; SSU: MH260346; TEF1: MH412778; ACT: MH412740; GPDH: MH412751; CHS-1: MH412746.
Notes: Neofusicoccum pandanicola was recovered from fallen dead and decaying leaves of Pandanus sp. in China. Neofusicoccum pandanicola shares a close phylogenetic affinity to N. algeriense Berraf-Tebbal & A.J.L. Phillips (CBS 137504) (Fig. 47). However, N. algeriense has fusiform conidia with a subtruncate to bluntly rounded base and 17.6 × 5.6 μm (Berraf-Tebbal et al. 2014), while N. pandanicola has ovoid to ellipsoid conidia rounded at both ends and 22 × 10 μm. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0184 is N. parvum with 100% identity to the strain OMNP10 (KY657475), while the closest matches with the TEF1 sequence were with 94% identical N. parvum strain JTTL3 (KP183195).
Phylogram generated from maximum likelihood analysis based on combined ITS, RBP2, TUB2 and TEF1 sequence data. Related sequences were obtained from Dissanayake et al. (2016). Forty-five strains are included in the combined sequence analysis, which comprise 1929 characters with gaps. Botryosphaeria dothidea (CBS 100564) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 7606.259934 is presented. The matrix had 592 distinct alignment patterns, with 22.67% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.217869, C = 0.290809, G = 0.275107, T = 0.216215; substitution rates AC = 1.027099, AG = 4.400483, AT = 0.842514, CG = 1.163706, CT = 9.026961, GT = 1.000000; gamma distribution shape parameter α = 0.248719. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Pseudofusicoccumaceae Yao Tan & Crous
Pseudofusicoccumaceae was erected by Tao Yang & Crous which is typified by Pseudofusicoccum Mohali, Slippers & M.J. Wingf. (Yang et al. 2017). The morphology this family is similar to Fusicoccum by differ in conidia tend to be more cylindrical in shape and are encased in a persistent mucoid sheath (Yang et al. 2017).
Pseudofusicoccum Mohali
Pseudofusicoccum was erected by Crous et al. (2006) with P. stromaticum (Mohali, Slippers & M.J. Wingf.) Mohali, Slippers & M.J. Wingf. as the type species. Pseudofusicoccum resembles Fusicoccum, but is distinct in conidia encased in a persistent mucous sheath (Crous et al. 2006). Pseudofusicoccum contains seven epithets are listed in Index Fungorum (2018). Pseudofusicoccum adansoniae is the first report of Pseudofusicoccum from Pandanaceae.
Pseudofusicoccum adansoniae Pavlic, T.I. Burgess & M.J. Wingf., Mycologia 100 (6): 855 (2008)
Facesoffungi number: FoF04569; Fig. 48
Phylogram generated from maximum likelihood analysis based on combined ITS and TEF1 sequence data. Related sequences were obtained from Dissanayake et al. (2016). Fifteen strains are included in the combined sequence analysis, which comprise 850 characters with gaps. Phyllosticta citricarpa (CBS102374) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 2202.181718 is presented. The matrix had 98 distinct alignment patterns, with 3.38% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.199529, C = 0.289959, G = 0.268934, T = 0.241578; substitution rates AC = 0.835955, AG = 4.035372, AT = 0.562345, CG = 0.351128, CT = 6.074342, GT = 1.000000; gamma distribution shape parameter α = 0.228580. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Saprobic on dead fruit of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 75–125 × 108–170 µm (\( \bar{x} \) = 92.5 × 134 µm, n = 5), scattered to gregarious, immersed, conspicuous on host surface, dark brown, shiny, solitary, uniloculate, globose to subglobose, ostiolate, without a papilla. Pycnidial wall 12–25 µm, composed of several layers, thick-walled, with hyaline to pale brown cells of textura angularis. Conidiogenous cells 6–13 × 1–3 μm (\( \bar{x} \) = 9 × 2.5 μm, n = 20), annellidic, cylindrical, thick-walled, smooth. Conidia 12–15 × 6.5–9 µm (\( \bar{x} \) = 14 × 8 µm, n = 20), cylindrical to ellipsoid, aseptate, hyaline to subhyaline, rounded at the ends, granular, without a mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, initially white, becoming black with age, dense, circular, with raised entire edge, velvety, black in reverse, with smooth margin.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, on dead fruit of Pandanus sp., 30 July 2015, S. Tibpromma SF15-013 (MFLU 16-0551, HKAS 96275); living culture, MFLUCC 16-0367.
GenBank numbers LSU: MH260317; ITS: MH275083; SSU: MH260357.
Notes: Pseudofusicoccum adansoniae is known from Acacia synchronica, Adansonia gibbosa, Eucalyptus sp., and Ficus opposite (Pavlic et al. 2008) but has never been reported from Pandanaceae. Our isolate (MFLUCC 16-0367) clustered with two P. adansoniae isolates from Adansonia gibbosa (Fig. 49). The morphology of these isolates is similar with our isolate by having ellipsoid conidia, rounded, smooth with granular, hyaline (Pavlic et al. 2008). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0367 is P. ardesiacum isolate with 99% identity to the strain CFE-11 (MH168331), while the closest matches with the TEF1 sequence were with 99% identical P. adansoniae strain B0341 (KM006477).
Pseudofusicoccum adansoniae (MFLU 16-0551). a Appearance of conidiomata on host substrate. b Section of conidioma. c Section through ostiole. d Section of pycnidial wall. e, f Conidiogenous cells producing conidia. g–i Conidia. j Germinating conidium. Scale bars: a = 200 μm, b = 50 μm, c–f = 10 μm, g–j = 5 μm
Tubeufiales Boonmee & K.D. Hyde
Tubeufiaceae M.E. Barr.
Tubeufiales was introduced by Boonmee et al. (2014) to accommodate the type family, Tubeufiaceae M.E. Barr. Tubeufiaceae was established in Pleosporales by Barr (1979) with Tubeufia as the type genus. The characteristic features of this family are uniloculate, superficial, pigmented ascomata with hyaline ascospores (sexual morph) and producing helicosporous conidia (asexual morph) (Tsui et al. 2006; Boonmee et al. 2011, 2014). There are 25 genera in the family (Wijayawardene et al. 2018). An updated phylogenetic analysis of Tubeufiaceae is presented (Fig. 50). Recenlty, Lu et al. (2018) provided an update of this family. Five new species and one known species collected on Pandanaceae are described based on morphology and phylogeny support.
Phylogram generated from maximum likelihood analysis based on combined ITS, LSU and TEF1 sequence data. Related sequences were obtained from Chaiwan et al. (2017). Eighty-eight strains are included in the combined sequence analysis, which comprise 2216 characters with gaps. Botryosphaeria dothidea (CBS 115476) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 21765.784174 is presented. The matrix had 1068 distinct alignment patterns, with 44.44% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.239982, C = 0.242120, G = 0.269480, T = 0.248419; substitution rates AC = 0.754032, AG = 3.061196, AT = 2.077623, CG = 0.823599, CT = 6.542463, GT = 1.000000; gamma distribution shape parameter α = 0.463512. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Helicoma Corda
Helicoma was established by Corda with the type species H. muelleri Corda. Helicoma has relatively short, erect, dark brown, holoblastic conidiogenous cells and helicoid, hyaline, brown to dark brown conidia and is a saprobe (Boonmee et al. 2011, 2014). There are 81 epithets are listed in Index Fungorum (2018). This is the first record of Helicoma from Pandanaceae.
Helicoma freycinetiae Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554502, Facesoffungi number: FoF04517; Fig. 51
Etymology: named after the host genus, Freycinetiae.
Holotype: MFLU 16-1903
Saprobic on dead root of Freycinetia javanica. Colonies on natural substratum effuse, subhyaline. Mycelium partly immersed in leaf substratum and partly superficial; composed of branched, septate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 160–250 × 6–12 μm, subhyaline, light brown at base, macronematous, septate, erect, conical at apex, smooth-walled. Conidiogenous cells monoblastic, holoblastic, integrated, denticulate. Conidia 44–57 μm, conidial filament 4–9 μm (\( \bar{x} \) = 52.5 × 7 μm, n = 20), 205–276 μm long, tightly coiled 3½ times, rounded at apex, solitary, up to 31-euseptate, slightly constricted at septa, subhyaline, smooth-walled, guttulate, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, surface rough, edge entire, yellow–brown to dark brown in MEA medium, yellow–brown in the middle, become dark brown at the margin. Mycelium superficial and partially immersed, raised.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Freycinetia javanica Blume, 17 December 2015, S. Tibpromma KB025 (MFLU 16-1903, holotype; HKAS 96252, isotype); ex-type living culture, MFLUCC 16-0363.
GenBank numbers LSU: MH260295; ITS: MH275062; SSU: MH260337; TEF1: MH412770.
Notes: Helicoma freycinetiae is introduced as a new species based on phylogenetic analysis (Fig. 50). It clustered with H. siamense Boonmee & K.D. Hyde (MFLUCC 10-0120) but is well-separated with high bootstrap support (100% in ML, 1 in BYPP, Fig. 50). Helicoma siamense has partly heavily pigmented conidia, with filaments 7–10 μm wide, coiled 2–3 times and darkened at septa (Boonmee et al. 2014), while H. freycinetiae has subhyaline conidia, filaments 4–9 μm wide and coiled 3½ times. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0363 is H. siamensewith 99% identity to the strain MFLUCC 10-0120 (JN865204), while the closest matches with the RPB2 sequence were with 85% identical Acanthohelicospora aurea strain GZCC 16-0060 (MF589911).
Neohelicomyces Z.L. Luo et al.
Neohelicomyces was erected by Luo et al. (2017) with N. aquaticus Z.L. Luo, Bhat & K.D. Hyde as type species. Its characteristics are similar to Helicomyces. There are three species for Neohelicomyces are listed in Index Fungorum (2018). This is the first report of Neohelicomyces from Pandanaceae.
Neohelicomyces pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554503, Facesoffungi number: FoF04512; Fig. 52
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 96202
Saprobic on dead roots of Pandanus sp. Colonies on substratum superficial, effuse, gregarious, subhyaline to pinkish. Mycelium partly immersed in leaf substratum and partly superficial, consisting of branched, septate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 110–220 × 3–6 μm, subhyaline to pale brown, macronematous, straight, simple or branched, septate, smooth-walled. Conidiogenous cells monoblastic, holoblastic, integrated, denticulate. Conidia 28–44 μm diam., conidial filament 60–123 μm long, 2–3 μm wide (\( \bar{x} \) = 106 × 2.5 μm, n = 20), filament coiled 2½-3½ times when tightly coiled, rounded at both ends, becoming loosely coiled in water, solitary, up to 10-euseptate, not constricted at septa, hyaline, smooth-walled, with numerous guttules, without mucilaginous sheath.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA, irregular, surface rough, pale brown to yellow–brown in PDA medium. Mycelium superficial and partially immersed raised.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 27 July 2016, S. Tibpromma NBH02 (HKAS 96202, holotype); ex-type living culture, KUMCC 16-0143; Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 2 August 2016, S. Tibpromma NBH07 (HKAS 96207, paratype).
GenBank numbers LSU: MH260307; ITS: MH275073; SSU: MH260347; TEF1: MH412779.
Notes: Neohelicomyces pandanicola was collected in China on Pandanus sp. In the phylogenetic analysis it clustered with N. submersus Z.L. Luo, Hong Y. Su & K.D. Hyde, collected on decaying wood from China, but well-separated as a distinct species with high bootstrap support (90% in ML, 1 in BYPP) (Fig. 50). Neohelicomyces submersus has hyaline to pale brown colonies with 142.5–207.5 μm long, pale brown conidia and filament coiled 3–3½ times (Luo et al. 2017), while N. pandanicola has subhyaline to pinkish colonies conidia 106 × 2.5 μm long, hyaline and filament coiled 2–3½ times. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0143 is N. grandisporus with 94% identity to the strain KUMCC 15-0470 (KX454173).
Tubeufia Penz. & Sacc.
Tubeufia was erected by Penzig and Saccardo (1898) with T. javanica Penz. & Sacc. as the type species, which was described from Bambusa emarcidis. There are 60 epithets are listed in Index Fungorum (2018). Only one species, Tubeufia helicoma has been reported previously on Pandanaceae (Freycinetia banksii) from New Zealand (Hughes 1978).
Tubeufia freycinetiae Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554504, Facesoffungi number: FoF04513; Fig. 53
Etymology: named after the host genus, Freycinetiae.
Holotype: MFLU 16-1897
Saprobic on dead roots of Freycinetia javanica. Colonies on natural substratum effuse, subhyaline. Mycelium partly immersed in leaf substratum and partly superficial, consisting of branched, aseptate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 6–36 × 1.5–5 μm, subhyaline, macronematous, slightly curved, unbranched, smooth. Conidiogenous cells monoblastic, integrated, each with single conidium. Conidia 24–46 μm diam., conidial filament 105–155 μm long, 3.5–7.5 μm wide (\( \bar{x} \) = 35.4 × 6 μm, n = 20), tightly coiled 1½–2½ times, with rounded apical end, up to 33-euseptate, slightly constricted at septa, hyaline, smooth-walled, with guttules, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, irregular, surface rough, edge undulate, brown to dark brown in MEA medium. Mycelium superficial and partially immersed raised.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Freycinetia javanica Blume., 17 December 2015, S. Tibpromma KB019 (MFLU 16-1897, holotype; HKAS 96246, isotype); ex-type living culture, MFLUCC 16-0252.
GenBank numbers LSU: MH260323; ITS: MH275089; SSU: MH260361; TEF1: MH412786.
Notes: Tubeufia helicoma have been reported from Pandanaceae (Hughes 1978). Tubeufia freycinetiae has hyaline, 105–155 μm long conidiophores with conidia 1½–2½ times coiled and up to 33–euseptate. In the phylogenetic tree, Tubeufia freycinetiae formed a clade between T. hyalospora Y.Z. Lu, Boonmee & K.D. Hyde. and T. filiformis Y.Z. Lu, Boonmee & K.D. Hyde. Tubeufia freycinetiae differs from T. hyalospora by having 16–33 μm diam conidia with conidial filament 110–225 μm long and 1½–3½ times coiled. Morphologically Tubeufia freycinetiae is similar to T. filiformis but the latter has pale brown conidiophores and conidia that are 110–225 μm long, tightly coiled 2½–3½ times and multi-septate (Hyde et al. 2016). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0252 is T. roseohelicospora with 94% identity to the strain MFLUCC 15-1247 (KX454177), while the closest matches with the TEF1 sequence were with 95% identical T. filiformis strain MFLUCC 16-1135 (KY117032).
Tubeufia inaequalis Y.Z. Lu, J.C. Kang & K.D. Hyde, Fungal Diversity 92: (2018)
Facesoffungi number: FoF04516; Fig. 54
Saprobic on dead leaves of Pandanus sp. Colonies on natural substratum effuse, subhyaline. Mycelium partly immersed in leaf substratum and partly superficial, consisting of branched, septate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 18–39 × 4–6 μm, hyaline, macronematous, slightly curved, simple or branched, smooth. Conidiogenous cells monoblastic, holoblastic, integrated, denticulate. Conidia 350–550 × 2.5–8 μm (\( \bar{x} \) = 478 × 6 μm, n = 20), coiled 2–2½ times, becoming uncoiled in water, rounded at both ends, solitary, up to 50-euseptate, not constricted at septa, hyaline, smooth-walled, with numerous guttules, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, irregular, surface rough, edge curled, grey in the middle with switch yellow–white and dark brown, in MEA medium. Mycelium superficial and partially immersed raised.
Material examined: THAILAND, Krabi Province, Tha Pom Klong Song Num, on Pandanus sp., 16 December 2015, S. Tibpromma KB021 (MFLU 16-1899; HKAS 96248); living culture, MFLUCC 16-0319.
GenBank numbers LSU: MH260326; SSU: MH260364.
Notes: Tubeufia inaequalis was previously known from on submerged decaying wood in a freshwater stream (Lu et al. 2018). The phylogenetic analysis showed that our isolate groups with T. inaequalis MFLUCC 17-0053 (98% in ML, 1 in BYPP, Fig. 50). The morphology of our isolate is similar to that of T. inaequalis (MFLUCC 17-0053). This is first report T. inaequalis from Pandanaceae.
Tubeufia pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554506, Facesoffungi number: FoF04515; Fig. 55
Tubeufia pandanicola (MFLU 16-1905, holotype). a Colonies on dead leaves of Pandanus sp. b Conidiophores, conidiogenous cells and conidia. c Conidiogenous cells with conidium. d Conidiophores and conidiogenous cells. e, f Conidia. g, h Colony on MEA from above and below. Scale bars: b–d = 20 μm, e, f = 10 μm
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1905
Saprobic on dead leaves of Pandanus sp. Colonies on natural substratum effuse, subhyaline. Mycelium partly immersed in leaf substratum and partly superficial, consisting of branched, septate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 90–145 × 5–6.5 μm, subhyaline at the tip, pale brown at base, macronematous, septate, erect, rounded at apex, smooth. Conidiogenous cells 4–15 × 2–5.5 μm (\( \bar{x} \) = 9 × 4 μm, n = 10), monoblastic, holoblastic, obclavate, hyaline. Conidia 30–45 μm diam., conidial filament 220–300 μm long, 3–4.5 μm wide (\( \bar{x} \) = 240 × 4 μm, n = 20), tightly coiled 3–3½ times, rounded at apex, solitary, up to 5-euseptate, not constricted at septa, hyaline, smooth-walled, guttulate, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, surface rough, edge entire, yellow–brown to dark brown in MEA medium, yellow–brown in the middle, become dark brown at the margin. Mycelium superficial and partially immersed raised.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB027 (MFLU 16-1905, holotype; HKAS 96254, isotype); ex-type living culture, MFLUCC 16-0321.
GenBank numbers LSU: MH260325; ITS: MH275091; SSU: MH260363.
Notes: Tubeufia pandanicola has conidiophores that are subhyaline at the apex and pale brown at the base, obclavate conidiogenous cells and up to 5-septate conidia that are 3–3½ times tightly coiled. In phylogeny, T. pandanicola clustered with T. guangxiensis Chaiwan, Boonmee, Y.Z. Lu & K.D. Hyde with 90% support in ML, and 0.99 in BYPP. However, T. guangxiensis has short, pale brown cylindrical conidiophores, with conidia that are 1½–2½ times coiled and up to 50-septate (Chaiwan et al. 2017). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0321 is T. roseohelicospora with 86% identity to the strain MFLUCC 15-1247 (KX454177).
Tubeufia parvispora Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555292, Facesoffungi number: FoF04514; Fig. 56
Etymology: “parvispora” referring to smaller-sized conidia compared to the other species of Tubeufia genus.
Holotype: MFLU 16-1911
Saprobic on dead leaves of Pandanus sp. Colonies on natural substratum effuse, subhyaline. Mycelium partly immersed in leaf substratum and partly superficial, consisting of branched, septate, subhyaline, smooth, thin-walled hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 44–58 × 1–2.5 μm, hyaline, macronematous, straight or slightly curved, unbranched, smooth. Conidiogenous cells monoblastic, holoblastic, integrated, denticulate. Conidia 11–15 μm, conidial filament 60–66 μm long, 1–2 μm wide (\( \bar{x} \) = 63 × 1.5 μm, n = 20), tightly coiled 2–3 times, rounded at both ends, becoming loosely coiled in water, solitary, up to 15-euseptate, not constricted at septa, hyaline, smooth-walled, without guttules and mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, surface rough with wrinkled, edge entire, grey to dark brown in MEA medium. Mycelium superficial and partially immersed raised.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 16 December 2015, S. Tibpromma KB033 (MFLU 16-1911, holotype; HKAS 96260, isotype); ex-type living culture, MFLUCC 16-0324.
GenBank numbers LSU: MH260324; ITS: MH275090; SSU: MH260362; TEF1: MH412787; RPB2: MH412761.
Notes: In our phylogenetic analysis Tubeufia parvispora clustered with T. chiangmaiensis Boonmee & K.D. Hyde with high support (66% in ML, 1 in BYPP). Sexual morph is known only for T. chiangmaiensis (Boonmee et al. 2014). When we compare nucleotides, there are 44 bp (8.81%) differences in 499 ITS (+5.8S) nucleotides and 34 bp (3.86%) differences in 879 TEF1 nucleotides between T. chiangmaiensis and T. parvispora. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLU 16-1911 is Helicoma sp. with 93% identity to the strain BCC 3512 (AY916484), while the closest matches with the TEF1 sequence were with 97% identical T. mackenziei strain MFLUCC 16-0222 (KY117031) and the closest matches with the RPB2 sequence were with 90% identical T. guangxiensis strain GZCC 16-0041 (MG012016).
Venturiales Y. Zhang ter et al.
Sympoventuriaceae Y Zhang ter et al.
Sympoventuriaceae was erected by Zhang et al. (2011) with Sympoventuria Crous & Seifert as the type genus. The family contains the asexual genus as hyphomycetes (Wijayawardene et al. 2017a). More details have been provided by Hyde et al. (2013). There are four genera in the family (Wijayawardene et al. 2018). We introduce the new genus, Yunnanomyces collected on Pandanaceae from China.
Yunnanomyces Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF555334, Facesoffungi number: FoF04518
Etymology: named after Yunnan Province, where the fungus was first discovered.
Type species: Yunnanomyces pandanicola Tibpromma & K.D. Hyde
Saprobic on decaying leaves or wood in terrestrial habitats. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, group together in small groups, blackish to brown, velvety, glistening, conidia readily liberated when disturbed. Mycelium immersed in the substrate, composed of branched, septate, hyaline to subhyaline hyphae. Conidiophores mononematous, fasciculate, septate, hyaline to subhyaline, branched or unbranched. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, cylindrical, hyaline to subhyaline. Conidia acrogenous, globose to broadly oval, flattened, one-cell thick, muriform, yellow–brown to brown.
Notes: Yunnanomyces is introduced to accommodate Y. pandanicola an asexual fungus with globose to broadly oval, yellow–brown, muriform conidia which was collected from Yunnan Province, China. It is morphologically similar to Pseudocoleodictyospora, reported from teak (Doilom et al. 2017), but in the phylogenic analysis Yunnanomyces does not cluster with Pseudocoleodictyosporaceae. It differs from Fusicladium and Verruconis by the presence of conidiophores and conidia (Fig. 57).
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU and RPB2 partial sequence data. Fifty-two strains are included in the sequence analysis, which comprise 3001 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Phaeotrichum benjaminii and Trichodelitschia bisporula are used as outgroup taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 13481.727487 is presented. The matrix had 1173 distinct alignment patterns, with 45.44% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.255891, C = 0.225681, G = 0.291461, T = 0.226966; substitution rates AC = 1.415398, AG = 3.201141, AT = 1.396732, CG = 1.092537, CT = 7.699345, GT = 1.000000; gamma distribution shape parameter α = 0.487049. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. The newly generated sequence is in red
Yunnanomyces pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555335, Facesoffungi number: FoF04519; Fig. 58
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 96206
Saprobic on dead leaves of Pandanus amaryllifolius. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, in small groups, blackish brown, velvety, glistening, conidia readily liberated when disturbed. Mycelium immersed in the substrate, composed of branched, septate, smooth, hyaline hyphae. Conidiophores mononematous, fasciculate, septate, hyaline, smooth, unbranched. Conidiogenous cells 2.5–5 × 1.5–5 μm (\( \bar{x} \) = 4 × 3.4 μm, n = 10), holoblastic, monoblastic, integrated, terminal, determinate, cylindrical, hyaline. Conidia 20–25 × 13–18 μm (\( \bar{x} \) = 22 × 15 μm, n = 20), acrogenous, solitary, globose to broadly oval, flattened, one-cell thick, thick-walled, muriform, consisting of 9–30 cells, yellow–brown.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA, circular, undulate with brown, velvety, floosy.
Material examined: CHINA, Yunnan Procince, Xishuangbanna, on fallen dead and decaying leaves of Pandanus amaryllifolius Roxb., 15 November 2016, T. Aluthwaththa XTBG03 (HKAS 99611, holotype); ex-type living culture, MFLUCC 17-2260.
GenBank numbers LSU: MH376743; ITS: MH388369; SSU: MH388333; RPB2: MH412736.
Notes: Based on the multi-gene sequence analyses, isolate MFLUCC 17-2260 clusters between Fusicladium sicilianum Koukol. (CBS 105.85) and F. ramoconidii Crous & de Hoog (CBS 462.82). However, Fusicladium morphologically differs from Yunnanomyces pandanicola by forming small sporodochial conidiomata with conidia amero- to phragmosporous (Seifert et al. 2011). In a BLASTn search on NCBI GenBank, the closest matches of LSU sequence of MFLUCC 17-2260 is Fusicladium ramoconidii with 95% identity to the strain CBS 462.82 (EU035439), while the closest matches with the ITS sequence were with 90% identical Fusicladium rhodense strain CPC 13156 (EU035440).
Class Lecanoromycetes O.E. Erikss. & Winka
Subclass Ostropomycetidae Reeb et al.
Ostropales Nannf.
Stictidaceae Fr.
Stictidaceae was erected by Fries (1849) with Stictis Pers. as type genus. Members of this family occur as saprobic discomycetes, but also as lichens, parasites, and endophytes (Gilenstam 1969; Fernández-Brime et al. 2011; Baloch et al. 2013; Aptroot et al. 2014; Jahn et al. 2017). Stictidaceae are characterised by deeply immersed apothecia, long, cylindrical asci and filiform, cylindrical or fusoid, septate ascospores sometimes fragmenting at maturity (Hawksworth et al. 1995). Stictidaceae is still very limited, and contains a small group of generally drought tolerant fungi, which are easily overlooked and rarely collected (Fernández-Brime et al. 2018). There are 25 genera in the family (Wijayawardene et al. 2018). We collected a Stictis species from Pandanaceae. Three species of Stictis are known on Pandanaceae (Whitton et al. 2012).
Stictis Pers.
Stictis was erected by Persoon (1780) with S. radiata (L.) Pers. as type species. The members can be found on many substrates, including decaying leaves, wood, bark, herbaceous stems, grass culms, fern rachises and many other hosts (Sherwood 1977; Johnston 1983; Kirk et al. 2008). Stictis contains 345 epithets (Index Fungorum 2018). Stictis can also be found on Pandanaceae hosts (S. carnea, S. pandani and S. subiculata) (Whitton et al. 2012).
Stictis pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554510, Facesoffungi number: FoF04520; Fig. 59
Phylogram generated from maximum likelihood analysis based on combined LSU, mt-SSU and ITS sequence data. Related sequences were obtained from Li and Hou (2016) and Fernández-Brime et al. (2018). Twenty-five strains are included in the combined sequence analysis, which comprise 2933 characters with gaps. Placopsis perrugosa (AFTOL-ID 383) is used as the outgroup taxon. Tree topology of theML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of - 14421.897248 is presented. The matrix had 1067 distinct alignment patterns, with 30.70% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.287575, C = 0.206010, G = 0.260355, T = 0.246059; substitution rates AC = 1.331310, AG = 2.504484, AT = 1.875317, CG = 1.124721, CT = 5.842916, GT = 1.000000; gamma distribution shape parameter a = 0.267597. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. The newly generated sequence is in red
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 96206
Saprobic on dead leaf of Pandanus sp. with immersed apothecia, later opening by a pore, with margin white-pruinose on host surface. Sexual morph Ascomata 350–410 × 520–650 µm (\( \bar{x} \) = 386 × 593 µm, n = 5), immersed, globose to subglobose, solitary, without papilla and ostiole, black. Peridium 27–46 µm wide, composed several layers of hyaline or colourless cells of textura epidermoidea. Hamathecium comprising 0.8–1.1 µm, numerous, filiform, hyaline, aseptate paraphyses. Asci 160–240 × 7.5–23 µm (\( \bar{x} \) = 212 × 17 µm, n = 20), unitunicate, (2–)8-spored, cylindrical to cylindric-clavate, with a short pedicel, apically thick and rounded. Ascospores 190–265 × 4–5 µm (\( \bar{x} \) = 241 × 4.4 µm, n = 10), overlapping uni- to bi-seriate, hyaline, fusiform, up to 40-septa, constricted at the central septum, smooth-walled, not surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 28 July 2016, S. Tibpromma NBH06 (HKAS 96206, holotype).
GenBank numbers LSU: MH260319; ITS: MH275085; mt-SSU: MH260359; TEF1: MH412782.
Notes: Stictis pandanicola has colonies on host very similar to those of S. carnea Seaver & Waterston. However, S. pandanicola has biger asci than S. carnea (S. pandanicola 160–240 × 7.5–23 vs. 118–165 × 4–5.6 µm S. carnea) (Seaver and Waterston 1941). Based on multi-gene phylogenetic analysis, S. pandanicola is distinct from other species of Stictis (0.99 in BYPP, Fig. 60). The placement of this genus requires resolving through additional collections and phylogenetic data. In a BLASTn search on NCBI GenBank, the closest matches of mt-SSU sequence of HKAS 96206 is S. confusum swith 88% identity to accession number DQ401141.
Class Leotiomycetes O.E. Erikss. & Winka
Rhytismatales M.E. Barr ex Minter
Rhytismataceae Chevall. (= Hypodermataceae Rehm; = Cryptomycetaceae Höhn. nom. inval. fide Jaklitsch et al. 2016)
Rhytismataceae was established by Chevallier (1826) with Rhytisma Fr. as the type genus and it is the largest family in order Rhytismatales (Johnston 2001). Forty-four genera have been are listed in Rhytismataceae by Lumbsch and Huhndorf (2010), while Wijayawardene et al. (2018) provided 50 genera in Rhytismataceae. Lophodermium is the largest genus with more than 100 species accepted (Lantz et al. 2011). A new species of Terriera, is introduced here and this is the first record of Rhytismataceae from Pandanaceae.
Terriera B. Erikss.
Eriksson (1970) introduced Terriera based on T. cladophila (Lév.) B. Erikss. (≡ Hysterium cladophilum Lév.). A modern description of T. cladophila was provided by Minter (1996). Terriera contains 37 epithets (Index Fungorum 2018), but only five species have sequences in GenBank. Terriera never have been recorded from Pandanaceae.
Terriera pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554511, Facesoffungi number: FoF04521; Fig. 61
Phylogram generated from maximum likelihood analysis based on LSU sequence data. Related sequences were obtained from Hyde et al. (2016). Twelve strains are included in the combined sequence analysis, which comprise 1206 characters with gaps. Cudonia lutea (C280) is used as the outgroup taxon. Tree topology of the ML analysis was similar to MP and BI. The best scoring RAxML tree with a final likelihood value of − 4022.180007 is presented. The matrix had 289 distinct alignment patterns, with 13.76% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.249070, C = 0.218694, G = 0.310904, T = 0.221332; substitution rates AC = 2.159879, AG = 3.488602, AT = 1.289966, CG = 1.444688, CT = 6.586959, GT = 1.000000; gamma distribution shape parameter α = 0.194634. Bootstrap support values for MP, ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. The newly generated sequence is in red
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1931
Saprobic on dead leaf of Pandanus sp. Sexual morph Ascomata hysterothecial in the surface view, 160–220 × 230–290 µm (\( \bar{x} \) = 190 × 264 µm, n = 5), elliptical, conspicuous at the surface, dull, with rounded to subacute ends, the central part of the ascomata strongly raising the surface of the substrate at maturity, opening by a longitudinal split that extends almost whole length of ascoma, rough. Peridium 15–37 µm wide, carbonaceous, hyaline cells of textura globulosa. Hamathecium composed of dense 1–2.6 µm, hyaline, aseptate, cylindrical, long, anastomosing paraphysate hyphae among asci. Asci 50–66 × 4–5 µm (\( \bar{x} \) = 57 × 4.5 µm, n = 20), 8-spored, unitunicate, cylindrical, apex obtuse to truncate, short-pedicellate, with a J-, apical ring. Ascospores 55–78 × 1–2 µm (\( \bar{x} \) = 66 × 1.6 µm, n = 10), overlapping bi-seriate, arranged in a fascicle, hyaline, filiform, tapering slightly towards each end, aseptate, guttulate, without a gelatinous sheath or appendages. Asexual morph Undetermined.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on dead leaf of Pandanus sp., 30 July 2015, S. Tibpromma & K.D Hyde SF15-025 (MFLU 16-1931, holotype).
GenBank numbers LSU: MH260320; ITS: MH275086; SSU: MH260360; TEF1: MH412783.
Notes: Terriera pandanicola is similar to T. aequabilis Qing Li & Y.R. Lin. Terriera. aequabilis has asci 75–105 × 4.5–5.5 µm, 8-spored, with ascospores 55–78 × 0.8–1 µm and covered by a 0.3–0.5 µm gelatinous sheath (Li et al. 2015), while T. pandanicola has asci 50–66 × 4–5 µm with ascospores 55–78 × 1–2 µm and, without a gelatinous sheath or appendages. Based on phylogenetic analysis, T. pandanicola is well-separated from other Terriera spp. (Fig. 62). Terriera has not been previously reported on Pandanaceae. In a BLASTn search on NCBI GenBank, the closest matches of LSU sequence of MFLU 16-1931 is T. minor with 98% identity to the strain ICMP 13974 (HM140571).
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Diaporthomycetidae Senan. et al.
Diaporthomycetidae families incertae sedis
Distoseptisporaceae K.D. Hyde & McKenzie
Distoseptisporaceae was introduces by Su et al. (2016) with Distoseptispora K.D. Hyde, McKenzie & Maharachch as the type genus. Morphologically, members of this family are sporidesmium-like taxa, a grouping that has been shown to be polyphyletic in the class Dothideomycetes (Shenoy et al. 2006; Su et al. 2016), with one family Distoseptisporaceae in Sordariomycetes. No sexual morph is known for this family. We follow Su et al. (2016) and Hyde et al. (2017) and introduce two new species in Distoseptispora from Pandanaceae and provide an updated tree for the family (Fig. 63).
Phylogram generated from maximum likelihood analysis based on combined ITS and LSU sequenced data. Related sequences were obtained from Su et al. (2016). Thirty strains are included in the combined sequence analysis, which comprise 1507 characters with gaps. Sordaria fimicola (CBS 508.50) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 7839.454976 is presented. The matrix had 649 distinct alignment patterns, with 39.30% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.243435, C = 0.245581, G = 0.299666, T = 0.211318; substitution rates AC = 1.626130, AG = 2.358078, AT = 1.438741, CG = 1.405567, CT = 7.750979, GT = 1.000000; gamma distribution shape parameter α = 0.334834. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in red
Distoseptispora K.D. Hyde et al.
Distoseptispora was introduced by Su et al. (2016) with D. aquatica Z.L. Luo, Hong Y. Su & K.D. Hyde as type species. Members of this genus have dark conidia with a slightly paler, rounded apex, and distinct basal cells. The conidia are of indeterminate length, while the conidiophores are relatively short. No sexual morph has been reported in this genus. Distoseptispora contains 13 epithets are listed in Index Fungorum (2018). We introduce two new species of Distoseptispora from Pandanaceae.
Distoseptispora thailandica Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554553, Facesoffungi number: FoF04562; Fig. 64
Distoseptispora thailandica (MFLU 16-0555, holotype). a Colony on dead leaves of Pandanus sp. b–d Conidiophores, conidiogenous cells and conidium. e Conidiogenous cells with conidiophores. f Apex of conidia. g Germinating conidium. h, i Colony on MEA from above and below. Scale bars: a–c = 50 μm, d, g = 20 μm, e, f = 10 μm
Etymology: named after Thailand,where the fungus was first discovered.
Holotype: MFLU 16-0555
Saprobic on dead leaves of Pandanus sp. Colonies on the substratum superficial, effuse, hairy or velvety, black. Mycelium mostly immersed, branched, septate, smooth, pale brown. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 15–26 × 3–6 μm, macronematous, mononematous, septate, unbranched, single or in groups, erect, straight or flexuous, smooth, brown to red-brown, cylindrical, thickened at the base. Conidiogenous cells monoblastic, integrated, determinate, terminal, red-brown, cylindrical. Conidia 130–230 × 13.5–17 μm (\( \bar{x} \) = 195 × 14.5 μm, n = 20), acrogenous, solitary, dry, oblong, obclavate, cylindrical or rostrate, straight or curved, truncate at the base, rounded at the apex, 35–52-distoseptate, reddish brown to brown, pale brown towards the apex, thick-walled. Conidial secession schizolytic.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, dark grey on the surface, dense, circular, with entire edge, velvety, raised, dark brown from below, with smooth margin.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-033 (MFLU 16-0555, holotype; HKAS 100838, isotype); ex-type living culture, MFLUCC 16-0270 = KUMCC 17-0306.
GenBank numbers LSU: MH260292; ITS: MH275060; SSU: MH260334; TEF1: MH412767.
Notes: Distoseptispora thailandica clusters with D. phangngaensis J. Yang, Maharachch. & K.D. Hyde but is separated as a distinct species with high bootstrap support (96% in ML, 0.97 in BYPP). Both species were collected in Thailand. Distoseptispora thailandica has reddish brown to brown conidia, remain pale brown towards the apex, measuring 130–230 × 13.5–17 μm, while D. phangngaensis has dark olivaceous to mid or dark brown conidia, with a subcylindrical basal cell and measuring 165–350 × 14–19 μm (Yang et al. 2018). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0270 is D. phangngaensis with 99% identity to the strain MFLU 17-0855 (MF077545), while the closest matches with the TEF1 sequence were with 99% identical D. phangngaensis strain MFLUCC 16-0857 (MF135653).
Distoseptispora xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554554, Facesoffungi number: FoF04563; Fig. 65
Distoseptispora xishuangbanna (HKAS 101809, holotype). a Colonies on dead leaf of Pandanus sp. b–e Conidia. f Conidiophores, conidiogenous cells and conidium. g Conidiophores and conidiogenous cells. h Germinating conidium. i, j Colony on MEA from above and below. Scale bars a = 100 μm, b–f, h = 50 μm, g = 10
Etymology: named after the location Xishuangbanna, the fungus was first discovered.
Holotype: HKAS 101809
Saprobic on dead leaf sheaths of Pandanus utilis. Colonies effuse, dark brown, hairy or velvety. Mycelium mostly immersed, composed of branched, septate, smooth, hyaline to pale brown hyphae. Asexual morph Hyphomycetous. Conidiophores 12–17 × 2–5 μm (\( \bar{x} \) = 14.4 × 4 μm, n = 10), macronematous, mononematous, solitary, 2–3-septate, straight or slightly flexuous, erect, slightly tapering distally, truncate at the apex. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, brown, determinate, cylindrical. Conidia 160–305 × 8–15 μm (\( \bar{x} \) = 244 × 11.5 μm, n = 10), acrogenous, solitary, cylindric-obclavate, up to 40-distoseptate, tapering towards apex, green–brown to brown, elongate, straight or slightly curved, rounded at apex, obconically truncate at base, thick-walled, smooth. Conidial secession schizolytic (Fig. 66).
One of the 10 most parsimonious trees obtained from a heuristic search of combined ITS, GPDH, CHS-1, HIS3, ACT and TUB2 sequence data of taxa from the gloeosporioides species complex. Fourty strains are included in the combined sequence analysis. Colletotrichum boninense (MAFF 305972) is used as the outgroup taxon. Tree topology of the MP analysis was similar to the ML and BYPP. The maximum parsimonious dataset consisted of 1840 characters, which 1280 were constant, 251 parsimony-informative and 306 parsimony uninformative. The parsimony analysis of the data matrix resulted in the maximum of 10 equally most parsimonious trees with a length of 961 steps (CI = 0.710, RI = 0.751, RC = 0.533, HI = 0.290) in the first tree. Bootstrap support values for MP, ML equal to or greater than 60%, BYPP equal to or greater than 0.95 are given above or below the nodes. The newly generated sequence is in red
Culture characteristics: Conidia germinating on PDA within 12 h. Germ tubes produced from both ends. Colonies on PDA reaching 9 cm diam., after 2 weeks at room temperature, white to cream, circular with undulate edge, velvety, dense, dark brown in reverse.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus utilis Bory., 28 April 2017, R. Phookamsak XTBG26 (HKAS 101809, holotype); ex-type living culture, KUMCC 17-0290; Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on Pandanus sp., 16 December 2017, S. Tibpromma P03 (MFLU 18-0022, paratype).
GenBank numbers LSU: MH260293; ITS: MH275061; SSU: MH260335; TEF1: MH412768; RPB2: MH412754.
Notes: The morphology of Distoseptispora xishuangbannaensis is very similar to Distoseptispora, Ellisembia and Sporidesmium (Shenoy et al. 2007; Su et al. 2016). However, based on DNA sequence analysis this strain belongs to Distoseptispora. Distoseptispora xishuangbannaensis clusters with D. leonensisi but is well-separated (99% in ML). There are more than 50 bp (> 4.42%) differences in 1130 RPB2 nucleotides between the two species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0290 is D. fluminicola with 94% identity to the strain MFLUCC 15-0417 (NR_154041), while the closest matches with the TEF1 sequence were with 97% identical D. tectonae strain MFLUCC 12-0291 (KX751710) and the closest matches with the RPB2 sequence were with 94% identical D. multiseptata strain MFLUCC 16-1044 (MF135644).
Subclass Hypocreomycetidae O.E. Erikss. & Winka
Glomerellales Chadef. ex Réblová et al.
Glomerellaceae Locq. ex Seifert & W. Gams
Glomerellaceae was informally erected by Locquin (1984) as Glomerella Spauld. & H. Schrenk as type genus. Uecker (1994) showed that Colletotrichum does not belong in the same order as Phyllachora based on preliminary sequence-based studies. Thus, Kirk et al. (2001) placed Glomerellaceae in an uncertain position in Sordariomycetidae. Zhang et al. (2006) validated the family Glomerellaceae with a Latin description placing this family in Hypocreomycetidae. Subsequently, Kirk et al. (2008) placed the family in an uncertain position in subclass Hypocreomycetidae. Réblová et al. (2011) validated the order Glomerellales with two new families (Australiascaceae and Reticulascaceae) based on analysis of combined sequence data (ITS, LSU, SSU and RPB2). Jayawardena et al. (2016b) provided a list of accepted species of Colletotrichum.
Colletotrichum Corda
Colletotrichum was erected by Corda (1831), for C. lineola Corda (Hyde et al. 2009) and comprises endophytes, pathogens and saprobes (Guo et al. 2001; Than et al. 2008; Hyde et al. 2009; Yang et al. 2011). Five species have been recorded on Pandanaceae: Colletotrichum dematium (Pers.) Grove, C. fructicola Prihast., L. Cai & K.D. Hyde, C. gloeosporioides (Penz.) Penz. & Sacc., C. pandani Syd. & P. Syd. and C. pandanicola Tibpromma S & Hyde KD (Whitton et al. 2012; Tibpromma et al. 2018). There are 869 epithets listed in Index Fungorum (2018).
Colletotrichum pandanicola Tibpromma & K.D. Hyde, Mycokeys 33: 47 (2018)
Facesoffungi number: FoF04534; Fig. 67
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata acervular, without setae, yellow to orange. Conidiophores hyaline to subhyaline, smooth-walled, aseptate. Conidiogenous cells 7–24.5 × 2–4 μm (\( \bar{x} \) = 13 × 3 μm, n = 20), hyaline to subhyaline, smooth-walled, phialidic, cylindrical, with distinct collarette, often extending to form new conidiogenous loci. Conidia 13–19 × 4.5–6 μm (\( \bar{x} \) = 15.5 × 5.4 μm, n = 40), oblong to cylindrical, hyaline to subhyaline, smooth-walled, aseptate, guttulate.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular with entire edge, grey in the central and white at the margin and become black with age, velvety.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Pandanus sp., 20 December 2017, S. Tibpromma P17 (MFLU 18-0036; HKAS 101802); living culture, MFLUCC 18-0118.
GenBank numbers LSU: MH376708; ITS: MH388336; SSU: MH388305; TEF1: MH388371; ACT: MH412714; TUB2: MH412718; GPDH: MH412726.
Notes: The gloeosporioides species complex are mainly known as plant pathogens although some can be found as endophytes (Weir et al. 2012; Liu et al. 2015a, b). We followed Jayawardena et al. (2016a, b) and identified our collection as C. pandanicola. Colletotrichum pandanicola is known as endophytic fungi which have been isolated from healthy leaves of Pandanus sp. in Thailand (Tibpromma et al. 2018). Our isolate is from the same host and from country, differing only in lifestyle. This fact supports the hypothesis that endophytes can change their life styles accordingly (Petrini 1991; Ghimire and Hyde 2004; Photita et al. 2004; Hyde et al. 2006).
Even though there is a short branch length difference in the phylogenetic tree, we were unable to find a reasonable base pair differences between our isolate and C. pandanicola type strain. The morphology of the two isolates also appear to be much more similar. Therefore, we prefer to keep this as C. pandanicola instead of describing it as a new species.
Malaysiascaceae Tibpromma & K.D. Hyde, fam. nov.
Index Fungorum number: IF554753, Facesoffungi number: FoF 04611
Saprobic on dead or decaying leaves, wood in terrestrial habitats. Sexual morph Undetermined. Asexual morph Conidiophores subcylindrical, unbranched, macronematous, erect, flexuous, thick-walled, rounded at apex, guttulate, multi-septate, pale brown to bark brown, pale brown towards slightly tapered apex, smooth-walled. Conidiogenous cells enteroblastic, phialidic, subcylindrical, subhyaline, terminal, integrated. Conidia solitary, cylindric-ellipsoid, rounded at apex, aseptate, hyaline, smooth-walled, guttulate, with dry in mass.
Type genus: Malaysiasca Crous & M.J. Wingf., Persoonia 36: 373 (2016)
Notes: Malaysiascaceae is introduced to accommodate the holomorphic genus Malaysiasca. Based on Bayesian analysis of LSU gene data, Malaysiasca is related to members of the order Glomerellales genera, incertae sedis, Sordariomycetes (Crous et al. 2016a). In the present phylogenetic analysis, Malaysiasca forms a well-supported clade (88% in ML, 1 in BYPP, Fig. 68) sister to Australiascaceae and Glomerellaceae. Morphologically Malaysiascaceae is similar to Australiascaceae; the asexual morph of the latter has phialidic conidiogenesis, with hyaline 0(–3)-septate conidia, aggregated in slime or in chains (Réblová et al. 2011). Malaysiascaceae is monotypic comprising only Malaysiasca phaii, which was found on leaves of Phaius reflexipetalus (Crous et al. 2016a) and now on Freycinetia javanica.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU and ITS sequence data. Related sequences were obtained from Crous et al. (2016a). Thirty-five strains are included in the combined sequence analysis, which comprise 2366 characters with gaps. Lasionectria mantuana (AR 4029) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 12724.747933 is presented. The matrix had 941 distinct alignment patterns, with 41.31% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.250455, C = 0.232927, G = 0.281835, T = 0.234784; substitution rates AC = 1.410406, AG = 2.256966, AT = 2.157471, CG = 1.313307, CT = 6.866289, GT = 1.000000; gamma distribution shape parameter α = 0.214370. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Malaysiasca Crous & M.J. Wingf.
Malaysiasca was erected by Crous et al. (2016a) to accommodate M. phaii Crous & M.J. Wingf. which first collected from Malaysia. The characteristics of this genus are perithecial ascomata, ellipsoid to oblong asci, hyaline, 1-septate ascospores, while the asexual morph forms a fascicle of long conidiophores, cylindrical phialides producing ellipsoidal to cylindrical-ellipsoid to somewhat clavate conidia in a slimy mass (Crous et al. 2016a).
Malaysiasca phaii Crous & M.J. Wingf., Persoonia 36: 373 (2016)
Facesoffungi number: FoF04538; Fig. 69
Malaysiasca phaii (MFLU 16-1910). a, b Colonies on dead leaf of Freycinetia sp. c Conidiophores, conidiogenous cells and conidia. d Conidiophores. e–g Conidiogenous cells and conidium. h, i Conidia. j, k Colony on MEA from above and below. Scale bars: a = 200 μm, b, c = 50 μm, d, e–g = 10 μm, h, i = 5 μm
Saprobic on dead leaves of Freycinetia javanica. Colonies on natural substratum effuse, brown to dark brown. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 160–240 × 4–5 μm, subcylindrical, unbranched, macronematous, erect, flexuous, thick-walled, round at the apex, guttulate, 5–7-septate, pale brown to dark brown, becoming somewhat pale brown towards slightly tapered apex, smooth-walled. Conidiogenous cells 50–100 × 4–7 μm (\( \bar{x} \) = 69.5 × 5 μm, n = 20), terminal, enteroblastic, phialidic, subcylindrical, subhyaline. Conidia 7–24 × 6–10 μm (\( \bar{x} \) = 14 × 8 μm, n = 20), solitary, cylindric-ellipsoid, rounded at apex, aseptate, hyaline, smooth-walled, guttulate, produced in a slimy mass.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, irregular, edge undulate, grey to dark grey in MEA medium. Mycelium superficial, flossy and raised.
Material examined: THAILAND, Krabi Province, Khlong Thom District, on dead leaves of Freycinetia javanica Blume., 15 December 2015, S. Tibpromma KB032 (MFLU 16-1910, HKAS 96259); living culture, MFLUCC 16-0256 = KUMCC 17-0285.
GenBank numbers LSU: MH260302; ITS: MH275069; SSU: MH260342; TEF1: MH412775.
Notes: In our phylogenetic tree, the Thai strain (MFLUCC 16-0256) clusters with Malaysiasca phaii Crous & M.J. Wingf. (CPC 27548). The new strain from Freycinetia javanica has 69.5 × 5 μm conidiogenous cells with 14 × 8 μm, cylindrical-ellipsoid, hyaline conidia, without a mucilaginous sheath. The type strain of M. phaii has conidiogenous cells 55–80 × 6 μm, which are similar to our strain but the conidia are slightly larger (16–)18–20(–24) × (8–)9–10(–11) μm (Crous et al. 2016a). This is the first record of Malaysiasca from Pandanaceae.
Plectosphaerellaceae W. Gams et al.
Plectosphaerellaceae was erected by Zare et al. (2007), with Plectosphaerella as the type genus. Hyde et al. (2017) updated phylogeny of this family and eleven genera are recognized in Plectosphaerellaceae. We introduce a new monotypic genus, Acremoniisimulans in Plectosphaerellaceae, and a new species of Musicillium on Pandanaceae (Fig. 70).
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF1 and ITS sequence data. Related sequences were obtained from Hyde et al. (2017). Forty-two strains are included in the combined sequence analysis, which comprise 3439 characters with gaps. Reticulascus clavatus (CBS 125296) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 13281.808204 is presented. The matrix had 852 distinct alignment patterns, with 50.50% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.229967, C = 0.276753, G = 0.279862, T = 0.213418; substitution rates AC = 0.717151, AG = 1.411613, AT = 1.372652, CG = 0.561709, CT = 4.777886, GT = 1.000000; gamma distribution shape parameter α = 0.463512. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in red
Acremoniisimulans Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF555329, Facesoffungi number: FoF04535
Etymology: refers to the morphology similar to Acremonium.
Type species: Acremoniisimulans thailandensis Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves, wood in terrestrial habitats. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, erect, straight or slightly flexuous, simple, blackish to brown. Mycelium immersed on the substrate, composed of septate, branched or unbranched, pale brown. Conidiophores macronematous, mononematous, scattered, brown with pale brown to hyaline apex, smooth, thick-walled, multi-septate, branched or unbranched, straight or slightly flexuous. Conidiogenous cells monophialidic, hyaline to subhyaline, cylindrical, slightly. Conidia solitary, hyaline or subhyaline to pale brown, oval, aseptate, rounded at end, slimy, with or without mucilaginous sheath.
Notes: We introduced a new genus, Acremoniisimulans having similar morphology with Acremonium, for a fungus isolated from dead leaves of Pandanaceae. Acremoniisimulans has similar conidia to Acremonium. The closest hits using a BLASTn search on NCBI GenBank of the LSU DNA sequence data are 99% similar to Stachylidium bicolor (GenBank GU180651), Wallrothiella gmelinae strain CBS 142520 (KY979808) and Acremonium hyalinulum strain CBS 652.96 (LN810512). The phylogenetic analysis of the combined LSU, SSU, ITS and TEF1 data set revealed that our novel taxon clusters independently in Plectosphaerellaceae. Our novel taxon includes different morphological characters from other members of Plectosphaerellaceae, in has brown with pale brown to hyaline at apex, septate, unbranched conidiophores with solitary, pale brown to brown, oval, aseptate conidia. However, Acremonium and Acremoniisimulans are phylogenetically distinct genera, well-segregated with high support (78% in ML and 0.95 in BYPP, Fig. 70) in our phylogeny.
Acremoniisimulans thailandensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555330, Facesoffungi number: FoF04536; Fig. 71
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 18-0012
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate consisting of erect conidiophores. Mycelium immersed in the substrate, composed of septate, branched, brown hyphae. Conidiophores 106–112 × 3–4.5 μm (\( \bar{x} \) = 109 × 4 μm, n = 10), macronematous, mononematous, scattered, brown, pale brown to subhyaline at apex, smooth, thick-walled, 3–4-septate, unbranched, straight or slightly flexuous. Conidiogenous cells 28.5–44 × 3–4.5 μm (\( \bar{x} \) = 37 × 3.5 μm, n = 20), monophialidic, subhyaline, cylindrical. Conidia 5.5–8 × 3–4 μm (\( \bar{x} \) = 6.5 × 3.5 μm, n = 30), solitary, pale brown to brown, oval, aseptate, rounded at each end, smooth-walled, slimy, without mucilaginous sheath.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA reaching 9 cm in 2 weeks at room temperature, circular, undulate with white to cream, raised on surface media, velvety.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-028 (MFLU 18-0012, holotype); ex-type living culture, MFLUCC 16-0372 = KUMCC 16-0159.
GenBank numbers LSU: MH260315; ITS: MH275081; SSU: MH260355.
Notes: Acremoniisimulans thailandensis is described as a unique new species based on our phylogenetic analysis where it formed a well-separated clade from other genera with high support. It has pale brown to brown, oval, aseptate conidia.
Musicillium Zare & W Gams
Musicillium is a monotypic genus which was erected by Zare et al. (2007) with M. theobromae (Turconi) Zare & W. Gams as the type species. This genus is a verticillium-like hyphomycete. We collected a new species of Musicillium from Pandanaceae in Thailand.
Musicillium pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555295, Facesoffungi number: FoF04537; Fig. 72
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0124
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies consisting of erect conidiophores. Mycelium immersed in the substrate, composed of septate, branched, brown hyphae. Conidiophores 500–578 × 6–9 μm (\( \bar{x} \) = 547 × 7 μm, n = 5), macronematous, mononematous, scattered, brown, pale brown at apex, smooth, thick-walled, multi-septate, branched, straight or slightly flexuous. Conidiogenous cells 15–23 × 1–5 μm (\( \bar{x} \) = 18 × 4.5 μm, n = 20), monophialidic, subhyaline, cylindrical or flask-shaped, slightly tapering. Conidia 4–5 × 1–3 μm (\( \bar{x} \) = 4.6 × 2.4 μm, n = 30), hyaline to subhyaline, oval, aseptate, rounded at both ends, smooth-walled, without slime.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA reaching 9 cm diam., in 2 weeks at room temperature, circular, entire edge with black-grey in the middle and white at the margin, raised on surface media, velvety.
Material examined: THAILAND, Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation (MRF), on Pandanus sp., 16 December 2017, S. Tibpromma P02 (MFLU 18-0124, holotype; HKAS 101790, isotype); ex-type living culture, MFLUCC 18-0109.
GenBank numbers LSU: MH260305; SSU: MH260345; TEF1: MH412777; RPB2: MH412757.
Notes: Musicillium was described from several hosts (Theobroma, Cacao and Musa). Phylogenetic analysis and a morphological comparison revealed that our new strain is closely related to M. theobromae (Turconi) Zare & W. Gams (CBS 548.51), but M. theobromae has conidiophores which are clearly distinct from the vegetative hyphae, brown 3–6 (rarely solitary), subhyaline, 17–35 × 1.5–2.7 μm phialides, and hyaline, cylindrical, symmetrically rounded, 4–6.5(− 9) × 1.0–1.7(− 2) μm conidia, adhering in slimy heads (Zare et al. 2007). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 18-0109 is M. theobromae with 98% identity to the strain NZD-mf44 (AJ292422).
Hypocreales Lindau
Bionectriaceae Samuels & Rossman
Bionectriaceae was erected by Rossman et al. (1999) with Bionectria Speg. as the type genus. This family is characterized by uniloculate perithecia or rarely cleistothecial ascomata lacking a stroma, and superficial or immersed in the substratum (Rossman et al. 1999). Rossman et al. (2001) included many genera and related asexual morph taxa and confirmed that Bionectriaceae is monophyletic within Hypocreales based on phylogenetic analysis. Thirty-nine genera are accepted in Bionectriaceae and we follow Wijayawardene et al. (2018) for the genera and provide an updated tree for members of Bionectriaceae collected from Pandanaceae.
Clonostachys Corda
Clonostachys was introduced with C. araucaria (Corda 1839). Clonostachys is an older, although asexual name for Bionectria. Thus, Rossman et al. (2013) synonymized Bionectria under Clonostachys. An update for Clonostachys was provided by Maharachchikumbura et al. (2015). The asexual morph of Clonostachys is characterized by penicillate, frequently sporodochial and, in many cases, dimorphic conidiophores (Schroers 2001). There are 83 epithets are listed in Index Fungorum (2018). Clonostachys compactiuscula (Sacc.) D. Hawksw. & W. Gams has been reported from Pandanaceae (Hawksworth and Punithalingam 1975; Dulymamode et al. 2001). We introduce a new species isolated from Pandanus in Thailand.
Clonostachys krabiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554527, Facesoffungi number: FoF04539; Fig. 73
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, cmdA, RPB2, TEF1 and TUB2 sequence data. Two hundred and eighty five strains are included in the combined sequence analysis, which comprise 4417 characters with gaps. Gelasinospora tetrasperma (AFTOL-ID 1287), Neurospora crassa (ICMP 6360) and Sordaria fimicola (AFTOL-ID 216) are used as the outgroup taxa. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 146671.299056 is presented. The matrix had 2852 distinct alignment patterns, with 46.08% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.239937, C = 0.264170, G = 0.277202, T = 0.21869; substitution rates AC = 1.466937, AG = 3.219738, AT = 1.533143, CG = 0.996922, CT = 6.732220, GT = 1.000000; gamma distribution shape parameter α = 0.371105. Bootstrap support values for ML equal to or greater than 50% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in red
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1907
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Setae 173–375 × 2–4 μm (\( \bar{x} \) = 252 × 3 μm, n = 20), hyaline, aseptate, cylindrical with conical apex. Conidiophores aggregated into sporodochia, with hyaline setae around the margin. Conidiogenous cells 10–13 × 1.5–2.5 μm (\( \bar{x} \) = 11.5 × 2 μm, n = 20), monophialidic, hyaline, subulate. Conidia 5–7 × 1–2 μm (\( \bar{x} \) = 6 × 1.6 μm, n = 40), slimy, solitary, aseptate, hyaline, cylindrical to oblong, smooth-walled.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, irregular, undulate edge with whorls, yellow–white, flat on media surface.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB029 (MFLU 16-1907, holotype; HKAS 96256, isotype); ex-type living culture, MFLUCC 16-0254.
GenBank numbers LSU: MH376707; ITS: MH388335; SSU: MH388304.
Notes: Based on phylogeny Clonostachys kraviensis clusters with C. byssicola Schroers (95% in ML, Fig. 74). However, C. byssicola has shorter conidia (frequently less than 5 μm long) with slightly curved, and a laterally displaced hilum (Schroers 2001). We also compared our fungus with Clonostachys compactiuscula but it has conidia produced in long chains up to 150 μm long (Hawksworth and Punithalingam 1975). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0254 is C. rosea with 99% identity to the strain FIS26 (KY378958), while the closest matches with the SSU sequence were with 99% identical C. rosea strain WY-1 (MH031706).
Lasionectria (Sacc.) Cooke
Lasionectria was erected by Cooke (1884) to accommodate L. mantuana (Sacc.) Cooke. Lasionectria occurs as saprobes in terrestrial and temperate habitats (Lumbsch and Huhndorf 2010). Lasionectria has 36 epithets are listed in Index Fungorum (2018). Lasionectria mantuana (Sacc.) Cooke and L. sylvana (Mouton) Rossman & Samuels have been reported from Pandanaceae (Whitton et al. 2012).
Lasionectria krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554528, Facesoffungi number: FoF04540; Fig. 75
Lasionectria krabiense (MFLU 16-0540 holotype). a Colonies on dead leaf of Pandanus sp. b Section of ascoma. c Papilla. d Paraphyses. e Peridium. f, g Asci. h–k Ascospores. l Germinating ascospore. m, n Colony on MEA from above and below. Scale bars: a = 200 μm, b, f = 20 μm, c–e = 5 μm, g = 10 μm, h–l = 2 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-0540
Saprobic on dead leaf of Pandanus sp. Sexual morph Ascomata 140–200 × 220–260 µm (\( \bar{x} \) = 168 × 236 µm, n = 5), solitary, superficial, globose to subglobose, with apex flattened with a minute papilla, orange to brownish orange, collapsing and becoming cupulate when dry, without ostiole. Peridium 15–30 μm wide, composed of hyaline to orange, hypha-like cells of textura prismatica. Hamathecium 1.5–3 µm wide, comprising numerous, dense, filiform, filamentous, branched, guttulate, septate paraphyses. Asci 30–55 × 8–11 µm (\( \bar{x} \) = 43 × 9 µm, n = 20), 6–8-spored, unitunicate, cylindrical to cylindric-clavate, short-pedicellate, apically rounded. Ascospores 15–20 × 3–5 µm (\( \bar{x} \) = 17 × 4 µm, n = 20), overlapping bi-seriate, fusoid-ellipsoidal, hyaline to subhyaline, 1-septate in the middle, rough, with longitudinal striations, granulate, not surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospore germinating on MEA within 12 h. Colonies on MEA, circular to irregular, undulate with white to yellow–white, flat on media, rough with wrinkles edge.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2014, S. Tibpromma SF14-022 (MFLU 16-0540 holotype; HKAS 100829, isotype); living culture, MFLUCC 15-0673; CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 27 July 2016, PE Mortimer NBH13 (HKAS 96213, paratype).
GenBank numbers LSU: MH376725; ITS: MH388352; SSU: MH388319; TEF1: MH388387.
Notes: On combined gene analysis, Lasionectria krabiense clusters with L. antillana (Lechat & Courtec.) Schroers, Ashrafi & W. Maier with strong bootstrap support (100% in ML, Fig. 74). The ascomata and ascospore morphology of both species is very similar, but L. antillana has 12.7 × 3.2 µm ascospores without longitudinal striations (Lechat and Courtecuisse 2010). There are more than 34 bp (6.77%) differences in the 502 ITS (+5.8S) nucleotides of L. antillana and L. krabiense which indicates they are distinct species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 15-0673 is L. antillana with 94% identity to the strain CBS 122797 (KY607537).
Paracylindrocarpon Crous et al.
Paracylindrocarpon was erected by Crous et al. (2016a) to accommodate P. aloicola Crous, Roets & L. Lombard. Paracylindrocarpon has morphological similarities to Cylindrocarpon with both having hyaline, smooth, conidiophores with hyaline granular, cylindrical, (0–)3-septate, conidia with obtuse apex and truncate base (Crous et al. 2016a). We describe three species of Paracylindrocarpon; this is the first report of Paracylindrocarpon from Pandanaceae.
Paracylindrocarpon nabanheensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554529, Facesoffungi number: FoF04541; Fig. 76
Etymology: named after Nabanhe, where the fungus was first discovered.
Holotype: HKAS 96210
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 214.5–346 × 320–367 µm (\( \bar{x} \) = 276.4 × 342.4 µm, n = 5), scattered to gregarious, globose to subglobose, superficial, flat at the base, conspicuous on host surface, easy to remove, orange, solitary, uniloculate, covered with hyaline papilla, with ostiole at the centre. Peridium 43–79 µm wide, composed of several layers, thick-walled hyaline to subhyaline cells of textura prismatica. Hamathecium comprising 7–9 µm wide, ellipsoid, cellular, unbranched, guttulate, septate paraphyses. Asci 58–81 × 9–14 μm (\( \bar{x} \) = 64.4 × 10.5 μm, n = 10), (6–)8-spored, unitunicate, cylindrical to cylindrical-clavate, with furcate pedicel, with J- apical ring. Ascospores 37–47 × 6–7.5 µm (\( \bar{x} \) = 40 × 6.5 µm, n = 10), fusiform, conical at both ends, 6-septate when mature, not constricted at the septa, hyaline to subhyaline, guttulate, without mucilaginous sheath, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, purple at the central and pink at the margin, with irregular, curled, with raised on media surface, dark-purple at the central with white at the margin in reverse.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 27 July 2016, PE Mortimer NBH10 (HKAS 96210, holotype); ex-type living culture, KUMCC 16-0147.
GenBank numbers LSU: MH376730; ITS: MH388356; SSU: MH388324; TEF1: MH388392.
Notes: Based on morphology Paracylindrocarpon nabanheensis is similar to P. xishuangbannaensis, which was collected from the same host and same location. Both species are known from only the sexual morph. In a comparison of the 506 ITS (+5.8S) nucleotides of P. nabanheensis and P. xishuangbannaensis reveals 20 (3.95%) nucleotide differences which justifies these two isolates as two distinct taxa and we also comparison ITS nucleotides of Paracylindrocarpon species (Table 3). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0147 is P. aloicola with 97% identity to the strain CPC 27362 (KX228277).
Paracylindrocarpon pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554530, Facesoffungi number: FoF04542; Fig. 77
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 100863
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium consisting of hyaline, smooth, branched, septate. Conidiophores sporodochial, solitary, hyaline, smooth, erect, straight to geniculate-sinuous, arising from superficial hyphae, unbranched or branched. Conidiogenous cells 26–38 × 2–3 μm (\( \bar{x} \) = 32.5 × 2 μm, n = 10), phialidic, hyaline, smooth, subcylindrical with slight apical taper, straight to slightly irregularly curved, terminal or lateral on conidiophores, apex with minute periclinal thickening. Conidia 15–22 × 2–4 μm (\( \bar{x} \) = 18 × 3 μm, n = 50), hyaline, smooth, granular, cylindrical, 2–3-septate, hyaline, filiform, slightly constricted at septa, slightly curved, thick-walled, distinctly guttulate, without appendages or mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, circular, edge entire, white to yellow.
Material examined: HONG KONG, Tai Tam Tuk Reservoir, on Pandanus sp., 21 September 2016, S. Tibpromma HK21 (HKAS 100863, holotype); ex-type living culture, KUMCC 17-0272 = MFLUCC 17-0639.
GenBank numbers LSU: MH376731; ITS: MH388357; SSU: MH388325; TEF1: MH388393.
Notes: Based on multi-gene analyses, Paracylindrocarpon pandanicola clusters with other Paracylindrocarpon spp. with strong bootstrap support (Fig. 74). Paracylindrocarpon pandanicola differs from P. aloicola Crous, Roets & L. Lombard has cylindrical conidia that have an obtuse apex, a truncate base and (0–)3-septa (Crous et al. 2016a). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0147 is P. aloicola with 94% identity to the strain CPC 27362 (KX228277).
Paracylindrocarpon xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554535, Facesoffungi number: FoF04543; Fig. 78
Paracylindrocarpon xishuangbannaensis (HKAS 96204, holotype). a, b Appearance of ascomata on host substrate. c Section of ascoma. d Section of peridium. e Hamathecium. f Papilla. g Ostiole. h, i Asci. j Ascus strained with cotton blue reagent. k–n Ascospores. o Germinating ascospore. Scale bars: c = 40 μm, d, f = 20 μm, e = 5 μm, g–o = 10 μm
Etymology: named after Xishuangbanna, where the fungus was first discovered.
Holotype: HKAS 96204
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 225–300 × 277.5–295 µm (\( \bar{x} \) = 259 × 288.5 µm, n = 5), scattered to gregarious, globose to subglobose, superficial, flat at the base, conspicuous on the host surface, easy to remove, orange, dull, solitary, uniloculate, with pale brown papilla, with central ostiole. Peridium 33–70.5 µm wide, composed of several layers of thick-walled, hyaline to pale brown cells of textura angularis. Hamathecium comprising 2.2–4 µm wide, cylindrical, filamentous, unbranched, guttulate, septate paraphyses. Asci 67–95 × 10–17 μm (\( \bar{x} \) = 72 × 12 μm, n = 20), 8-spored, unitunicate, cylindrical-clavate, with short pedicel, with J- apical ring. Ascospores 27–41 × 3–8 µm (\( \bar{x} \) = 34.5 × 5.6 µm, n = 20), fusiform, conical towards both ends, 3–5-septate, not constricted at septa, hyaline, guttulate without mucilaginous sheath, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 12 h. Colonies on PDA, yellow–white, with irregular whorls, raised and rough on media surface, yellow–brown in reverse.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 27 July 2016, PE Mortimer NBH04 (HKAS 96204, holotype); ex-type living culture, KUMCC 16-0144 = MFLUCC 17-0557.
GenBank numbers LSU: MH376732; ITS: MH388358; SSU: MH388326.
Notes: Here we provide first record sexual morphs of Paracylindrocarpon. Based on morphology P. xishuangbannaensis and P. nabanheensis both have fusiform, 3–5-septate, hyaline ascospores, that are not constricted at septa, while P. xishuangbannaensis has 72 × 12 μm, 8-spored asci, with 34.5 × 5.6 µm ascospores, but P. nabanheensis has smaller (6–)8-spored, 64.4 × 10.5 μm asci, with larger ascospores (41 × 6.5 µm). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0144 is Hyaloseta nolinae with 90% identity to the strain CBS 109837 (NR_156291).
Nectriaceae Tul. & C. Tul.
Nectriaceae was erected by Tulasne and Tulasne (1865) with Nectria (Fr.) Fr. as the type genus. Nectriaceae is characterized by uniloculate, pigmented ascomata and phialidic amerosporous to phragmosporous conidia (Rossman et al. 1999; Rossman 2000; Lombard et al. 2015). Members of this family can be found as pathogens, saprobes, or are fungicolous or insecticolous (Rossman et al. 1999; Rossman 2000; Chaverri et al. 2011; Schroers et al. 2011; Hyde et al. 2014). In previous studies, the placement of Nectriaceae was not stable and it was placed in different orders such as Hypocreales (Seaver 1909a, b, 1910a, b, 1911) and Sphaeriales (Munk 1957; Dennis 1960). Petch (1938) accepted Nectriaceae as a separate family in Hypocreales and later, Miller (1949), Bessy (1950), Luttrell (1951), Dingley (1951a, b, 1952a, b, 1953, 1954, 1956), von Arx and Müller (1954), Müller and von Arx (1962), Gäumann (1964), Rogerson (1970), and Barr (1990b) placed Nectriaceae synonyms under Hypocreaceae as one family. The family was later resolved by Lumbsch and Huhndorf (2010), Maharachchikumbura et al. (2015) and Lombard et al. (2015). There are 66 genera in the family (Wijayawardene et al. 2018). In this paper, we introduce new taxa to Nectriaceae based on phylogenetic evidence and morphology.
Cylindrocladiella Boesew.
Cylindrocladiella was erected by Boesewinkel (1982) to accommodate C. parva (P.J. Anderson) Boesew. Members of Cylindrocladiella spp. are found as soil-borne fungi, pathogens and/or saprobes of various plant hosts and substrates in temperate, sub-tropical and tropical regions. Cylindrocladiella has 43 epithets are listed in Index Fungorum (2018). We describe a new species from China.
Cylindrocladiella xishuangbannaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554536, Facesoffungi number: FoF04546; Fig. 79
Etymology: named after Xishuangbanna, where the fungus was first discovered.
Holotype: HKAS 96209
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores dimorphic, penicillate and subverticillate, mononematous, hyaline, penicillate, 456–476 × 6–7 μm (\( \bar{x} \) = 469 × 6.4 μm, n = 5), conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth; stipe extension aseptate, straight, thick-walled with one basal septum, terminating in a thin-walled, cylindrical vesicle. Penicillate conidiogenous apparatus 18–47.5 × 2.5–4 μm (\( \bar{x} \) = 30 × 3.6 μm, n = 10), with primary branches aseptate, secondary branches aseptate, each terminal branch producing 2–4 phialides; phialides doliiform to cymbiform, hyaline, aseptate, apex with minute periclinal thickening and collarette. Subverticillate 6–12 × 2–4 μm (\( \bar{x} \) = 9 × 3 μm, n = 20), conidiophores in moderate numbers, comprising a septate stipe, primary and secondary branches terminating in 1–3 phialides; primary branches straight, hyaline, 0–1-septate. Conidia 52.5–60.5 × 3–6 μm (\( \bar{x} \) = 57 × 5 μm, n = 40), cylindrical, rounded at both ends, straight, 1-septate, frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA, circular, entire edge with cinnamon, raised on surface media, produce pigment on the media.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on Pandanus sp., 28 July 2016, S. Tibpromma NBH09 (HKAS 96209, holotype); ex-type living culture, KUMCC 16-0146 = MFLUCC 17-0559.
GenBank numbers LSU: MH376709; ITS: MH388337; SSU: MH388306; RPB2: MH412727.
Notes: This species morphologically fits within the generic concepts of Cylindrocladiella. Based on the multi-gene sequence analyses, C. xishuangbannaensis clusters with C. camelliae (Venkataram. & C.S.V. Ram) Boesew. (Fig. 74). We compare the morphology with C. camelliae, but they differ. Cylindrocladiella camelliae has a slight difference in terminal vesicle shape with conidia size 15–26 × 2–3.5 μm (Boesewinkel 1982). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0146 is C. solicola with 99% identity to the strain CMW47198 (MH017021), while the closest matches with the RPB2 sequence were with 99% identical C. camelliae strain CPC 234 (KM232304).
Pandanaceomyces Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF554537, Facesoffungi number: FoF04544
Etymology: named after the host family, Pandanaceae.
Type species: Pandanaceomyces krabiensis Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves and wood in terrestrial habitats. Sexual morph Ascomata scattered to gregarious, superficial or semi-immersed with flat on the base, conspicuous on host surface, pigments, solitary, uniloculate, oval, papillate with ostiole. Peridium composed of several layers, thin-walled of textura prismatica. Hamathecium cellular, comprising branched or unbranched, guttulate, septate paraphyses. Asci 6–8-spored, unitunicate, cylindrical to cylindrical-clavate, with furcated pedicel, apically with J- or J + apical ring. Ascospores fusiform, curved towards both ends, 1-septate, not constricted at septa, hyaline to subhyaline, with or without guttulate and mucilaginous sheath. Asexual morph Undetermined.
Notes: Pandanaceomyces bears a close morphological similarity with Nectria which has light to bright coloured, soft-textured, superficial, uniloculate ascomata and unitunicate asci (Hirooka et al. 2012). A new genus Pandanaceomyces, is therefore introduced to accommodate the present collection. However, Pandanaceomyces based on DNA data it can be distinguished from other genera (Fig. 74) and no high similarity (> 99%) hits were obtained when the LSU gene sequences were blasted against NCBI GenBank nucleotide database.
Pandanaceomyces krabiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554538, Facesoffungi number: FoF04545; Fig. 80
Pandanaceomyces pandanicola (MFLU 16-1909, holotype). a Appearance of ascomata on host substrate. b Ascoma. c Section of ascoma. d Section of peridium. e Papilla. f Ostiole. g–j Asci. k-n Ascospores. o, p Colony on MEA from above and below. Scale bars: a = 200 μm, b = 50 μm, c, j = 20 μm, d, e, k–n = 5 μm, f–i = 10 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1909
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 115–133 × 100–120 µm (\( \bar{x} \) = 125 × 109 µm, n = 5), scattered to gregarious, superficial to erumpent, with flat base, conspicuous on the host surface, easy to remove, yellow–orange, dull, solitary, uniloculate, oval, with hyaline papilla, with ostiole at centre. Peridium 7–10 µm wide, composed of several layers of thin-walled, yellow cells of textura prismatica. Hamathecium 8.5–11 µm wide, oval to ellipsoid, composed of cellular, unbranched, guttulate, septate paraphyses. Asci 37.5–48 × 6–11 μm (\( \bar{x} \) = 43 × 7.5 μm, n = 20), (6–)8-spored, unitunicate, cylindrical to cylindrical-clavate, with short, furcate pedicel, with J- apical ring. Ascospores 11–16 × 2–3 µm (\( \bar{x} \) = 13 × 2.5 µm, n = 20), fusiform, curved towards both ends, 1-septate, not constricted at the septum, hyaline to subhyaline, guttulate, without mucilaginous sheath, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospore germinating on MEA within 12 h. Colonies on MEA, purple at the central and pink at the margin, with irregular whorls, raised on media surface, dark-purple at the central with white at the margin from below.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB031 (MFLU 16-1909, holotype; HKAS 96258, isotype); ex-type living culture, MFLUCC 16-0323.
GenBank numbers LSU: MH376729; ITS: MH388355; SSU: MH388323; TEF1: MH388391.
Notes: Phylogenetic analysis of combined sequence data indicated that Pandanaceomyces krabiensis clusters in a separate clade from other members of Nectriaceae with 73% ML bootstrap support (Fig. 74). Pandanaceomyces krabiensis has fusiform, 1-septate, not constricted at septa, hyaline to subhyaline ascospores curved towards both ends.
Pseudoachroiostachys Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF554547, Facesoffungi number: FoF04554
Etymology: name refers to the morphology being similar to Achroiostachys.
Type species: Pseudoachroiostachys krabiense Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves, wood in terrestrial habitats. Mycelium superficial, hyaline, rough, unbranched, septate, pale orange. Sexual morph Undetermined. Asexual morph Conidiophores macronematous, mononematous, unbranched, erect, straight, multi-septate, slightly thick-walled towards the base, hyaline and glassy, bearing a whorl of 5–6 conidiogenous cells. Conidiogenous cells terminal, phialidic, cylindrical, hyaline, smooth, narrowing to a short neck. Conidia ovoid, aseptate, sometimes flattened on one side, smooth, hyaline to subhyaline, rounded apex, in slimy masses, with or without mucilaginous sheath.
Notes: Combined rDNA gene sequence data revealed that Pseudoachroiostachys belongs to the family Stachybotryaceae and is closely related to Pandanaceomyces (Fig. 74). Pseudoachroiostachys is also similar to Achroiostachys, but the conidia of Achroiostachys are ellipsoidal, limoniform, globose to subglobose and contain 1–2 large or several small guttules (Lombard et al. 2016), while Pseudoachroiostachys has ovoid slimy conidia.
Pseudoachroiostachys krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554548, Facesoffungi number: FoF04555; Fig. 81
Pseudoachroiostachy krabiense (MFLU 16-1915, holotype). a Colonies on dead leaf of Pandanus sp. b Conidiophores, conidiogenous cells and conidia. c Conidiogenous cells and conidia. d–f Conidia. g Germinating conidium. h, i Colonies on MEA from above and below. Scale bars: a = 50 μm, b, c, g = 5 μm, d–f = 2 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1915
Saprobic on dead leaves of Pandanus sp. Mycelium superficial, hyaline, rough, unbranched, septate, aggregated in dense fascicles, pale orange. Sexual morph Undetermined. Asexual morph Conidiophores 40–50 × 3.5–4.5 µm (\( \bar{x} \) = 46 × 4 μm, n = 20), macronematous, mononematous, single, unbranched, erect, straight, 1–3-septate, slightly thick-walled towards the base, smooth, hyaline and glassy, bearing a whorl of 5–6 conidiogenous cells. Conidiogenous cells 6–8 × 1–2.5 μm (\( \bar{x} \) = 7 × 2 μm, n = 20), terminal, phialidic, discrete, cylindrical, hyaline, smooth, 10–13 × 3.5–4.5 μm, narrowing to a short neck about 1 μm. Conidia 5–9 × 3–4 μm (\( \bar{x} \) = 7 × 3.5 μm, n = 40), ovoid, in a group, aseptate, sometimes flattened on one side, smooth, hyaline, with rounded apex, slimy in mass, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular with whorls, undulate, white, smooth and raised on surface media, velvety.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 15 December 2015, S. Tibpromma KB037 (MFLU 16-1915, holotype; HKAS 96264, isotype); ex-type living culture, MFLUCC 16-0325.
GenBank numbers LSU: MH376736; ITS: MH388362; SSU: MH388328; TEF1: MH388397.
Notes: Achroiostachys aurantispora L. Lombard & Crous has similar morphology with Pseudoachroiostachys krabiense in having similar colonies on host plant and conidiophores. Achroiostachys aurantispora has elongate ampulliform to ventricose or clavate conidiogenous cells and ellipsoidal conidia (Lombard et al. 2016) while Pseudoachroiostachys krabiense has cylindrical conidiogenous cells and ovoid conidia.
Volutella Fr.
Volutella was erected by Fries (1832) to accommodate V. ciliata (Alb. & Schwein.) Fr. Volutella is characterised by discoid sporodochia with marginal setae, simple to verticillate conidiophores, compact and phialidic conidiogenous cells, and 1-celled, ovoid to oblong conidia (Gräfenhan et al. 2011; Luo and Zhuang 2012; Lombard et al. 2015). There are 144 epithets are listed in Index Fungorum (2018), but only eight species have available sequences in GenBank. Volutella mellea J.F. Clark and Volutella sp. have been reported from Pandanaceae (Dingley et al. 1981; Thongkantha et al. 2008).
Volutella krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554539, Facesoffungi number: FoF04547; Fig. 82
Volutella krabiense (MFLU 16-1932, holotype). a, b Sporodochial conidiomata on dead leaves of Pandanus sp. c, d Conidiophores, conidiogenous cells and conidium. e Conidia strain with cotton blue. f Germinating conidium. g, h Colonies on MEA from above and below. Scale bars: b = 50 μm, c = 20 μm, d–f = 5 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1932
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Setae 60–113 × 2–4 μm (\( \bar{x} \) = 92 × 3 μm, n = 10), hyaline, aseptate, cylindrical with conical apex. Conidiophores aggregated into sporodochia, with hyaline, stiff setae around the margin of conidiomata. Conidiogenous cells 2–4 × 2–3.5 μm (\( \bar{x} \) = 3 × 2.5 μm, n = 20), monophialidic, hyaline, subulate. Conidia 3–4 × 1–2 μm (\( \bar{x} \) = 3.4 × 1.5 μm, n = 30), slimy, aseptate, hyaline, oblong, guttulate, without mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, irregular, undulate edge with curled, white, flat on media surface.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma SF15-030 (MFLU 16-1932, holotype; HKAS 96281, isotype); ex-type living culture, MFLUCC 16-0268 = KUMCC 17-0305.
GenBank numbers LSU: MH376741; ITS: MH388367; SSU: MH388331; TEF1: MH388401.
Notes: Volutella krabiense has a close phylogenetic affinity to V. thailandensis in multi-gene phylogenetic analysis (Fig. 74). Volutella krabiense has smaller conidia than V. thailandensis (4 × 1.3 vs. 5 × 2 μm) and conidiomata of V. thailandensis lack setae. In a BLASTn search on NCBI GenBank, the closest matches of SSU sequence of MFLUCC 16-0268 is V. aeria with 99% identity to the strain CGMCC 3.17945 (KY883302).
Volutella thailandensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554540, Facesoffungi number: FoF04548; Fig. 83
Volutella thailandensis (MFLU 18-0014, holotype). a Appearance of ascomata on host substrate. b Ascomata. c Ascoma wall. d Hamathecium. e Asci. f Asci stained with cotton blue reagent. g Papilla. h, i Ascospores stained with cotton blue reagent. j, k Culture growing on MEA media. l Sporodochia formed in culture. m Conidiogenous cells and conidia formed in culture. n Conidia formed in culture. Scale bars: b = 50 μm, c, e, f, m, n = 5 μm, d, h, i = 2 μm, g = 10 μm, l = 200 μm
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 18-0014
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 115–140 × 100–130 µm (\( \bar{x} \) = 128 × 114 µm, n = 5), scattered to gregarious, superficial, flat at the base, conspicuous on host surface, easy to remove, black, dull, solitary, uniloculate, black with conspicuous setae, without ostiole. Peridium composed of several layers of thin-walled subhyaline to pale brown cells of textura prismatica. Hamathecium composed of 2–9.4 µm wide, cylindrical, cellular, unbranched, guttulate, septate paraphyses. Asci 25–40 × 6–9 μm (\( \bar{x} \) = 36 × 7.4 μm, n = 20), (6–)8-spored, unitunicate, cylindrical to cylindrical-clavate, with short furcate pedicel. Ascospores 10–16 × 2–5 µm (\( \bar{x} \) = 14 × 3 µm, n = 20), fusiform, curved towards both ends, 1-septate, constricted at septum, hyaline to subhyaline, guttulate, with mucilaginous sheath. Asexual morph Hyphomycetous. Conidiophores aggregated into sporodochia or synnemata, without hyaline setae around the margin of conidiomata. Conidiogenous cells 7–15 × 1–2.5 μm (\( \bar{x} \) = 12 × 2 μm, n = 20), monophialidic, hyaline, subulate. Conidia 3–8 × 1–3 μm (\( \bar{x} \) = 5 × 2 μm, n = 40), slimy, aseptate, hyaline, oblong, guttulate, without a mucilaginous sheath.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, filamentous, hyaline, smooth and flat on surface media.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-034 (MFLU 18-0014, holotype; HKAS 100839, isotype); ex-type living culture, MFLUCC 16-0366 = MFLUCC 16-0368 = KUMCC 16-0160.
GenBank numbers LSU: MH376742; ITS: MH388368; SSU: MH388332; TEF1: MH388402.
Notes: The sexual and asexual morphs of Volutella thailandensis fits well within the holomorphconcept for Volutella (Grafenhan et al. 2011; Seifert et al. 2011). In the phylogenetic tree (Fig. 74) based on the sequences of multi-gene, V. thailandensis clusters with V. krabiense, but there are more than 30 bp (5.83%) differences in 514 ITS (+5.8S) nucleotides between the two species and thus indicating they are distinct species. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0366 is Volutella sp. with 99% identity to the strain DoF21 (JQ388266).
Niessliaceae Kirschst.
Niessliaceae was introduced by Kirschstein (1939) with Niesslia Auersw. as the type genus. Members of this family are saprobic or parasitic on wood, leaves or lichens (Maharachchikumbura et al. 2016). The family was accepted in a narrow sense by Barr (1990b) and was separated from other families: Sphaeriaceae (Müller and von Arx 1962, 1973) and Trichosphaeriaceae (Barr 1983; Hawksworth et al. 1983). Eriksson and Hawksworth (1993), Hawksworth et al. (1995) and Samuels and Barr (1997) and accepted this family in the Hypocreales based on morphological features. Lumbsch and Huhndorf (2010) accepted 17 genera in the family and Jaklitsch and Voglmayr (2012) confirmed the placement of Niessliaceae in the Hypocreales by molecular data. An update for this family is provided in Maharachchikumbura et al. (2015, 2016) and Wijayawardene et al. (2018). We introduce a new monotypic genus in this family.
Pseudohyaloseta Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF554542, Facesoffungi number: FoF04549
Etymology: name refers to the characteristics similar to Hyaloseta.
Type species: Pseudohyaloseta pandanicola Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves or wood in terrestrial habitats. Sexual morph Ascomata scattered to gregarious, superficial or semi-immersed, flat at the base, conspicuous on host surface, pigmented, shiny, usually flattened when dry, solitary or in small groups, globose to subglobose, deflated at the middle, with hyaline papilla, ostiole. Peridium composed of several layers, thin-walled; outer layers comprising thin-walled, yellow–brown to brown cells of textura prismatica and inner layer of larger, thin-walled, lightly pigmented or hyaline cells of textura angularis. Hamathecium of filiform, filamentous, unbranched, guttulate, septate paraphyses. Asci 6–8-spored, unitunicate, obclavate, pedicellate, with J-, apical ring. Ascospores cylindrical, curved towards both ends, 1-septate, slightly constricted at septum, hyaline to subhyaline, rough at the margin, guttulate, with a mucilaginous sheath. Asexual morph Undetermined.
Notes: The morphology of Pseudohyaloseta is similar to that of Hyaloseta. Based on phylogenic analysis, Pseudohyaloseta is well-separated from Hyaloseta (100% in ML, Fig. 74). There are more than 30 bp (> 5.60%) differences in 535 ITS (+5.8S) nucleotides and 14 bp (1.70%) in 819 LSU nucleotides between Hyaloseta and Pseudohyaloseta. Therefore, Pseudohyaloseta is introduced here as a new genus in Niessliaceae based on phylogenetic and morphological evidences.
Pseudohyaloseta pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554543, Facesoffungi number: FoF04550; Fig. 84
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1882
Saprobic on dead roots of Freycinetia javanica. Sexual morph Ascomata 100–200 × 60–100 µm (\( \bar{x} \) = 139 × 80 µm, n = 5), scattered to gregarious, superficial, flat at the base, conspicuous on host surface, black, shiny, solitary, uniloculate, globose to subglobose, compressed at the middle, with a hyaline papilla, ostiole. Peridium 9–15 µm wide, composed of several layers; outer layers comprising thin-walled, yellow–brown to brown of textura prismatica and inner layer of larger, thin-walled, lightly pigmented or hyaline cells of textura angularis. Hamathecium with 1–3.8 µm wide, filiform, filamentous, unbranched, guttulate, septate paraphyses. Asci 45–60 × 9–16 μm (\( \bar{x} \) = 52 × 12 μm, n = 20), (6–)8-spored, unitunicate, obclavate, short-pedicellate, with J- apical ring. Ascospores 9–17 × 3–4 µm (\( \bar{x} \) = 15 × 3.5 µm, n = 40), cylindrical, curved towards both ends, 1-septate, slightly constricted at septum, hyaline to subhyaline, rough at the margin, guttulate, with mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, dark green to dark on the surface, dense, circular, with entire edge, raised, dark brown from below, with smooth margin.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Freycinetia javanica Blume., 17 December 2015, S. Tibpromma KB004 (MFLU 16-1882, holotype; HKAS 96233, isotype); ex-type living culture, MFLUCC 16-0316; Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on Pandanus sp., 16 December 2017, S. Tibpromma P09 (MFLU 18-0028, paratype).
GenBank numbers LSU: MH376737; ITS: MH388363; TEF1: MH388398; RPB2: MH412733.
Notes: Pseudohyaloseta pandanicola is similar to Hyaloseta nolinae A.W. Ramaley, but can be differentiated from H. nolinae has 28–48 × 3.2–4.8 μm, oblong asci, and 5.6–7.2(9.6) × 2.4 μm, oblong-ellipsoid ascospores (Ramaley 2001). In the phylogenetic analysis, based on the multi-gene sequence analyses, P. pandanicola formed a separate branch from H. nolinae.
Stachybotryaceae L. Lombard & Crous
Stachybotryaceae was introduced by Crous et al. (2014) to accommodate Myrothecium, Peethambara and Stachybotrys. Castlebury et al. (2004) and Summerbell et al. (2011) showed those three genera form a monophyletic lineage, distinct from other families in the Hypocreales. There are 33 genera in the family (Wijayawardene et al. 2018). We collected taxa of Stachybotriaceae from Pandanaceae and confirm their identity in the order. In this paper, we introduce one new genus, Pseudoornatispora and a new species of Parasarcopodium and Sirastachys with an updated phylogeny (Fig. 74).
Parasarcopodium Mel’nik et al.
Parasarcopodium is a hyphomycetous genus, which was erected by Mel’nik et al. (2004) with P. ceratocaryi Mel’nik, S.J. Lee & Crous as the type species. Parasarcopodium was earlier placed in family Bionectriaceae (Mel’nik et al. 2004), but recently included in Stachybotryaceae (Maharachchikumbura et al. 2015). At present there are two epithets are listed in Index Fungorum (2018). Parasarcopodium pandanicola Tibpromma & K.D. Hyde has been previously recorded from Pandanaceae (Tibpromma et al. 2016a).
Parasarcopodium hongkongensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554546, Facesoffungi number: FoF04553; Fig. 85
Etymology: named after Hong Kong, where the fungus was first discovered.
Holotype: HKAS 100859
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiomata sporodochia superficial on host surface scattered, solitary, orange, with long papilla. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–8.5 × 2–3 μm (\( \bar{x} \) = 7.5 × 2.5 μm, n = 10), simple, smooth, enteroblastic, phialidic, ampulliform, hyaline, smooth-walled. Conidia 12–18 × 1.5–3 μm (\( \bar{x} \) = 15 × 2 μm, n = 40), hyaline, filiform, straight to slightly curved, aseptate, smooth, thick-walled, distinctly guttulate, with an appendage at the base.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, circular, edge entire, white with brown, with dense, aerial mycelium on surface, rough with wrinkled, with small-dots, black; reverse of culture brown to black.
Material examined: HONG KONG, Tai Tam Tuk Reservoir, on Pandanus sp., 21 September 2016, S. Tibpromma HK02 (HKAS 100859, holotype); ex-type living culture, KUMCC 17-0267 = MFLUCC 17-0632; THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB034 (MFLU 16-1912, paratype).
GenBank numbers LSU: MH376733; ITS: MH388359; SSU: MH388327; TEF1: MH388394.
Notes: Parasarcopodium hongkongensis has hyaline, filiform, straight to slightly curved, aseptate, basally appendaged conidia which are similar to the conidia of P. pandanicola Tibpromma & K.D. Hyde. However, they differ in size of conidiogenous cells and conidia. Parasarcopodium hongkongensis has conidiogenous cells 6–8.5 × 2–3 μm with conidia 12–18 × 1.5–3 μm, while P. pandanicola has conidiogenous cells 5–14 × 3–4 μm with conidia 10–20 × 2–3 μm (Tibpromma et al. 2016a). Comparing SSU nucleotides of P. hongkongensis and P. pandanicola we found 29 bp (2.68%) differences in 1080 SSU nucleotides. Based on phylogeny P. hongkongensis is distinct from other species of Parasarcopodium with high support (98% in ML, Fig. 74). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0267 is P. ceratocaryi with 92% identity to the strain CBS 110664 (NR_154283).
Pseudoornatispora Tibpromma & K.D. Hyde, gen. nov.
Index Fungorum number: IF555336, Facesoffungi number: FoF04551
Etymology: name refers to the characteristics similar to Ornatispora.
Type species: Pseudoornatispora krabiense Tibpromma & K.D. Hyde
Saprobic on dead or decaying leaves, wood in terrestrial habitats. Sexual morph Ascomata scattered or gregarious, superficial or semi-immersed with flat base, conspicuous on host surface, back, shiny, globose to subglobose, covered by dark brown, papilla, with or without ostiole, with hyaline setae-like periphyses. Peridium composed of several layers; outer layers comprising thick-walled, pale brown to brown cells of textura angularis; inner layers of larger, thin-walled, lightly pigmented or hyaline cells of textura angularis. Hamathecium with filiform, cellular, branched or unbranched, guttulate, aseptate paraphyses. Asci 6–8-spored, unitunicate, cylindrical-clavate, with club shape pedicel, J- apical ring. Ascospores fusiform, curved towards both end, 1-septate, constricted at septum, hyaline to subhyaline, rough at the margin, guttulate, with mucilaginous sheath. Asexual morph Hyphomycetous. Conidiophores aggregated in dense fascicles. Conidiogenous cells holoblastic, phialidic, discrete, cylindrical, hyaline, conical at apex. Conidia ellipsoid, hyaline to subhyaline, tapering to conical apex, aseptate, guttules, without mucilaginous sheath, smooth-walled.
Notes: Pseudoornatispora is introduced as a monotypic genus to accommodate P. krabiense based on morphology (sexual and asexual morphs) and phylogenetic analysis. The sexual morph Pseudoornatispora is similar to Ornatispora (≡ Stachybotrys) K.D. Hyde, Goh, Joanne E. Taylor, J. Fröhl. in ascomata and ascospores characters, which are black ascomata with seta and ostioles, and hyaline, 1-septate and verrucose ascospores (Hyde et al. 1999), but this genus has never been sequenced and is probably polyphyletic. The asexual morph of Pseudoornatispora is morphologically similar to Peethambara Subram. & Bhat, but these two genera are phylogenetically apart in multi-gene phylogenetic analysis. Fresh collections are needed.
Pseudoornatispora krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF555337, Facesoffungi number: FoF04552; Figs. 86, 87
Pseudoornatispora krabiense (MFLU 16-1892, holotype). a, b Appearance of ascomata on host substrate. c Ascoma. d Section of ascoma. e Section of peridium. f Ostiole. g Papilla. h–i Asci. j Paraphyses. k–n Ascospores. o Germinating ascospore. p, q Colonies on MEA from above and below. Scale bars: a, b = 100 μm, c, d = 50 μm, e, g–i = 10 μm, f = 20 μm, j–o = 5 μm
Pseudoornatispora krabiense (MFLUCC 16-0318). a Colonies on dead leaves of Pandanus sp. b, c Conidiogenous cells and conidia. d Conidiogenous cells. e–g Conidia. h Conidium strained in cotton blue reagent. i Germinating conidium. j, k Colonies on MEA from above and below. Scale bars: a = 200 μm, b, c = 20 μm, d–i = 5 μm
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1892
Saprobic on dead leaves of Pandanus sp. Sexual morph Ascomata 150–200 × 165–180 µm (\( \bar{x} \) = 172 × 172.5 µm, n = 5), scattered to gregarious, superficial, flat on the base, conspicuous on host surface, back, shiny, solitary, uniloculate, globose to subglobose, covered by dark septate setae, papillate, centrally located ostiole with pore-like opening and with hyaline periphyses. Peridium 20–30 µm wide, composed of several layers; outer layers comprising thick-walled, pale brown to brown of textura angularis and inner layers of larger, thin-walled, lightly pigmented or hyaline cells of textura angularis. Hamathecium composed of 4–5 µm wide, filiform, cellular, unbranched, guttulate, aseptate paraphyses. Asci 70–130 × 15–20 μm (\( \bar{x} \) = 89.5 × 17 μm, n = 10), (6–)8-spored, unitunicate, cylindrical-clavate, with club-shaped pedicel, with J- apical ring. Ascospores 26–43 × 5–17 µm (\( \bar{x} \) = 34.5 × 9.5 µm, n = 30), fusiform, curved towards both ends, 1-septate, constricted at septum, hyaline to subhyaline, rough at the margin, guttulate, with mucilaginous sheath. Asexual morph Hyphomycetous. Conidiophores aggregated in dense fascicles, sporodochial. Conidiogenous cells 9–15 × 3–4 µm (\( \bar{x} \) = 11 × 3.5 µm, n = 10), holoblastic, phialidic, discrete, cylindrical, smooth, hyaline, with conical apex. Conidia 10–17 × 4–6 μm (\( \bar{x} \) = 14.6 × 5 μm, n = 30), ellipsoid, solitary, hyaline, tapering to conical apex and base, aseptate, smooth, guttulate, slimy, without a mucilaginous sheath, smooth-walled.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA, orange pink, with irregular, curled, flat and dry on media surface, orange pink in reverse.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 15 December 2015, S. Tibpromma KB014 (MFLU 16-1892, holotype; HKAS 96242, isotype); ex-type living culture, MFLUCC 16-0317; Krabi Province, Mueang Krabi District, on Pandanus sp., 15 December 2015, S. Tibpromma KB015 (MFLU 16-1893; HKAS 96243, paratype); living culture, MFLUCC 16-0318 = KUMCC 16-0139.
GenBank numbers MFLUCC 16-0317 LSU: MH376721; ITS: MH388348; SSU: MH388315; TEF1: MH388383; RPB2: MH412729. MFLUCC 16-0318 LSU: MH376722; ITS: MH388349; SSU: MH388316; TEF1: MH388384; RPB2: MH412730.
Notes: The sexual morph of Pseudoornatispora krabiense is similar to Stachybotrys which has superficial, globose ascomata covered with numerous setae, clavate, pedicellate asci, and ellipsoidal, 1-septate and verrucose ascospores (Corda 1837; Wang et al. 2015). However, P. krabiense differs as ascospores are rough at the margin and guttulate and have a mucilaginous sheath. The sexual morph of Pseudoornatispora krabiense is similar to Stachybotrys in having superficial, globose ascomata, covered with numerous setae, clavate, pedicellate asci, and ellipsoidal, 1-septate and verrucose ascospores (Corda 1837). Pseudoornatispora krabiense is similar to Ornatispora novae-zelandiae Whitton, K.D. Hyde & McKenzie, but O. novae-zelandiae has narrowly clavate, 8-spored asci, with an apical refractive ring and discoid, narrowly ellipsoid to ellipsoid or fusoid and verrucose ascospores (Whitton et al. 2012), while P. krabiense differs in having cylindrical-clavate, (6–)8-spored asci, with a club-shaped pedicel and J- apical ring, and fusiform ascospores, which are rough at the margin, guttulate, and with mucilaginous sheath.
Sirastachys L. Lombard & Crous
Sirastachys was introduced by Lombard et al. (2016) to accommodate S. phaeospora L. Lombard & Crous. Sirastachys is a stachybotrys-like fungus that forms synnemata in culture. Phylogenetic analyses showed that Sirastachys forms a well-supported clade distantly related to Stachybotrys (Lombard et al. 2016). At present, there are eight epithets are listed in Index Fungorum (2018). Sirastachys pandanicola L. Lombard & Crous has been reported from Singapore on a decaying leaf of Pandanus sp. (Lombard et al. 2016).
Sirastachys phangngaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554549, Facesoffungi number: FoF04556; Fig. 88
Etymology: named after Phang Nga Province, where the fungus was first discovered.
Holotype: MFLU 16-0544
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium composed of septate, branched, smooth, subhyaline, hyphae. Conidiophores 35–46 × 2–3 µm (\( \bar{x} \) = 42.6 × 2.4 µm, n = 10), wider at the base, determinate, macronematous, solitary or in groups, erect, unbranched, septate, hyaline, smooth-walled. Conidiogenous cells 4–7 × 1.5–2 µm (\( \bar{x} \) = 5.6 × 1.7 µm, n = 10), phialidic, ellipsoid, discrete, light pink, often with conspicuous collarettes and producing conidia singly or successively in basipetal succession. Conidia 3.5–4 × 1.5–2 µm (\( \bar{x} \) = 3.6 × 1.7 µm, n = 30), blastic-phialidic, ovoid to ellipsoidal, aseptate, guttulate, reddish–brown to green–brown, rough-walled, slimy (Fig. 89).
Phylogram generated from maximum likelihood analysis of ITS sequenced data. Related sequences were obtained from Lackner and de Hoog (2011). Fourty-nine strains are included in the combined sequence analysis, which comprise 589 characters with gaps. Microascus rigonosporus var. rigonosporuss (CBS 665.71) is used as the outgroup taxon. The best scoring RAxML tree with a final likelihood value of − 3685.984786 is presented. The matrix had 330 distinct alignment patterns, with 16.30% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.227200, C = 0.292880, G = 0.246760, T = 0.233160; substitution rates AC = 1.819950, AG = 3.705991, AT = 2.078108, CG = 1.765696, CT = 5.084148, GT = 1.000000; gamma distribution shape parameter α = 0.382819. Bootstrap support values for ML equal to or greater than 60% is given above the nodes. The newly generated sequence is in red
Culture characteristics: on MEA reaching 9 cm diam., after 2 weeks at room temperature, circular with filiform, flat on media surface, cream with rough and wrinkled.
Material examined: THAILAND, Phang Nga Province, Mueang Phang Nga District, on dead leaf of Pandanus sp., 6 December 2014, S. Tibpromma & K.D. Hyde SF14-039 (MFLU 16-0544, holotype; HKAS 100832, isotype); ex-type living culture, MFLUCC 15-0680.
GenBank numbers LSU: MH376739; ITS: MH388365; TEF1: MH388400; RPB2: MH412735.
Notes: In blast homology searches of the ITS sequences, Sirastachys phangngaensis matches with S. phaeospora L. Lombard & Crous (CBS 100155) with 98% identity. Sirastachys phaeospora has hyaline to pale olivaceous brown to dark brown conidia, smooth to verrucose, ellipsoidal to obovoid to cylindrical (Lombard et al. 2016). We also compared Sirastachys phangngaensis with S. pandanicola morphologically. They have similar conidiogenous cells size 6–9 × 2–4 μm with conidia 3–4 × 2–3 μm, obovoid to ellipsoidal, darkly olivaceous (Lombard et al. 2016). A synopsis of Sirastachys species are shown in Table 4. Sirastachys can form synnemata in culture but Sirastachys phangngaensis does not show synnematous conidiophores habit in culture. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 15-0680 is S. phaeospora with 98% identity to the strain CBS 136167 (KU846667).
Microascales Luttr. ex Benny & Kimbr.
Microascaceae Luttr. ex Malloch
Microascaceae was proposed by Luttrell (1951) with validly published by Latin descriptions by Malloch (1970) and Benny and Kimbrough (1980). The sexual and asexual characteristics were reviewed by Malloch (1970). The taxonomic placement of Microascaceae using molecular data is shown by Réblová et al. (2011) and Maharachchikumbura et al. (2015). There are 21 genera in the family (Wijayawardene et al. 2018). We collected Parascedosporium putredinis from Pandanaceae in Thailand and provided new molecular data for this species.
Parascedosporium Gilgado et al.
Parascedosporium was introduced by Gilgado et al. (2007) to accommodate P. tectonae (C. Booth) Gilgado, Gené, Cano & Guarro. Parascedosporium shows morphology similar to Scedosporium (Gilgado et al. 2007). Later, Parascedosporium was segregated from Scedosporium and its synnematous synanamorph Graphium on the basis of morphology (Lackner and de Hoog 2011). The sexual morph of this genus is unknown (Seifert et al. 2011). Parascedosporium has twelve epithets are listed in Index Fungorum (2018) and this is the first report of Parascedosporium from Pandanaceae.
Parascedosporium putredinis (Corda) Lackner and de Hoog, IMA Fungus 2 (1): 44 (2011)
Facesoffungi number: FoF04557; Fig. 90
Parascedosporium putredinis (MFLU 16-0545). a, b Synnemata on host substrate. c Conidiogenous cells with conidiophores and conidia. d Close up conidiogenous cells with conidia. e Conidiophores. f, g Conidiogenous cells with conidia. h–j Conidia. k Germinating conidium. Scale bars: a, b = 200 μm, c = 100 μm, d = 10 μm, e, f–g = 2 μm, h, i, k = 1 μm, j = 5 μm
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Synnemata 540–600 × 7.5–15 μm (\( \bar{x} \) = 587 × 8.5 μm, n = 5), scattered but abundant, arising from the host plant. Stipes pale to dark brown, (68–)88–140(–151) × (23–)60–97 µm. Conidiophores 450–500 × 9–13 μm (\( \bar{x} \) = 469.6 × 11 μm, n = 5), with multi-branches. Conidiogenous cells annellate, brown, septate. Conidia 3–5 × 1–3 μm (\( \bar{x} \) = 4 × 2 μm, n = 40), aseptate, hyaline, cylindrical to obovoid, hyaline, produced in mucilaginous mass on synnemata.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA circular, entire edge with white to cream, raised on surface media. Sporulating in culture after 3 months which produce conidia similar in shape to those recorded on natural dead leaves.
Material examination: THAILAND, Krabi Province, Mueang Krabi District, on dead leaf of Pandanus sp., 8 December 2014, S. Tibpromma & KD Hyde SF14-044 (MFLU 16-0545, HKAS 100834); living culture, MFLUCC 16-0373 = KUMCC 16-0157.
GenBank numbers LSU: MH260311; ITS: MH275077; SSU: MH260351.
Notes: Parascedosporium putredinis (MFLUCC 16-0373) from dead leaf of Pandanus sp. showed morphological similarities to P. putredinis (CBS 108.10) (Lackner and de Hoog 2011). Phylogenetic analysis confirmed our collection is P. putredinis.
Subclass Savoryellomycetidae Hongsanan et al.
Savoryellales Boonyuen et al.
Savoryellaceae Jaklitsch & Réblová
Savoryellaceae (Halosphaeriales) was invalidly erected by Ranghoo (1998) in her PhD thesis, then Jaklitsch and Réblová (2015) formally introduced Savoryellaceae (Savoryellales) with Savoryella E.B.G. Jones & R.A. Eaton as the type genus. There are six genera in the family (Wijayawardene et al. 2018). We describe two new species of Canalisporium and provide an updated tree for the family (Fig. 91).
Phylogram generated from maximum likelihood analysis of combined ITS, LSU and SSU sequence data. Related sequences were obtained from Sri-indrasutdhi et al. (2010a, b) and Boonyuen et al. (2011). Thirty-three strains are included in the combined sequence analysis, which comprise 2936 characters with gaps. Niesslia exilis (CBS560.74) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 16595.878216 is presented. The matrix had 1187 distinct alignment patterns, with 19.20% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.231716, C = 0.252553, G = 0.301318, T = 0.214412; substitution rates AC = 1.494718, AG = 2.678433, AT = 1.590940, CG = 1.066185, CT = 5.489636, GT = 1.000000; gamma distribution shape parameter α = 0.258630. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in red
Canalisporium Nawawi & Kuthub.
Canalisporium was introduced with C. caribense as the type species (Nawawi and Kuthubutheen 1989). Twelve epithets are listed in Index Fungorum (2018), but only seven species have sequences available in GenBank. Three species of Canalisporium have been found previously on Pandanaceae. Canalisporium caribense var. caribense (Hol.-Jech. & Mercado) Nawawi & Kuthub. and C. elegans Nawawi & Kuthub. were recorded on Freycinetia and Pandanus species (McKenzie and Hyde 1997; Goh et al. 1998; Whitton et al. 2012), while C. exiguum Goh & K.D. Hyde was found on Pandanus penetrans in Thailand (Thongkantha et al. 2008). Whitton et al. (2012) provided a key to the genus and a synoptic table.
Canalisporium krabiense Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554550, Facesoffungi number: FoF04558; Fig. 92
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1888
Conidiomata sporodochia on natural substrate scattered on leaf sheath of Pandanus sp. in small group, punctiform, pulvinate, granular, black, velvety. Mycelium immersed in the substrate, composed of branched, septate, smooth, hyaline hyphae. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores 2–20 × 3–6 μm (\( \bar{x} \) = 10 × 5 μm, n = 20), micronematous or semi-macronematous, mononematous, fasciculate, aseptate, hyaline, smooth. Conidiogenous cells holoblastic, integrated, terminal, determinate. Conidia 27–50 × 22–32 μm (\( \bar{x} \) = 40 × 27.4 μm, n = 40), acrogenous, solitary, broadly ellipsoidal to obovoid in surface view, fusiform to obclavate in lateral view, flattened, one-cell thick, muriform with 8–12 cells, smooth, olivaceous brown to brown, with 1 straight vertical septum and 4–6 rows of transverse septa, slightly constricted at septa, dark and thickly banded at septa, canals in the septa obscured by dark pigmentation in face view, visible in side view; basal cell subhyaline to pale brown, cuneiform, thin-walled. Conidial secession rhexolytic.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on leaf sheath of Pandanus sp., 16 December 2017, S. Tibpromma KB010 (MFLU 16-1888, holotype; HKAS 96238, isotype).
GenBank numbers LSU: MH260283; ITS: MH275051.
Notes: Canalisporium krabiense clustered with C. thailandensis and C. exiguum. Canalisporium krabiense differs from C. exiguum in conidial size, number of rows of cells and colour. Conidia of C. krabiense are 27–50 × 22–32 μm, with 4–6 rows of cells, while those of C. exiguum are 18–25 × 13–15 × 5–8 µm, with 2–3(–4) rows of cells (Goh et al. 1998). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLU 16-1888 is C. exiguum with 99% identity to the strain SS00809 (GQ390296).
Canalisporium thailandensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554551, Facesoffungi number: FoF04559; Fig. 93
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 16-1900
Saprobic on dead leaf sheath of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, in small groups, blackish brown, velvety, glistening, conidia readily liberated when disturbed. Mycelium immersed in the substrate, composed of branched, septate, smooth, hyaline hyphae. Conidiophores 39–60 × 13–17.5 μm (\( \bar{x} \) = 51.5 × 15 μm, n = 20), mononematous, fasciculate, septate, deeply constricted at septa, hyaline, smooth. Conidiogenous cells 11–13 × 9–13 μm (\( \bar{x} \) = 12 × 11 μm, n = 20), holoblastic, monoblastic, integrated, terminal, determinate, globose to oval connected in a chain, with guttules, hyaline. Conidia 22.5–31 × 17–22 μm (\( \bar{x} \) = 28 × 19 μm, n = 20), acrogenous, solitary, broadly ellipsoidal to obovoid, flattened, one-cell thick, muriform with 8–10 cells, smooth, yellow–brown to brown, with 1 straight column of vertical septa and 4–5 rows of cells, slightly constricted at septa, dark and thickly banded at septa, septa obscured by dark pigmentation in face view and visible in side view; basal cell subhyaline to pale brown, cuneiform, thin-walled.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on dead leaf sheath of Pandanus sp., 16 December 2017, S. Tibpromma KB022 (MFLU 16-1900, holotype; HKAS 96249, isotype).
GenBank numbers LSU: MH260284; ITS: MH275052.
Notes: Canalisporium thailandensis differs from other Canalisporium species by globose to oval conidiogenous cells connected in a chain, which is a unique character for Canalisporium (Sri-indrasutdhi et al. 2010a, b). Phylogenetic analysis also supported this as a distinct new species with high bootstrap support (100% in ML, 1.00 in BYPP, Fig. 91). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLU MFLU 16-1900 is C. exiguum with 99% identity to the strain SS00809 (GQ390296).
Subclass Sordariomycetidae O.E. Erikss & Winka (= Meliolomycetidae P.M. Kirk & K.D. Hyde)
Chaetosphaeriales Huhndorf et al.
Chaetosphaeriaceae Réblová et al.
Chaetosphaeriaceae was erected by Réblová et al. (1999) with Chaetosphaeria Tul. & C. Tul. as the type genus (Tulasne and Tulasne 1863). Several authors have carried out morphological studies of the genus (Munk 1953; Booth 1957; Müller and von Arx 1962; Saccardo 1883; Samuels et al. 1997; Réblová et al. 1999). Maharachchikumbura et al. (2016) listed 37 genera in this family and mentioned that the limits of the family are confused and more work with molecular support is needed to resolve the accepted genera. We introduce new species of Chaetosphaeriaceae in the genera Dictyochaeta, Menisporopsis and Thozetella, which were collected on Pandanaceae from Thailand and China.
Dictyochaeta Speg.
Dictyochaeta was erected by Spegazzini (1923) with D. fuegiana Speg. as type species. It is characterised by conidia that are hyaline, smooth, typically falcate but can be ellipsoidal, clavate, fusoid, or cylindrical, 0–1(–3)-septate, and often with setulae (Gamundi et al. 1977; Kuthubutheen and Nawawi 1991a, b, c, d). Index Fungorum (2018) listed 101 epithets for Dictyochaeta. Several species (Dictyochaeta fertilis, D. parva, D. renispora and D. simplex) have been recorded on Pandanaceae (Whitton et al. 2012). An updated phylogenetic tree for the family Dictyochaetaceae is presented (Fig. 94).
Phylogram generated from maximum likelihood analysis based on combined LSU, and ITS partial sequence data. Eighty-nine strains are included in the sequence analysis, which comprise 1803 characters with gaps. Single gene analysis was carried out and compared with each species, to compare the topology of the tree and clade stability. Sordaria fimicola (Sordariaceae) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 18027.584016 is presented. The matrix had 880 distinct alignment patterns, with 27.96% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.233040, C = 0.260279, G = 0.300714, T = 0.205967; substitution rates AC = 1.320651, AG = 2.076856, AT = 1.962948, CG = 0.818781, CT = 6.443592, GT = 1.000000; gamma distribution shape parameter α = 0.238273. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in red
Dictyochaeta pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554520, Facesoffungi number: FoF04530; Fig. 95
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 101807
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium partly superficial composed of septate, branched, smooth, hyaline hyphae. Conidiophores 65–260 × 5–10 µm (\( \bar{x} \) = 142.5 × 7 µm, n = 10), erect, straight or slightly flexuous, in groups of 4–6 arising from common basal stroma, brown to dark brown at base, hyaline to pale brown towards apex, septate, smooth. Conidiogenous cells 15–33 × 6–8 µm (\( \bar{x} \) = 25.5 × 7 µm, n = 20), mono- or polyphialidic, terminal, integrated, cylindrical, hyaline to pale brown, smooth, with conspicuous, flared collarettes. Conidia 16–26 × 4–6 µm (\( \bar{x} \) = 23 × 4.4 µm, n = 20), allantoid, conical at ends, hyaline, guttulate, with one setula at each end, 8–13.5 μm long.
Culture characteristics: Colonies on PDA attaining 9 cm diam., within 2 weeks at room temperature, grey to dark grey, irregular, undulate edge with curled on media surface, raised on media.
Material examined: CHINA, Yunnan Province, Xishuangbanna, Nabanhe, on fallen dead and decaying leaves of Pandanus sp., 2 August 2016, S. Tibpromma NBH22 (HKAS 101807, holotype); ex-type living culture, KUMCC 16-0153 = MFLUCC 17-0563.
GenBank numbers LSU: MH376710; ITS: MH388338; SSU: MH388307; TEF1: MH388373.
Notes: In our phylogenetic analysis, Dictyochaeta pandanicola is well-separated from other Dictyochaeta species with high bootstrap support (100% in ML, 1 in BYPP, Fig. 94). Morphologically, D. pandanicola is similar to D. siamensis J. Yang, K.D. Hyde & J.K. Liu. but differs in D. siamensis has setae, 4–6 septate conidiophores with 17.5 × 3 μm conidia, aggregated in slimy mass at the apex of the conidiophore, with 7–12 μm long setulae (Liu et al. 2016). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 16-0153 is D. siamensis with 99% identity to the strain MFLU 15-1149 (NR_154016).
Dictyochaeta siamensis J. Yang, K.D. Hyde & J.K. Liu, Mycological Progress 15 (10): 1159 (2016)
Facesoffungi number: FoF04531; Fig. 96
Dictyochaeta siamensis (MFLU 16-0553). a, b Colonies on dead leaves of Pandanus sp. c–f Conidiogenous cells with conidiophores. d Setae-walled. e–i Conidiogenous cells with conidiophores and conidia. j–n Conidia. o Germinating conidium. Scale bars: a, b = 200 μm, c = 100 μm, d = 2 μm, e–i = 10 μm, j–o = 5 μm
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium partly superficial, composed of septate, branched, smooth, hyaline hyphae. Setae up to 400 µm long, erect, straight or slightly flexuous, simple, smooth, septate, brown to dark brown, terminating with an acute tip, unbranched. Conidiophores 43.5–87 × 2.5–6 µm (\( \bar{x} \) = 67 × 4 µm, n = 10), erect, straight or slightly flexuous, in groups of 4–6 arising from common basal stroma, dark brown at base, pale brown towards apex, 4–6 septate, unbranched. Conidiogenous cells mono- or polyphialidic, terminal, integrated, cylindrical, smooth, with conspicuous, flared collarettes. Conidia 8–17 × 2–5 µm (\( \bar{x} \) = 13 × 3 µm, n = 20), allantoid, cylindrical or long fusiform, conical at both ends, hyaline, aseptate, guttulate, with one setula at each end 1–10 μm long.
Culture characteristics: Colonies on MEA attaining 9 cm diam., within 2 weeks at room temperature, dark green, circular, entire edge with convex on media surface, flossy with velvety.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on dead leaf of Pandanus sp., 30 July 2015, S. Tibpromma & K.D Hyde SF15-022 (MFLU 16-0553; HKAS 96279); living culture, MFLUCC 16-0371.
GenBank numbers LSU: MH376711; ITS: MH388339; SSU: MH388308; TEF1: MH388374.
Notes: In the phylogenetic analysis our isolate grouped with Dictyochaeta siamensis with high bootstrap support (100% in ML, 1 in BYPP, Fig. 94). Our isolate shares similar morphology to D. siamensis (MFLU 15-1149) in septate, unbranched setae, 4–6-septate, unbranched conidiophores and hyaline, aseptate, cylindrical or long fusiform conidia, with setulae at both ends (Liu et al. 2016).
Menisporopsis S. Hughes
Menisporopsis was introduced with single species M. theobromae S. Hughes (Hughes 1952). Menisporopsis is characterized by synnematous conidiophores that surround a single sterile, erect seta with falcate conidia has one or more setulae at each end (Hughes 1952; Pirozynski and Hodges 1973; Varghese and Rao 1978; Rao and de Hoog 1986). There are ten epithets are listed in Index Fungorum (2018). Two species, M. pirozynskii Varghese & V.G. Rao and M. theobromae S. Hughes have been reported from Pandanaceae (Whitton et al. 2012).
Menisporopsis pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554522, Facesoffungi number: FoF04533; Fig. 97
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 100862
Saprobic on dead leaf of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Mycelium partly superficial, composed of septate, branched, smooth, hyaline hyphae. Setae 344–375 × 7–10.5 µm, erect, straight or slightly flexuous, simple, smooth, septate, brown to dark brown at the base, pale brown towards the apex. Conidiophores 85–100 × 14.5–23 µm (\( \bar{x} \) = 93 × 19 µm, n = 5), synnematous encircling the setae, erect, straight or slightly flexuous, pale brown, septate, smooth. Conidiogenous cells 8–29 × 0.5–2 µm (\( \bar{x} \) = 17 × 1.5 µm, n = 20), polyphialidic, terminal, integrated, cylindrical, smooth. Conidia 17–22 × 2–3 µm (\( \bar{x} \) = 19 × 2.5 µm, n = 40), lunate, conical at both ends, aseptate, guttulate, aggregated into a slimy mass, hyaline, with (1–)2 setula at each end, 4–12 µm long.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies growing show on MEA, circular, entire edge with white to cream, raised on surface media.
Material examined: HONG KONG, Lantau Island, Pui O Beach, on Pandanus sp., 20 September 2016, S. Tibpromma HK011 (HKAS 100862, holotype); ex-type living culture, KUMCC 17-0271 = MFLUCC 17-0638.
GenBank numbers LSU: MH376726; ITS: MH388353; SSU: MH388320; TEF1: MH388388; RPB2: MH412732.
Notes: In the phylogenic tree Menisporopsis pandanicola clustered with M. theobromae (MFLU 15-1168) (100% in ML, 1 in BYPP). Menisporopsis pandanicola has conidia 19 × 3 µm, with one or two setulae at each end measuring 4–12 µm long, while M. theobromae has conidia 17 × 2.5 μm, setula with a single setula at each end measuring 6–7.5 μm long (Liu et al. 2016). We also compared ITS and LSU nucleotides and found that they are different 20 bp (4.08%) in 490 ITS (+5.8S) nucleotides and 10 bp (1.23%) in 808 LSU nucleotides. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0271 is M. theobromae with 95% identity to the strain MFLUCC 15-0055 (KX609957).
Thozetella Kuntze
Thozetella was introduced with T. nivea (Kuntze 1891) as the type species. The Thozetella is characterised by conidiophores that are grouped into short conspicuous sporodochia or short synnemata and terminated by phialidic conidiogenous cells. There are 22 epithets are listed in Index Fungorum (2018). Whitton et al. (2012) provided a synoptic table to 17 species of Thozetella. Thozetella serrata Whitton, McKenzie & K.D. Hyde has been reported from Pandanaceae (Whitton et al. 2012).
Thozetella pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554521, Facesoffungi number: FoF04532; Fig. 98
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 16-1898
Saprobic on dead leaves of Pandanus sp. Mycelium superficial, white to yellow–white. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–6 × 2–3.5 µm (\( \bar{x} \) = 4.7 × 2.4 µm, n = 10), enteroblastic, phialidic, doliiform, smooth, hyaline. Conidia 17–21 × 2–3 μm (\( \bar{x} \) = 19 × 2.4 μm, n = 30, fusiform, hyaline, acute apex, aseptate, smooth, guttulate, with flexuous appendage at each end 6–9 μm long, without a mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, entire with curled, white to cream, smooth and raised on surface media. Mycelium superficial, flossy.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 16 December 2017, S. Tibpromma KB020 (MFLU 16-1898, holotype; HKAS 96247, isotype); ex-type living culture, MFLUCC 16-0253.
GenBank numbers LSU: MH376740; ITS: MH388366; SSU: MH388330.
Notes: Thozetella serrata has been recorded on decaying leaves of Pandanus furcatus in Hong Kong. Thozetella serrata has fusoid almost straight conidia, 11.5–17 × 2–3 μm (Whitton et al. 2012), while T. pandanicola has fusiform conidia, 17–21 × 2–3 μm. The phylogeny showed T. pandanicola well-separated from other species in Thozetella (0.98 in BYPP, Fig. 94). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0253 is Thozetella sp. with 96% identity to the strain C20-79 (KJ439080).
Sordariales Chadef. ex D. Hawksw. & O.E. Erikss.
Chaetomiaceae G. Winter
Chaetomiaceae was erected by Winter (1885a, b) (as Chaetomiea) with Chaetomium Kunze. as the type genus. Members of this family are ubiquitous, commonly residing in soil, on lignin and cellulosic or similar materials and some species are also human opportunistic pathogens (von Arx et al. 1986; Mukerji and Manoharachary 2010; Ahmed et al. 2015b). The placement of this family is not clear and it has been placed in various orders such as Chaetomiales (Ames 1961; Alexopoulos 1962; Mukerji 1968), Sphaeriales (Barr 1976; Müller and von Arx 1973), and later Sordariales (Hawksworth and Wells 1973; Mehrotra and Aneja 1990). Huhndorf et al. (2004), Liu (2011) and Maharachchikumbura et al. (2015) used molecular data to resolve the relationship of Chaetomiaceae in the order Sordariales and there are twenty-six genera in the family (Wijayawardene et al. 2018). We collected the type species, Chaetomium globosum from Pandanus sp. in Thailand and provide illustrations and a phylogenic analysis (Fig. 99).
Phylogram generated from maximum likelihood analysis of combined ITS and LSU sequenced data. Related sequences were obtained from Wang et al. (2016). Thirty-two strains are included in the sequence analysis, which comprise 1113 characters with gaps. Achaetomium strumarium (CBS 333.67) is used as the outgroup taxon. The best scoring RAxML tree with a final likelihood value of − 2044.666920 is presented. The matrix had 68 distinct alignment patterns, with 28.05% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.228029, C = 0.270995, G = 0.282117, T = 0.218859; substitution rates AC = 3.702498, AG = 4.553243, AT = 4.549924, CG = 8.172223, CT = 30.986343, GT = 1.000000; gamma distribution shape parameter α = 0.020000. Bootstrap support values for ML equal to or greater than 65% is given above the nodes. The newly generated sequence is in red
Chaetomium Kunze
Chaetomium globosum Kunze is the type species of Chaetomium. The special characteristic of Chaetomium is its ascoma covered with hairs or setae (Hawksworth and Wells 1973). Many Chaetomium species are potential biological control agents and can produce bioactive metabolites (Soytong et al. 2001; Pieckova 2003; Wang et al. 2012; Li et al. 2014). Currently, there are 435 epithets are listed in Index Fungorum (2018). We report Chaetomium globosum from Pandanaceae.
Chaetomium globosum Kunze ex Fr., Systema Mycologicum 3: 255 (1829)
Facesoffungi number: FoF04560; Fig. 100
Chaetomium globosum (MFLU 18-0015). a Colony on dead leaf of Pandanus sp. b Ascomata. c Upper part of terminal ascomatal hairs. d Structure of ascomatal wall in surface view. e Basal part of terminal ascomatal hair. f, g Asci. h–k Ascospores. l Germinating ascospore. m Colony on MEA. n, o Asci formed in culture (stained with congo red reagent). Scale bars: b = 100 μm, c, f, n = 50 μm, d, g, o = 10 μm, e, h–l = 5 μm
Saprobic on dead leaf of Pandanus sp. with immersed perithecia and later opening by a pore with white-pruinose margin on host surface. Sexual morph Ascomata 600–710 × 425–480 µm (\( \bar{x} \) = 666 × 460.5 µm, n = 5), superficial to semi-immersed with thick aerial hyphae or exposed, ostiolate, covered with black ascomatal hairs, oval or ellipsoid, without papilla. Ascomatal wall brown to black, composed of hypha-like cells, textura intricata in surface view. Terminal hairs 3–7 μm, finely punctate to verrucose, pale brown, hypha-like, flexuous or undulate, sometimes geniculate. Asci 16–29 × 4–6 µm (\( \bar{x} \) = 21 × 5 µm, n = 10), 8-spored, cylindrical to cylindric-clavate, long-pedicel, apically rounded. Ascospores 4–5 × 2–4 µm (\( \bar{x} \) = 4.5 × 3 µm, n = 40), overlapping, globose to subglobose, hyaline, becoming brown with age, aseptate, rough, not surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospore germinating on MEA within 12 h. Colonies on MEA, circular, undulate with white to yellow–white, smooth and raised on surface media. Mycelium superficial, velvety.
Material examined: THAILAND, Prachuap Khiri Khan Province, Bang Saphan District, Sai Khu Waterfall, on Pandanus sp., 30 July 2015, S. Tibpromma SF15-039 (MFLU 18-0015; HKAS 100841); living culture, KUMCC 17-0307.
GenBank numbers LSU: MH260286; ITS: MH275054; SSU: MH260332.
Notes: Chaetomium is a cosmopolitan genus with more than 150 species (Asgari and Zare 2011a, b; Zhang et al. 2012). Chaetomium globosum has been isolated previously from Pandanus odoratissimus and P. penetrans in Thailand (Thongkantha et al. 2008). Chaetomium globosum is commonly found in natural environments worldwide and can be a causal agent of emerging fungal infections (Sugiyama et al. 2008). The closest hits using a BLASTn search on NCBI GenBank of the ITS sequence were 99% similar to Chaetomium globosum (Accession numbers KT780353; KY132157; KY132151).
Subclass Xylariomycetidae O.E. Erikss & Winka
Amphisphaeriales D. Hawksw. & O.E. Erikss.
Beltraniaceae Nann.
Beltraniaceae was established by Nannizzi (1934) to accommodate a single genus Beltrania Penzig. Maharachchikumbura et al. (2016) accepted Beltrania, Beltraniella, Beltraniomyces, Beltraniopsis, Parapleurotheciopsis, Porobeltraniella, Pseudobeltrania and Subramaniomyces in the family. Later, Rajeshkumar et al. (2016) accepted an additional genus Hemibeltrania in the family and Lin et al. (2017) introduced Subsessila to this family. Species in Beltraniaceae are generally hyphomycetous and commonly isolated as saprobes (Seifert et al. 2011; Crous et al. 2015b; Maharachchikumbura et al. 2016). We describe two new species of Beltraniella and a new Beltrania species, collected on Pandanaceae in Thailand.
Beltrania Penz.
Beltrania was established with B. rhombic Penz. as the type species, found on Citrus limonum in Italy by Penzig (1882). The characteristics of this genus are setae with radially lobed basal cells, conidiophores with separating cells and biconic conidia with a hyaline transverse band and an apical tubular appendage. Pirozynski (1963) provided an illustrated monograph. Beltrania rhombica was found on Pandanus sp. from Mauritius (Dulymamode et al. 2001). There are 18 epithets for Beltrania are listed in Index Fungorum (2018).
Beltrania krabiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554512, Facesoffungi number: FoF04523; Fig. 101
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data. Related sequences were obtained from Lin et al. (2017). Forty-three strains are included in the combined sequence analysis, which comprise 1365 characters with gaps. Anthostomella leucospermi (CBS 110126) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 6451.337345 is presented. The matrix had 478 distinct alignment patterns, with 20.17% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.249145, C = 0.216405, G = 0.265430, T = 0.269020; substitution rates AC = 1.149297, AG = 2.533695, AT = 2.034519, CG = 1.003899, CT = 5.840755, GT = 1.000000; gamma distribution shape parameter α = 0.167365. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1913
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Setae erect, brown, thick-walled, indistinctly septate, straight to somewhat flexuous, conical at the apex, globose at basal cell. Conidiophores 43–86 × 2–4 µm, aggregated in dense fascicles, pale brown, cylindrical, septate, unbranched, straight to variously curved, proliferating sympodially at apex. Conidiogenous cells terminal, subhyaline, smooth, holoblastic, polyblastic, with several flat tipped denticles. Separating cells 7–8.5 × 3–4 μm (\( \bar{x} \) = 8 × 3.4 μm, n = 10), subhyaline, finely roughened, with several apical, flat-tipped denticles. Conidia 17–23 × 5–8 μm (\( \bar{x} \) = 20 × 7.5 μm, n = 20, including apical appendage), biconic, aseptate, solitary, subhyaline to pale brown, with distinct granules, without median transverse band, apical appendage 4–8 μm long, tapering to an acutely rounded tip, smooth, without a mucilaginous sheath.
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, filamentous with curled edge, raised on the media surface with white, smooth and flossy. Mycelium superficial, velvety.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 4 December 2017, S. Tibpromma KB035 (MFLU 16-1913, holotype; HKAS 96262, isotype); ex-type living culture, MFLUCC 16-0257.
GenBank numbers LSU: MH260280; ITS: MH275048; SSU: MH260328.
Notes: Based on phylogenetic analysis, Beltrania krabiensis (69% in ML, 0.90 in BYPP) and clustered with B. pseudorhombica Crous & Y. Zhang. Beltrania krabiensis has 7–8.5 × 3–4 μm separating cells, subhyaline to pale brown conidia, without median transverse band, 17–23 × 5–8 μm, 4–8 μm apical appendages, while B. pseudorhombica has 7–12 × 5–6 μm separating cells, pale brown conidia with a distinct median transverse band of lighter pigment, (20–)22–25(–26) × (7–)8(–9) μm, 7–11 × 1 μm apical appendages. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0257 is Beltrania sp. with 99% identity to the strain 114.2.1 (KP133187).
Beltraniella Subram.
Subramanian (1952) proposed Beltraniella, with B. odinae Subram. as the type species. Beltraniella is characterised by setiform conidiophores and polyblastic, sympodial, denticulate conidiogenous cells, lageniform conidia with a truncate base, rostrate apex, and a hyaline transverse band at the equatorial zone (Subramanian 1952). There are 25 epithets are listed in Index Fungorum (2018). Beltraniella has not been previously reported on Pandanaceae.
Beltraniella pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554513, Facesoffungi number: FoF04524; Fig. 102
Beltrania krabiensis (MFLU 16-1913, holotype). a Colonies on dead leaves of Pandanus sp. b Conidiogenous cells with conidiophores and conidia. c Seta. d Conidiogenous cells. e–h Conidia. i Germinating conidium. j, k Colony on MEA from above and below. Scale bars: a = 100 μm, b, c = 10 μm, d–i = 5 μm
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0039
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Setae 114–200 × 5–6 µm, erect, cylindrical, dark brown, 2–4-septate, thick-walled, straight to flexuous, conical at the tip, unbranched, smooth. Conidiophores 20–42 × 2–5 μm (\( \bar{x} \) = 26 × 4 μm, n = 10), subcylindrical, pale brown, smooth, arising from base of setae, straight to flexuous, septate. Conidiogenous cells 6–10 × 3–4.5 μm (\( \bar{x} \) = 8 × 4 μm, n = 10), terminal or lateral, discrete, subcylindrical to somewhat clavate, subhyaline to pale brown, smooth, polyblastic, with 2–4 denticulate conidiogenous loci. Conidia 14–24 × 4–6 μm (\( \bar{x} \) = 17.6 × 5 μm, n = 20), solitary, hyaline to subhyaline, biconic, aseptate, round at attenuated base, smooth-walled, without a transverse band and mucilaginous sheath or appendage.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular with filamentous-like, filamentous edge, flay on surface media, powdery, white to grey.
Material examined: THAILAND, Phuket Province, Thalang District, on fallen dead and decaying leaves of Pandanus sp., 19 December 2017, N. Chaiwan P20 (MFLU 18-0039, holotype); ex-type living culture, MFLUCC 18-0121.
GenBank numbers LSU: MH260281; ITS: MH275049; SSU: MH260329.
Notes: Beltraniella pandanicola is group together with B. portoricensis (F. Stevens) Piroz. & S.D. Patil and B. acacia Crous. Beltraniella portoricensis has subhyaline to olivaceous conidia with a hyaline transverse band (Rajeshkumar et al. 2016), while B. acacia has conidia with median transverse bands of lighter pigments and apical appendages tapering to an acutely rounded tip (Crous et al. 2016b). The conidia of B. pandanicola lack both an apical appendage and a median transverse band. In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 18-0121 is B. fertilis with 99% identity to the strain MRC 4-1 (MF580248).
Beltraniella thailandicus Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554514, Facesoffungi number: FoF04522; Fig. 103
Etymology: named after Thailand, where the fungus was first discovered.
Holotype: MFLU 16-1878
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Hyphomycetous. Setae 100–118 × 3–4 µm, erect, cylindrical, dark brown, 1–2-septate, thick-walled, straight to flexuous, conical at the tip, unbranched. Conidiophores subcylindrical, pale brown, smooth, arising base of setae, straight to flexuous, aseptate. Conidiogenous cells 4.5–7.5 × 2–3 μm (\( \bar{x} \) = 6 × 2.6 μm, n = 10), terminal or lateral, discrete, subcylindrical to somewhat clavate, pale brown, smooth, polyblastic, sympodial, with 1–4 denticulate conidiogenous loci. Conidia 10–25 × 3–6 μm (\( \bar{x} \) = 16 × 4 μm, n = 30), solitary, hyaline to subhyaline, kite-like, aseptate, smooth, without a hyaline transverse band, mucilaginous sheath or appendage.
Culture characteristics: Conidia germinating on MEA within 24 h. Colonies on MEA, circular, entire edge with curled, raised on surface media, rough, grey with black at the curled, hard.
Material examined: THAILAND, Chonburi Province, Bang Lamung District, on Pandanus sp., 18 July 2016, W. Jaidee PTY03 (MFLU 16-1878, holotype; HKAS 96272, isotype); ex-type living culture, MFLUCC 16-0377 = KUMCC 17-0300.
GenBank numbers LSU: MH260282; ITS: MH275050; SSU: MH260330.
Notes: In the phylogenetic analyses, Beltraniella thailandicus is distinct from other species of Beltraniella (0.97 in BYPP, Fig. 101). It shares similar conidial morphology to B. portoricensis but differs by absence of a hyaline transverse band in the conidia (Rajeshkumar et al. 2016). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of MFLUCC 16-0377 is Beltraniella sp. with 98% identity to the strain 111.3.3 (KP133179).
Sporocadaceae Corda
Sporocadaceae was established by Corda (1842) with Sporocadus Corda (1839) as type genus. Sporocadus was synonymized under Seimatosporium by Maharachchikumbura et al. (2016). Maharachchikumbura et al. (2015) and Senanayake et al. (2015) updated the family. Members of the family are saprobes, endophytes, or foliar pathogens in tropical and temperate regions (Nag Raj 1993; Tanaka et al. 2011). There are 22 genera in the family (Wijayawardene et al. 2018). We describe three new species of Neopestalotiopsis and two new species of Pestalotiopsis collected from Pandanaceae.
Neopestalotiopsis Maharachch. et al.
Neopestalotiopsis was erected by Maharachchikumbura et al. (2014) with N. protearum (Crous & L. Swart) Maharachch., K.D. Hyde & Crous As the type species. The characteristics of this genus are similar to Pestalotiopsis but can be distinguished from the latter by distinct phylogeny and versicolourous median cells (Maharachchikumbura et al. 2014). There are 31 epithets listed in Index Fungorum (2018). This is the first report of Neopestalotiopsis species from Pandanaceae.
Neopestalotiopsis chiangmaiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554515, Facesoffungi number: FoF04525; Fig. 105
Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2 and TEF1 sequence data. Thirty-six strains are included in the combined sequence analysis, which comprise 1452 characters with gaps. Pestalotiopsis trachicarpicola (IFRDCC 2440) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 5392.679460 is presented. The matrix had 396 distinct alignment patterns, with 12.70% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.229675, C = 0.268221, G = 0.215256, T = 0.286848; substitution rates AC = 1.007965, AG = 3.841487, AT = 1.520713, CG = 0.561002, CT = 4.529758, GT = 1.000000; gamma distribution shape parameter α = 0.234364. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Chiang Mai Province, where the fungus was first discovered.
Holotype: MFLU 18-0026
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata sporodochia on surface of substrate, scattered, dark brown, dull. Conidiophores reduced to conidiogenous cells. Conidiogenous cells holoblastic, filiform, smooth, thin-walled, hyaline, short. Conidia fusoid to ellipsoid, straight to slightly curved, 4-septate, 18–22 × 8–11 μm (\( \bar{x} \) = 20 × 9 μm, n = 20); basal cell conic to obconic with obtuse end, subhyaline, thin-walled and verruculose at the basal cell, 4–5 μm long (\( \bar{x} \) = 4.2 μm); three median cells, doliiform, concolorous, yellow to brown, together 12–17 μm long (\( \bar{x} \) = 14 μm); second cell from base, yellow, 3.5–5 μm long (\( \bar{x} \) = 4 μm); third cell, yellow–brown, 3.5–6 μm long (\( \bar{x} \) = 4.6 μm); fourth cell, brown, 3–5 μm long (\( \bar{x} \) = 4.4 μm); apical cell hyaline to subhyaline, conic, 3–5 μm long (\( \bar{x} \) = 4 μm), with (2–)3 tubular apical appendages; appendages unbranched, 4–28 μm long (\( \bar{x} \) = 14 μm); basal appendage present, 3–5 μm long (\( \bar{x} \) = 4 μm).
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, edge undulate, whitish, aerial mycelium on surface, floosy, velvety; reverse of culture yellow.
Material examined: THAILAND, Chiang Mai Province, Mae Taeng District, Mushroom Research Foundation, on Pandanus sp., 16 December 2017, S. Tibpromma P07 (MFLU 18-0026, holotype; HKAS 101795, isotype); ex-type living culture, MFLUCC 18-0113.
GenBank numbers TEF1: MH388404; TUB2: MH412725.
Notes: Based on multi-gene phylogenetic analysis, Neopestalotiopsis chiangmaiensis clusters with N. pandanicola and N. cubana Maharachch., K.D. Hyde & Crous (Fig. 106). Neopestalotiopsis pandanicola has 2(–3) tubular apical appendages 9.5–26 μm long, while N. cubana has 2–4 tubular apical appendages (19–)21–27(–28) μm long (Maharachchikumbura et al. 2014). Neopestalotiopsis chiangmaiensis has (2–)3 tubular apical appendages; appendages unbranched, 4–28 μm long. In a comparison of the 512 TEF1 nucleotides of N. chiangmaiensis and N. cubana differed 6 bp (1.17%) and 441 TUB2 nucleotides differed 3 bp (0.68%). In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of MFLUCC 18-0113 is N. clavispora with 98% identity to the strain TOR-802-803-804 (KU096881), while the closest matches with the TUB2 sequence were with 99% identical N. clavispora strain JT11 (MG740724).
Neopestalotiopsis pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554516, Facesoffungi number: FoF04526; Fig. 107
Neopestalotiopsis pandanicola (HKAS 99617, holotype). a Appearance of conidiomata on dead leaves of Pandanus sp. b Section of conidiomata. c Conidiogenous cells with conidiophores and conidia. d–g Conidia. h Germinating conidium. i, j Colonies on MEA from above and below. Scale bars: b = 100 μm, c–h = 5 μm
Etymology: named after the host genus, Pandanus.
Holotype: HKAS 99617
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiomata 280–450 μm, acervulus, subepidermal in origin, with basal stroma, thin-walled comprising hyaline to pale brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells oval, smooth, thin-walled, hyaline, short. Conidia fusoid to ellipsoid, straight to slightly curved, 4-septate, 27–35 × 7.5–11 μm (\( \bar{x} \) = 31 × 9 μm, n = 20); basal cell conic to obconic with obtuse end, subhyaline to pale brown, thin-walled and verruculose, 3.5–8 μm long (\( \bar{x} \) = 6 μm); three median cells, doliiform, concolorous, brown, periclinal walls darker than rest of the cell, together 17–25 μm long (\( \bar{x} \) = 21 μm); second cell from base 4.5–12.5 μm long (\( \bar{x} \) = 8 μm); third cell 5–8 μm long (\( \bar{x} \) = 7 μm); fourth cell 6–10 μm long (\( \bar{x} \) = 8 μm); apical cell hyaline, conic to cylindrical 3–6 μm long (\( \bar{x} \) = 4 μm), with 2(–3) tubular apical appendages; unbranched, 9.5–26 μm long (\( \bar{x} \) = 17 μm); with single basal appendage 3–6 μm long (\( \bar{x} \) = 5 μm), rarely absent, tubular, unbranched, centric.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies on PDA reaching 9 cm diam., after 7 days at room temperature, edge undulate, whitish, aerial mycelium on surface, floosy, velvety; reverse of culture yellow–white.
Material examined: CHINA, Yunnan Province, Xishuangbanna, on fallen dead and decaying leaves of Pandanus sp., 12 November 2016, T. Aluthwaththa XTBG09 (HKAS 99617, holotype); ex-type living culture, KUMCC 17-0175 = MFLUCC 17-2261.
GenBank numbers LSU: MH376727; SSU: MH388321; TEF1: MH388389; TUB2: MH412720.
Notes: Neopestalotiopsis pandanicola is morphologically similar to N. javaensis Maharachch., K.D. Hyde & Crous. Conidiogenous cells of N. pandanicola are filiform and short, while the conidiogenous cells of N. javaensis are ampulliform to lageniform and rugose-walled. Neopestalotiopsis pandanicola is phylogenetically closely related to N. chiangmaiensis (Fig. 106). However, this species differs from N. pandanicola by has smaller conidia (20 × 9 μm) and apical appendages. In a comparison of the 490 TEF1 nucleotides of N. pandanicola and N. chiangmaiensis differed 23 bp (4.69%). In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of KUMCC 17-0175 is N. clavispora with 99% identity to the strain TOR-802-803-804 (KU096881), while the closest matches with the TUB2 sequence were with 99% identical N. clavispora strain NM16311a (LC209221).
Neopestalotiopsis phangngaensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554517, Facesoffungi number: FoF04527; Fig. 108
Etymology: named after Phang Nga Province, where the fungus was first discovered.
Holotype: MFLU 18-0037
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, annellidic, filiform, ampulliform, smooth, thin-walled, hyaline, short. Conidia fusoid to ellipsoid, straight to slightly curved, 4-septate, 18–25 × 6–7.5 μm (\( \bar{x} \) = 21 × 6.6 μm, n = 20); basal cell conic to obconic with obtuse end, hyaline, verruculose, 3–5 μm long (\( \bar{x} \) = 4.3 μm); three median cells doliiform, concolorous, hyaline to subhyaline, becoming brown with age, septa and periclinal walls darker than rest of cells, together 13–15 μm long (\( \bar{x} \) = 14 μm); second cell from base 3.5–5 μm long (\( \bar{x} \) = 4.6 μm); third cell 2.5–5 μm long (\( \bar{x} \) = 4.5 μm); fourth cell 4–5 μm long (\( \bar{x} \) = 4.5 μm); apical cell hyaline, conic 3–4.5 μm long (\( \bar{x} \) = 3.5 μm), with 3 tubular apical appendages; appendages unbranched, 16–24.5 μm long (\( \bar{x} \) = 18 μm); basal appendage present 3–6 μm long (\( \bar{x} \) = 3.9 μm), rarely absent, tubular, unbranched, centric.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, edge undulate, whitish, aerial mycelium on surface, floosy, velvety; reverse of culture yellow–white.
Material examined: THAILAND, Phang Nga Province, Thap Put District, on Pandanus sp., 20 December 2017, N. Chaiwan P18 (MFLU 18-0037, holotype; HKAS 101796, isotype); ex-type living culture, MFLUCC 18-0119.
GenBank numbers LSU: MH376728; ITS: MH388354; SSU: MH388322; TEF1: MH388390; TUB2: MH412721.
Notes: Neopestalotiopsis phangngaensis is well-separated from other species of Neopestalotiopsis in the combined gene phylogenetic analyses (Fig. 106). Neopestalotiopsis phangngaensis is similar to N. eucalypticola isolated from Eucalyptus globules in country name. However, N. eucalypticola has 1–2, long tubular apical appendages, which are sometimes branched (Maharachchikumbura et al. 2016), while N. phangngaensis has 5 tubular apical appendages which are unbranched. In a comparison of the 512 TEF1 nucleotides of N. eucalypticola and N. phangngaensis differed 5 bp (0.97%) and 441 TUB2 nucleotides differed 26 bp (5.89%). In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of KUMCC 17-0175 is Neopestalotiopsis sp. with 90% identity to the strain MMf-0011 (LC184190), while the closest matches with the TUB2 sequence were with 99% Pestalotiopsis sp. strain HGUP4077 (KF179298).
Pestalotiopsis Steyaert
Pestalotiopsis was erected by Steyaert (1949) with P. guepinii (Desm.) Steyaert as the type species. Pestalotiopsis are opportunistic pathogens, endophytes and saprobes (Hu et al. 2007; Maharachchikumbura et al. 2011, 2012, 2014). Steyaert (1949) split Pestalotia into three genera, namely Pestalotia, Pestalotiopsis and Truncatella based on conidial morphology. Maharachchikumbura et al. (2014), provided morphological and phylogeny. There are 309 epithets listed in Index Fungorum (2018). Pestalotiopsis funerea (Desm.) Steyaert and P. guepinii (Desm.) Steyaert have been reported from Pandanaceae (Whitton et al. 2012).
Pestalotiopsis krabiensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554518, Facesoffungi number: FoF04528; Fig. 109
Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2 and TEF1 sequence data. Eighty-four strains are included in the combined sequence analysis, which comprise 1577 characters with gaps. Pestalotiopsis trachicarpicola (IFRDCC 2440) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 11764.880209 is presented. The matrix had 766 distinct alignment patterns, with 11.72% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.234091, C = 0.292387, G = 0.213607, T = 0.259915; substitution rates AC = 1.039520, AG = 3.238137, AT = 1.214322, CG = 0.889565, CT = 3.748492, GT = 1.000000; gamma distribution shape parameter α = 0.310200. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in red
Etymology: named after Krabi Province, where the fungus was first discovered.
Holotype: MFLU 16-1919
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiophores short, unbranched, reduced to conidiogenous cells. Conidiogenous cells discrete, holoblastic, simple, filiform, smooth, thin-walled, hyaline. Conidia fusoid to ellipsoid, straight to slightly curved, 4-septate, 19–25 × 4–6 μm (\( \bar{x} \) = 21 × 5 μm, n = 30), basal cell conic to obconic with obtuse end, hyaline, thin-walled, verruculose, 3.5–5 μm long (\( \bar{x} \) = 4 μm); three median cells, doliiform, concolorous, hyaline becoming yellow–white with age, septa and periclinal walls darker than rest of the cell, together 13–15 μm long (\( \bar{x} \) = 13.7 μm); second cell from base 3–5 μm long (\( \bar{x} \) = 4 μm); third cell 4–5.5 μm long (\( \bar{x} \) = 4.6 μm); fourth cell 4–5 μm long (\( \bar{x} \) = 4.3 μm); apical cell hyaline, conic 4–5.5 μm long (\( \bar{x} \) = 4.5 μm), with 2(–3) tubular apical appendages; appendages arising from the apex of the apical cell, unbranched, 11–19 μm long (\( \bar{x} \) = 15 μm); single basal appendage usually present, 2–4 μm long (\( \bar{x} \) = 2.6 μm), tubular, unbranched, centric.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, edge undulate, whitish, aerial mycelium on surface, curled, fruiting bodies black after 2 months, concentric, floosy, velvety; reverse of culture yellow–white.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB041 (MFLU 16-1919, holotype; HKAS 96268, isotype); ex-type living culture, MFLUCC 16-0260 = KUMCC 16-0141.
GenBank numbers LSU: MH376734; ITS: MH388360; TEF1: MH388395; ACT: MH412715; TUB2: MH412722.
Notes: In the phylogenetic analysis, Pestalotiopsis krabiensis forms a sister group to P. pandanicola. Pestalotiopsis krabiensis has larger conidia (21.1 × 5.2 μm) with mostly 2 apical appendages, while P. pandanicola has smaller conidia (15.1 × 3.6 μm) with 3 apical appendages. In a comparison of the 517 TEF1 nucleotides of P. pandanicola and P. krabiensis differed 1 bp (0.19%), 475 TUB2 nucleotides differed 17 bp (3.57%) and 577 ITS (+5.8S) nucleotides differed 1 bp (0.17%) which justifies these two isolates as two distinct taxa. In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of MFLUCC 16-0260 is P. adusta strain with 99% identity to the strain MFLUCC 10-0146 (JX399071), while the closest matches with the TUB2 sequence were with 99% P. adusta strain LPJZ02 (KJ885549).
Pestalotiopsis pandanicola Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554519, Facesoffungi number: FoF04529; Fig. 110
Pestalotiopsis krabiensis (MFLU 16-1919, holotype). a, b Colonies on dead leaves of Pandanus sp. c–f Conidiogenous cells with conidiophores and conidia. g–i Conidia. j Germinating conidium. k, l Colonies on MEA from above and below. m, n Conidiogenous cells and conidia formed in culture. Scale bars: a = 100 μm, b = 200 μm, c, d, m, n = 10 μm, e–j = 5 μm
Etymology: named after the host genus, Pandanus.
Holotype: MFLU 18-0039
Saprobic on dead leaves of Pandanus sp. Sexual morph Undetermined. Asexual morph Conidiophores reduced to conidiogenous cells. Conidiogenous cells discrete, holoblastic, simple, filiform, smooth, thin-walled, hyaline, and long. Conidia fusoid to ellipsoid, straight to slightly curved, 4-septate, 13–18 × 2.5–4.5 μm (\( \bar{x} \) = 15.1 × 3.6 μm, n = 30), basal cell conic to obconic with obtuse end, hyaline, thin and verruculose, 2–4 μm long (\( \bar{x} \) = 3.1 μm), with three median cells, doliiform, concolorous, yellow to pale brown, septa, together 8–11 μm long (\( \bar{x} \) = 9.5 μm); second cell from base, pale brown, 2–4 μm long (\( \bar{x} \) = 3 μm); third cell, pale brown, 2.5–4 μm long (\( \bar{x} \) = 3 μm); fourth cell, pale brown, 5–6.5 μm long (\( \bar{x} \) = 3.1 μm), apical cell hyaline, conic to sometimes cylindrical, 2–3 μm long (\( \bar{x} \) = .4 μm), with (2–)3 tubular apical appendages; appendages arising from apex of the apical cell, unbranched, 3.5–8 μm long (\( \bar{x} \) = 6.7 μm); basal appendage present, 1–4 μm long (\( \bar{x} \) = 2.7 μm), single, tubular, centric.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA reaching 9 cm diam., after 7 days at room temperature, circular, edge entire, whitish to pale yellow, with dense, aerial mycelium on surface, fruiting bodies black; reverse of culture whitish to pale yellow.
Material examined: THAILAND, Krabi Province, Mueang Krabi District, on Pandanus sp., 14 December 2015, S. Tibpromma KB030 (MFLU 16-1908, holotype; HKAS 96257, isotype); ex-type living culture, MFLUCC 16-0255 = KUMCC 16-0140.
GenBank numbers LSU: MH376735; ITS: MH388361; TEF1: MH388396; ACT: MH412716; TUB2: MH412723.
Notes: Pestalotiopsis pandanicola clusters with P. krabiensis in the phylogenetic analysis (Fig. 111), which has been described from the same location. Therefore, we propose P. pandanicola as a new species in Pestalotiopsis based on morphology and phylogenic analysis which discussion at above. In a BLASTn search on NCBI GenBank, the closest matches of TEF1 sequence of MFLUCC 16-0255 is P. adusta strain with 94% identity to the strain MFLUCC 10-0146 (JX399071), while the closest matches with the TUB2 sequence were with 99% P. uvicola strain HHL-AG (HM573238).
Pestalotiopsis pandanicola (MFLU 16-1908, holotype). a Colonies on dead leaves of Pandanus sp. b, c Conidiogenous cells with conidiophores and conidia. d–h Conidia. i Germinating conidium. j, k Colony on MEA from above and below. l Conidiogenous cells with conidia formed in culture. m Conidia formed in culture. Scale bars: b–d = 10 lm, e–i, l, m = 5 lm
Sordariomycetes orders incertae sedis
Vermiculariopsiellales Hern.-Restr. et al.
Vermiculariopsiellaceae Hern.-Rest. et al.
Vermiculariopsiellaceae was erected by Hernández-Restrepo et al. (2017) with Vermiculariopsiella Bender as the type genus based on phylogenetic analysis. Echinosphaeria macrospora Gawas, Bhat & K.D. Hyde. and E. pteridis S. Dharg. & Bhat. have been reported as sexual morphs of Vermiculariopsiella species based on culture techniques (Gawas et al. 2006; Dhargalkar and Bhat 2009), but need to be confirmed as the type species of Echinosphaeria is related to the family Helminthosphaeriaceae in the order Chaetosphaeriales (Miller and Huhndorf 2004; Miller et al. 2014). Morphological details for Helminthosphaeriaceae are provided by Hernández-Restrepo et al. (2017). We introduced a new species of Vermiculariopsiella collected from Pandanus sp.
Vermiculariopsiella Bender
Vermiculariopsiella spp. have been reported as asexual morphs of Echinosphaeria macrospora Gawas, Bhat & K.D. Hyde. and E. pteridis S. Dharg. & Bhat based on culture techniques (Gawas et al. 2006; Dhargalkar and Bhat 2009). However, Miller and Huhndorf (2004) and Miller et al. (2014) suggested that this relationship needed to be confirmed using molecular studies. Maharachchikumbura et al. (2016) showed that Vermiculariopsiella belonged to Microascales, genera incertae sedis. However, Hernández-Restrepo et al. (2017) introduced Vermiculariopsiella to Vermiculariopsiellaceae. Vermiculariopsiella has sporodochia with brown, erect setae dispersed throughout, and subhyaline conidiophores that give rise to phialidic conidiogenous cells with prominently curved apices, and hyaline, aseptate conidia (Crous et al. 2017). Vermiculariopsiella has 24 epithets are listed in Index Fungorum (2018). Vermiculariopsiella never has been reported from Pandanaceae.
Vermiculariopsiella hongkongensis Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF554552, Facesoffungi number: FoF04561; Fig. 112
Etymology: named after Hong Kong, where the fungus was first discovered.
Holotype: HKAS 100861
Saprobic on dead leaves of Pandanus sp. Colonies on natural substrate minute, brown, with erect, thick-walled, roughened, straight to flexuous, cylindrical and brown setae dispersed throughout sporodochium. Sexual morph Undetermined. Asexual morph Hyphomycetous. Conidiophores subcylindrical, septate, hyaline to subhyaline and dense. Conidiogenous cells 3.5–6 × 0.5–1.5 μm, terminal, subcylindrical, hyaline to subhyaline. Conidia 50–100 × 4–7.5 μm (\( \bar{x} \) = 70 × 5 μm, n = 20), solitary, hyaline, guttulate, straight to slightly curved, oblong with conical at both end, smooth-walled, without mucilaginous sheath. Setae 150–175 × 1.5–3 μm, cylindrical, conical at the apex, pale brown, septate.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA, circular, edge entire, white to yellow with white flossy in some part, velvety, with raised on media.
Material examined: HONG KONG, around Tai Tam Tuk Reservoir Dam, on Pandanus sp., 21 September 2016, S. Tibpromma HK09 (HKAS 100861, holotype); ex-type living culture, KUMCC 17-0270, MFLUCC 17-0636.
GenBank numbers LSU: MH260327; ITS: MH275092; SSU: MH260365.
Notes: Vermiculariopsiella has never been reported from Pandanaceae. Based on phylogenetic analysis, V. hongkongensis is well-separated from other species of Vermiculariopsiella (0.99 in BYPP, Fig. 68). However, we compare the morphology V. hongkongensis with V. acaciae Crous & M.J. Wingf. and V. immersa (Desm.) Bender. but V. acacia has dimorphic conidia (Crous et al. 2016a), while V. immerse has fusoid to cylindrical conidia (Bender 1932). In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 17-0270 is V. dichapetali culture with 99% identity to the strain CBS:143440 (MH107924).
References
Ahmed SA, Desbois N, Quist D, Miossec C, Atoche C, Bonifaz A, De Hoog GS (2015a) Phaeohyphomycosis caused by a novel species, Pseudochaetosphaeronema martinelli. J Clin Microbiol 53:2927–2934
Ahmed SA, Khan Z, Wang XW, Moussa TA, Al-Zahrani HS, Almaghrabi OA, de Hoog GS (2015b) Chaetomium-like fungi causing opportunistic infections in humans: a possible role for extremotolerance. Fungal Divers 76:1–16
Ahmed SA, van de Sande WW, Stevens DA, Fahal A, Van Diepeningen AD, Menken SB, de Hoog GS (2014) Revision of agents of black-grain eumycetoma in the order pleosporales. Persoonia 33:141–154
Alexopoulos CJ (1962) Introductory mycology, 2nd edn. Wiley, New York
Alves A, Crous PW, Correia A, Phillips AJL (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers 28:1–13
Ames LM (1961) A monograph of the Chaetomiaceae. US Army Res Dev Ser, No. 2. Washington DC
Aptroot A (2006) Mycosphaerella and its anamorphs 2. Conspectus of Mycosphaerella. CBS Biodivers Ser 5:1–231
Aptroot A, Parnmen S, Lücking R, Baloch E, Jungbluth P, Caceres ME, Lumbsch HT (2014) Molecular phylogeny resolves a taxonomic misunderstanding and places Geisleria close to Absconditella s. str. (Ostropales: Stictidaceae). Lichenologist 46:115–128
Aptroot A (2004) Two new ascomycetes with long gelatinous appendages collected from monocots in the tropics. Stud Mycol 50:307–311
Ariyawansa HA, Maharachchikumbura SSN, Karunarathne SC, Chukeatirote E, Bahkali AH, Kang JC, Bhat JD, Hyde KD (2013) Deniquelata barringtoniae from Barringtoniaasiatica, associated with leaf spots of Barringtonia asiatica. Phytotaxa 105:11–20
Ariyawansa HA, Tanaka K, Thambugala KM, Phookamsak R, Tian Q, Camporesi E, Hongsanan S, Monkai J, Wanasinghe DN, Mapook A, Chukeatirote E (2014) A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Divers 68:69–104
Ariyawansa HA, Thambugala KM, Manamgoda DS, Jayawardena R, Camporesi E, Boonmee S, Wanasinghe DN, Phookamsak R, Hongsanan S, Singtripop C (2015a) Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers 71:85–139
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala KM, Voigt K, Zhao RL, Li GJ, Doilom M, Boonmee S, Yang ZL, Cai Q, Cui YY, Bahkali AH, Chen J, Cui BK, Chen JJ, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, Hashimoto A, Hongsanan S, Jones EBG, Larsson E, Li WJ, Li QR, Liu JK, Luo ZL, Maharachchikumbura SSN, Mapook A, McKenzie EHC, Norphanphoun C, Konta S, Pang KL, Perera RH, Phookamsak R, Phukhamsakda C, Pinruan U, Randrianjohany E, Singtripop C, Tanaka K, Tian CM, Tibpromma S, Abdel-Wahab MA, Wanasinghe DN, Wijayawardene NN, Zhang JF, Zhang H, Abdel-Aziz FA, Wedin M, Westberg M, Ammirati JF, Bulgakov TS, Lima DX, Callaghan TM, Callac P, Chang CH, Coca LF, Dal-Forno M, Dollhofer V, Fliegerová K, Greiner K, Griffith GW, Ho HM, Hofstetter V, Jeewon R, Kang JC, Wen TC, Kirk PM, Kytovuori I, Lawrey JD, Xing J, Li H, Liu ZY, Liu XZ, Liimatainen K, Thorsten Lumbsch H, Matsumura M, Moncada B, Nuankaew S, Parnmen S, Santiago ALCMA, Sommai S, Song Y, de Souza CAF, de Souza-Motta CM, Su HY, Suetrong S, Wang Y, FongWS YH, Zhou LW, Réblová M, Fournier J, Camporesi E, Luangsa-ard JJ, Tasanathai K, Khonsanit A, Thanakitpipattana D, Somrithipol S, Diederich P, Millanes AM, Common RS, Stadler M, Yan JY, Li XH, Lee HW, Nguyen TTT, Lee HB, Battistin E, Marsico O, Vizzini A, Vila J, Ercole E, Eberhardt U, Simonini G, Wen HA, Chen XH, Miettinen O, Spirin V, Hernawati H (2015b) Fungal diversity notes 111–252 taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75:27–274
Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol 58:57–93
Asgari B, Zare R (2011a) The genus Chaetomium in Iran, a phylogenetic study including six new species. Mycologia 103:863–882
Asgari B, Zare R (2011b) A contribution to the taxonomy of the genus Coniocessia (Xylariales). Mycol Prog 10:189–206
Ávila A, Groenewald JZ, Trapero A, Crous PW (2005) Characterisation and epitypification of Pseudocercospora cladosporioides, the causal organism of Cercospora leaf spot of olives. Mycol Prog 109:881–888
Baloch E, Gilenstam G, Wedin M (2013) The relationships of Odontotrema (Odontotremataceae) and the resurrected Sphaeropezia (Stictidaceae)-new combinations and three new Sphaeropezia species. Mycologia 105:384–397
Barr ME (1976) Perspectives in the Ascomycotina. Mem N Y Bot Gard 28:1–8
Barr ME (1979) A classification of Loculoascomycetes. Mycologia 71:935–957
Barr ME (1983) The ascomycete connection. Mycologia 75:1–13
Barr ME (1990a) Melanommatales (Loculoascomycetes). N Am Flora Ser II Part 13:1–129
Barr ME (1990b) Prodromus to nonlichenized, pyrenomycetous members of class Hymenoascomycetes. Mycotaxon 39:98–100
Barr ME (1994) Notes on the Amphisphaeriaceae and related families. Mycotaxon 51:191–224
Bender HB (1932) The genera of Fungi Imperfecti. Mycologia 24(4):410–412
Benny GL, Kimbrough JW (1980) A synopsis of the orders and families of Plectomycetes with keys to genera. Mycotaxon 12:1–91
Berlese AN (1896) Icones Fungorum. Pyrenomycetes 2:1–216
Berraf-Tebbal A, Guereiro MA, Phillips AJ (2014) Phylogeny of Neofusicoccum species associated with grapevine trunk diseases in Algeria, with description of Neofusicoccum algeriense sp. nov. Phytopathol Mediterr 53:416–427
Bessy EA (1950) Morphology and taxonomy of fungi. Blakiston Co., Philadelphia
Boedijn KB (1933) Über einige phragmosporen Dematiazen. Bull Jardin Bot Buitenzorg 13(1):120–134
Boesewinkel HJ (1982) Cylindrocladiella, a new genus to accommodate Cylindrocladium parvum and other small-spored species of Cylindrocladium. Can J Bot 60:2288–2294
Boonmee S, D’souza MJ, Luo Z, Pinruan U, Tanaka K, Su H, Bhat JD, McKenzie EHC, Jones EBG, Taylor JE, Phillips AJL, Hirayama K, Eungwanichayapant PD, Hyde KD (2016) Dictyosporiaceae fam. nov. Fungal Divers 80:457–482
Boonmee S, Rossmana AY, Liu JK, Li WJ (2014) Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers 68:239–298
Boonmee S, Zhang Y, Chomnunti P, Chukeatirote E (2011) Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers 51:63–102
Boonyuen N, Chuaseeharonnachai C, Suetrong S, Sri-Indrasutdhi V, Sivichai S, Jones EBG, Pang KL (2011) Savoryellales (Hypocreomycetidae, Sordariomycetes): a novel lineage of aquatic ascomycetes inferred from multiple-gene phylogenies of the genera Ascotaiwania, Ascothailandia, and Savoryella. Mycologia 103(6):1351–1371
Booth C (1957) Studies on pyrenomycetes I. and II. Mycol Pap 68:1–27
Braun U, Crous PW, Dugan F, Groenewald JZ, de Hoog GS (2003) Phylogeny and taxonomy of Cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycol Prog 2:3–18
Bussaban B, Lumyong S, Lumyong P, Mckenzie EHC, Hyde KD (2001) A synopsis of the genus Berkleasmium with two new species and new records of Canalisporium caribense from Zingiberaceae in Thailand. Fungal Divers 8:73–85
Cai L, Jeewon R, Hyde KD (2006) Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol Res 110:137–150
Callmander MW, Lowry PP II, Forest F, Devey DS, Beentje H, Buerki S (2012) Benstonea Callm. & Buerki (Pandanaceae): characterization, circumscription, and distribution of a new genus of screw-pines, with a synopsis of accepted species. Candollea 67(2):323–345
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Castlebury LA, Rossman AY, Sung G-H, Hyten AS, Spatafora JW (2004) Multigene phylogeny reveals new lineages for Stachybotrys chartarum, the indoor air fungus. Mycol Prog 108:864–872
Chaiwan N, Lu YZ, Tibpromma S, Bhat DJ, Hyde KD, Boonmee S (2017) Neotubeufia gen. nov. and Tubeufia guangxiensis sp. nov. (Tubeufiaceae) from freshwater habitats. Mycosphere 8(9):1443–1456
Chang HS (1995) Notes on Taiwan dematiaceous hyphomycetes, some species of the genera Exserticlava, Craspedodidymum and Hermatomyces. Bot Bull Acad Sinica (Taiwan) 36:243–246
Chang TC, Salvucci A, Crous PW, Stergiopoulos I (2016) Comparative genomics of the Sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLoS Genet 12(8):e1005904
Chaverri P, Salgado C, Hirooka Y, Rossman A, Samuels G (2011) Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol 68:35–56
Chen JL, Hwang CH, Tzean SS (1991) Dictyosporium digitatum, a new hyphomycete from Taiwan. Mycol Res 95:1145–1149
Chevallier FF (1826) Flore Générale des Environs de Paris, vol. 1. Ferra Jeune, Paris, pp 1–674
Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu Z, Stadler M, Hyde KD (2014) The sooty moulds. Fungal Divers 66:1–36
Churchill ACL (2010) Mycosphaerella fijiensis, the black leaf streak pathogen of banana: progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol Plant Pathol 12:307–328
Clark JF (1899) A new Volutella. Bull Torrey Bot Club 26:617–620
Clendenin I (1896) Lasiodiplodia Ellis & Everh. n. gen. Bot Gaz Crawfordsville 21:92–93
Cooke MC, Plowright CB (1879) British Sphaeriacei. Grevillea 7(43):77–89
Cooke MC (1884) Synopsis Pyrenomycetum. Grevillea 12(64):102–113
Corda ACI (1829) Deutschl. Fl., 3 Abt. Sturm, Nürnberg 3(2):71
Corda ACI (1831) Die Pilze Deutschlands. In: Sturm J (ed) Deutschlands Flora in Abbildungen nach der Natur mit Beschreibungen, vol 12(3). Sturm, Nurnberg, pp 33–64
Corda ACJ (1837) Icones fungorum hucusque cognitorum, vol 1. J.G. Calve, Prague, pp 1–32
Corda ACJ (1839) Icones fungorum hucusque cognitorum, vol 3. J.G. Calve, Prague, pp 1–55
Corda ACJ (1842) Icones fungorum hucusque cognitorum, vol 5. J.G. Calve, Prague, pp 1–92
Correia KC, Câmara MPS, Barbosa MAG, Sales Jr-R, Agustí-Brisach C, Gramaje D, Maela L, José GJ, Paloma AC, Josep A, Michereff SJ (2013) Fungal trunk pathogens associated with table grape decline in North-eastern Brazil. Phytopathol Mediterr 52(2):380–387
Coutinho IBL, Freire FCO, Lima CS, Lima JS, Gonçalves FJT, Machado AR, Silva AMS, Cardoso JE (2017) Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in northeastern Brazil. Plant Pathol 66:90–104
Crous PW, Braun U, Hunter GC, Wingfield MJ, Verkley GJM, Shin HD, Nakashima C, Groenewald JZ (2013) Phylogenetic lineages in Pseudocercospora. Stud Mycol 75:37–114
Crous PW, Braun U, Wingfield MJ, Wood AR, Shin HD, Summerell BA, Alfenas AC, Cumagun CJR, Groenewald JZ (2009) Phylogeny and taxonomy of obscure genera of microfungi. Persoonia 22:139–161
Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, Hernández-Restrepo M, Wood AR (2015a) The genera of fungifixing the application of the type species of generic names–G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula and Wojnowicia. IMA Fungus 6(1):163–198
Crous PW, Shivas RG, Quaedvlieg W, van der Bank M, Zhang Y, Summerell BA, Guarro J, Wingfield MJ, Wood AR, Alfenas AC, Braun U, Cano-Lira JF, García D, Marin-Felix Y, Alvarado P, Andrade JP, Armengol J, Assefa A, den Breeÿen A, Camele I, Cheewangkoon R, De Souza JT, Duong TA, Esteve-Raventós F, Fournier J, Frisullo S, García-Jiménez J, Gardiennet A, Gené J, Hernández-Restrepo M, Hirooka Y, Hospenthal DR, King A, Lechat C, Lombard L, Mang SM, Marbach PAS, Marincowitz S, Marin-Felix Y, Montaño-Mata NJ, Moreno G, Perez CA, Pérez Sierra AM, Robertson JL, Roux J, Rubio E, Schumacher RK, Stchigel AM, Sutton DA, Tan YP, Thompson EH, van der Linde E, Walker AK, Walker DM, Wickes BL, Wong PTW, Groenewald JZ (2014) Fungal Planet description sheets: 214–280. Persoonia 32:184–306
Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ (2006) Phylogenetic lineages in the Botryosphaeriaceae. Stud Mycol 55:235–253
Crous PW, Wingfield MJ, Burgess TI, Hardy GESTJ, Crane C, Barrett S, Cano-Lira JF, Leroux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara J-P, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da Silva SS, Daniel R, de Beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suarez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta CM, Stephenson SL (2016a) Fungal Planet description sheets: 469–557. Persoonia 37:218–403
Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, de Jesús Yáñez-Morales M, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ (2015b) Fungal Planet description sheets: 371–399. Persoonia 35:264–327
Crous PW, Wingfield MJ, Richardson DM, Leroux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernández-Restrepo M, Kolařík M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Čmoková A, Dimitrov RA, Dyakov MYu, Dueñas M, Dutta AK, Esteve-Raventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GESTJ, Held BW, Jurjević Ž, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin H-D, Silva BDB, Silva GA, Smith MTH, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ (2016b) Fungal Planet description sheets: 400–468. Persoonia 36:316–458
Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESTJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustin Colmán A, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello Rodríguez F, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MTH, Tkalčec Z, Valenzuela-Lopez N, Van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ (2017) Fungal Planet description sheets: 625–715. Persoonia 39:270–467
da Cunha KC, Sutton DA, Fothergill AW, Gené J, Cano J, Madrid H, de Hoog S, Crous PW, Guarro J (2013) In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn Microbiol Infect Dis 76(2):168–174
Dai DQ, Bhat DJ, Liu JK, Chukeatirote E, Zhao RL, Hyde KD (2012) Bambusicola a new genus from bamboo with asexual and sexual morphs. Cryptogam Mycol 33:363–379
Dai DQ, Bahkali AH, Li WJ, Bhat DJ, Zhao RL, Hyde KD (2015) Bambusicola loculata sp. nov. (Bambusicolaceae) from bamboo. Phytotaxa 213:122–130
Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E (2017) Bambusicolous fungi. Fungal Divers 82:1–105
de Aldana BRV, Bills G, Zabalgogeazcoa I (2013) Are endophytes an important link between airborne spores and allergen exposure? Fungal Divers 60:33–42
de Wit PJ (2016) Cladosporium fulvum effectors: weapons in the arms race with tomato. Annu Rev Phytopathol 54:1–23
Dennis RWG (1960) British cup fungi and their allies. British cup fungi and their allies. An introduction to the Ascomycetes, Bernard Quaritch Ltd London, p 280
Dhargalkar S, Bhat DJ (2009) Echinosphaeria pteridis sp. nov. and its Vermiculariopsiella anamorph. Mycotaxon 108:115–122
Dingley JM (1951a) The Hypocreales of New Zealand I. Trans R Soc N Z 79:55–61
Dingley JM (1951b) The Hypocreales of New Zealand II. The genus Nectria. Trans R Soc N Z 79:177–202
Dingley JM (1952a) The Hypocreales of New Zealand III. The genus Hypocrea. Trans R Soc N Z 79:323–337
Dingley JM (1952b) The Hypocreales of New Zealand IV. The genera Calonectria, Gibberella and Thyronectria. Trans R Soc N Z 79:403–411
Dingley JM (1953) The Hypocreales of New Zealand V. The genera Cordyceps and Torrubilla. Trans R Soc N Z 81:329–343
Dingley JM (1954) The Hypocreales of New Zealand VI. The genera Hypocrella, Barya, Claviceps and Podonectria. Trans R Soc N Z 83:643–662
Dingley JM (1956) The Hypocreales of New Zealand VII. A revision of records and species in the Hypocreaceae. Trans R Soc N Z 81:329–343
Dingley JM, Fullerton RA, McKenzie EHC (1981) Records of fungi, bacteria, algae, and angiosperms pathogenic on plants in Cook islands, Fiji, Kiribati, Niue, Tonga, Tuvalu, and western Samoa (UNDP/FAO/SPEC Survey of Agricultural Pests and Diseases 2. UNDP/FAO/SPEC, Rome, p 485
Dissanayake AJ, Camporesi E, Hyde KD, Yan JY, Li XH (2017) Saprobic Botryosphaeriaceae including Dothiorella italica sp. nov. associated with urban and forest trees in Italy. Mycosphere 8:1157–1176
Dissanayake AJ, Phillips AJL, Hyde KD, Li XH (2016) Botryosphaeriaceae: current status of genera and species. Mycosphere 7:1001–1073
Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EHC, Hyde KD (2017) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers 82:107–182
Dou ZP, He W, Zhang Y (2017) Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp. nov. Mycosphere 8(2):1014–1027
Dulymamode R, Cannon PF, Peerally A (1998a) Fungi from Mauritius: Linocarpon species on Pandanus. Mycol Prog 102:1331–1337
Dulymamode R, Cannon PF, Peerally A (1998b) Fungi from Mauritius: Anthostomella species on Pandanaceae. Mycol Prog 102:1319–1324
Dulymamode R, Cannon PF, Peerally A (1998c) Fungi from Mauritius: three Astrocystis species from Pandanus. Mycol Prog 102:1325–1330
Dulymamode R, Cannon PF, Peerally A (2001) Fungi on endemic plants of Mauritius. Mycol Prog 105:1472–1479
Dulymamode R, Minter DW, Peerally A (1998d) Fungi from Mauritius: Rubikia splendida sp. nov. a coelomycete with unusual features. Mycol Prog 102:1242–1244
Dulymamode R, Wu W, Peerally A (1998e) Three new hyphomycetes on Pandanus leaves from Mauritius. Mycoscience 39:285–291
Ellis MB (1971) Dematiaceous hyphomycetes. Commonwealth, Mycological Institute Kew
Ellis MB (1976) More dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, p 507
Eriksson OE, Hawksworth DL (1993) Outline of the ascomycetes-1993. Syst Ascomycetum 12:51–257
Fernández-Brime S, Llimona X, Molnar K, Stenroos S, Högnabba F, Bjork C, Lutzoni F, Gaya E (2011) Expansion of the Stictidaceae by the addition of the saxicolous lichen-forming genus Ingvariella. Mycologia 103:755–763
Fernández-Brime S, Olariaga I, Baral HO, Friebes G, Jaklitsch W, Senn-Irlet B, Wedin M (2018) Cryptodiscus muriformis and Schizoxylon gilenstamii two new species of Stictidaceae (Ascomycota). Mycol Prog 17:295–305
Fries EM (1832) Systema Mycologicum, vol 3. Moritz, Greifswald, pp 261–524
Fries EM (1849) Summa vegetabilium Scandinaviae, vol 2. Typographia Academica, Uppsala, pp 259–572
Fuckel KWGL (1863) Fungi Rhenani exsiccati, Fasc. I–IV. Hedwigia 2:132–136
Gamundi IJ, Arambarri AM, Giaiotti AL (1977) Micro flora dela Hojarasca de Nothofagus dombeyi. Darwiniana 21:81–114
Gawas P, Shenoy BD, Hyde KD, Bhat DJ (2006) Echinosphaeria macrospora sp. nov., teleomorph of Vermiculariopsiella endophytica sp. nov. Cryptogam Mycol 27:11–20
Gäumann E (1964) Die Pilze. Birkhäuser Verlag, Basel
Ghimire SR, Hyde KD (2004) Fungal endophytes. In: Varma A, Abbott L, Werner D, Hampp R (eds) Plant Surface Microbiology. Springer-Verlag, Berlin, pp 281–292
Gilenstam G (1969) Studies in the genus Conotrema. ArkBot 7:149–179
Gilgado F, Gene J, Cano J, Guarro J (2007) Reclassification of Graphium tectonae as Parascedosporium tectonae gen. nov., comb. nov., Pseudallescheria africana as Petriellopsis africana gen. nov., comb. nov. and Pseudallescheria fimeti as Lophotrichus fimeti comb. nov. Int J Syst Evol Microbiol 57:2171–2178
Gräfenhan T, Schroers HJ, Nirenberg HI, Seifert KA (2011) An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol 68:79–113
Groenewald JZ, Nakashima C, Nishikawa J, Shin HD, Park JH, Jama AN, Groenewald M, Braun U, Crous PW (2013) Species concepts in Cercospora: spotting the weeds among the roses. Stud Mycol 75:115–170
Goh TK, Ho WH, Hyde KD, Whitton SR, Umali TE (1998) New records and species of Canalisporium (hyphomycetes) with a revision of the genus. Can J Bot 76:142–152
Goh TK, Hyde KD, Ho WH (1999) A revision of the genus Dictyosporium, with descriptions of three new species. Fungal Divers 2:65–100
Guo LD, Hyde KD, Liew ECY (2001) Detection and taxonomic placement of endophytic fungi within frond tissues of Livistona chinensis based on rDNA sequences. Mol Phylogenet Evol 20:1–13
Hall T (2004) BioEdit version 607. http://www.mbio-ncsuedu/bioedit/bioedit.html
Hashimoto A, Matsumura M, Hirayama K, Tanaka K (2017) Revision of Lophiotremataceae (Pleosporales Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae and Hermatomycetaceae fam. nov. Persoonia 39:51–73
Hawksworth DL, Wells H (1973) Ornamentation on the terminal hairs in Chaetomium Kunze ex Fr. and some allied genera. Commonwealth Mycological Institute, Kew
Hawksworth DL, Punithalingam E (1975) New and interesting microfungi from Slapton, South Devonshire: Deuteromycotina II. Trans Br Mycol Soc 64:89–99
Hawksworth DL, Sutton BC, Ainsworth GC (eds) (1983) Ainsworth & Bisby’s Dictionary of the Fungi, 7th edn. CABI, Kew
Hawksworth DL, Kirk PM, Sutton BC, Pegler DN (1995) Ainsworth & Bisby’s dictionary of the fungi, 8th edn. CAB International, Wallingford
Hawksworth DL (2000) The magnitude of fungal diversity: the 15 million species estimate revisited. Paper presented at the Asian Mycological Congress 2000 (AMC 2000) incorporating the 2nd Asia-Pacific Mycological Congress on Biodiversity and Biotechnology and held at the University of Hong Kong on 9–13 July 2000. Mycol Prog 105:1422–1432. https://doi.org/10.1017/S0953756201004725
Hawksworth DL (2002) Why study tropical fungi. Trop Mycol 2:1–11
Hawksworth DL, Lücking R (2017) Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr. https://doi.org/10.1128/microbiolspecFUNK-0052-2016
Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J (2017) Phylogeny of saprobic microfungi from Southern Europe. Stud Mycol 86:53–97
Hirooka Y, Rossman AY, Samuels GJ, Lechat C, Chaverri P (2012) A monograph of Allantonectria, Nectria, and Pleonectria (Nectriaceae, Hypocreales, Ascomycota) and their pycnidial, sporodochial, and synnematous anamorphs. Stud Mycol 71:1–210
Hosagoudar VB (2012) Asterinales of India. Mycosphere 2:617–852
Hu HL, Jeewon R, Zhou DQ, Zhou TX, Hyde KD (2007) Phylogenetic diversity of endophytic Pestalotiopsis species in Pinus armandii and Ribes spp.: evidence from rDNA and beta-tubulin gene phylogenies. Fungal Divers 24:1–22
Hughes SJ (1952) Fungi from the Gold Coast. I. Mycol Pap 48:1–91
Hughes SJ (1953) Conidiophores conidia and classification. Can J Bot 31:577–659
Hughes SJ (1978) New Zealand fungi 25 Miscellaneous species. N Z J Bot J 16:311–370
Huhndorf SM, Miller AN, Fernandez FA (2004) Molecular systematic of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96:368–387
Hyde KD (1994) Fungi from Pandanus I. Pandanicola gen. nov. from Australia and the Philippine Islands. Sydowia 46:35–40
Hyde KD (1997) Additions to the genus Linocarpon (Ascomycetes: Hyponectriaceae). Bot J Linn Soc 123:109–131
Hyde KD, Hawksworth DL (1997) Measuring and monitoring the biodiversity of microfungi. In: Hyde KD (ed) Biodiversity of tropical microfungi. Hong Kong University Press, Hong Kong, pp 11–28
Hyde KD, Goh TK, Taylor JE, Fröhlich J (1999) Byssosphaeria, Chaetosphaeria, Niesslia and Ornatispora gen. nov., from palms. Mycol Res 103(11):1423–1439
Hyde KD, Bussaban B, Paulus B, Crous PW, Lee S, McKenzie EHC, Photita W, Lumyong S (2006) Biodiversity of saprobic fungi. Biodivers Conserv 16:7–35
Hyde KD, Cai L, McKenzie EHC, Yang YL, Zhang JZ, Prihastuti H (2009) Colletotrichum: a catalogue of confusion. Fungal Divers 39:1–17
Hyde KD, McKenzie EHC, Koko TW (2011) Towards incorporating anamorphic fungi in a natural classification–checklist and notes for 2010. Mycosphere 2:1–88
Hyde KD, Jones EBG, Liu JK, Ariyawansa H, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai DQ, Diederich P, Dissanayake A, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li YM, Liu YX, Lücking R, Monka J, Muggia L, Nelsen MP, Pang KL, Phookamsak R, Senanayake IC, Shearer CA, Suetrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu HX, Zhang Y, Begoña AH, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat DJ, Camporesi E, Chukea E, Gueidan C, Hawksworth DL, Hirayama K, Hoog SD, Kang JK, Knudsen K, Li WJ, Li XH, Liu ZY, Mapook A, Mckenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu JC, Yacharoen S, Yan JY, Zang M (2013) Families of Dothideomycetes. Fungal Divers 63:1–313
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence MH, Lévesque CA, Li XH, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawłowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JC, Yan JY, Zhou N (2014) One stop shop: backbones trees for important phytopathogenic genera: I. Fungal Divers 67:21–125
Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de Santiago ALCMA, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu HX, Yang J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar AM, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev S, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XZ, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JZ, Zhu L (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270
Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He MQ, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kušan I, Lee H, Li J, Lin CG, Liu NG, Lu YZ, Luo ZL, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Abdel-Aziz FA, Li WJ, Senanayake IC, Shang QJ, Daranagama DA, De Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones GEB, Kang JC, Karunarathna SC, Lim YW, Liu JK, Liu ZY, Plautz HL Jr, Lumyong S, Maharachchikumbura SSN, Matocˇec N, Mckenzie EHC, Meśič A, Miller D, Pawłowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su HY, Suetrong S, Tkalčec Z, Vizzini A, Wen TC, Wisitrassameewong K, Wrzosek M, Xu JC, Zhao Q, Zhao RL, Mortimer PE (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers 87:1–235
Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva NI, Dissanayake AJ, Ekanayaka AH, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena R, Jiang HB, Karunarathna A, Lin CG, Liu JK, Liu NG, Lu YZ, Luo ZL, Maharachchimbura SSN, Manawasinghe IS, Pem D, Perera RH, Phukhamsakda C, Samarakoon MC, Senwanna C, Shang QJ, Tennakoon DS, Thambugala KM, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Zhang JF, Zhang SN, Bulgakov TS, Bhat DJ, Cheewangkoon R, Goh TK, Jones EBG, Kang JC, Jeewon R, Liu ZY, Lumyong S, Kuo CH, McKenzie EHC, Wen TC, Yan JY, Zhao Q (2018) Mycosphere notes 169–224. Mycosphere 9(2):271–430
Index Fungorum (2018) http://www.indexfungorumorg/Names/Namesasp
Ismail AM, Cirvilleri G, Polizzi G, Crous PW, Groenewald JZ, Lombard L (2012) Lasiodiplodia species associated with dieback disease of mango (Mangifera indica) in Egypt. Australas Plant Pathol 41:649–660
Jahn L, Schafhauser T, Pan S, Weber T, Wohlleben W, Fewer D, Sivonen K, Flor L, van Pée KH, Caradec T, Jacques P, Huijbers M, van Berkel W, Ludwig-Müller J (2017) Cyanodermella asteris sp. nov. (Ostropales) from the inflorescence axis of Aster tataricus. Mycotaxon 132:107–123
Jaklitsch WM, Réblová M (2015) Savoryellaceae. Jaklitsch & Réblová. Index Fungorum 209:1
Jaklitsch WM, Voglmayr H (2012) Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Divers 52:75–98
Jaklitsch W, Baral HO, Lucking R, Lumbsch HT (2016) Ascomycota. In: Frey W (ed) Syllabus of plant families, 23rd edn. Borntraeger Science Publishers, Stuttgart
Jaklitsch WM, Voglmayr H (2016) Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud Mycol 85:35–64
Jayasiri SC, Hyde KD, Abd-Elsalam KA, Abdel-Wahab MA, Ariyawansa HA, Bhat J, Buyck B, Dai YC, Ertz D, Hidayat I, Jeewon R, Jones EBG, Karunarathna SC, Kirk P, Lei C, Liu JK, Maharachchikumbura SSN, McKenzie E, Ghobad-Nejhad M, Nilsson H, Pang KL, Phookamsak R, Rollins AW, Romero AI, Stephenson S, Suetrong S, Tsui CKM, Vizzini A, Wen TC, De Silva NI, Promputtha I, Kang JC (2015) The Facesoffungi database: fungal names linked with morphology, molecular and human attributes. Fungal Divers 74:3–18
Jayawardena RS, Hyde KD, Damm U, Cai L, Liu M, Li XH, Zhang W, Zhao WS, Yan JY (2016a) Notes on currently accepted species of Colletotrichum. Mycosphere 7:1192–1260
Jayawardena RS, Hyde KD, Jeewon R, Li XH, Liu M, Yan JY (2016b) Mycosphere essay 6: why is it important to correctly name Colletotrichum species. Mycosphere 7:1076–1092
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7:1669–1677
Johnston PR (1983) Stictis and its anamorphs in New Zealand. N Z J Bot J 21:249–279
Johnston PR (2001) Monograph of the monocotyledon-inhabiting species of Lophodermium. Mycol Pap 176:1–239
Katoh K, Standley DM (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32:1933–1942
Kirk PM, Cannon PF, David JC, Stalpers JA (2001) Ainsworth and Bisby’s Dictionary of the Fungi, 8th edn. CABI, London
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth & Bisby’s dictionary of the fungi, 10th edn. CABI, Wallingford
Kirschstein W (1939) Über neue seltene und kritische. Ascomyceten und Fungi Imperfecti II. Ann Mycol 37:88–140
Koukol O, Delgado G, Hofmann TA, Piepenbring M (2018) Panama, a hot spot for Hermatomyces (Hermatomycetaceae, Pleosporales) with five new species, and a critical synopsis of the genus. IMA Fungus 9(1):107–141
Kuntze O (1891) Revisio generum plantarum, vol 2. A. Felix, Leipzig, pp 375–1011
Kuthubutheen AJ, Nawawi A (1991a) Dictyochaeta macrospora sp. nov.: a litter-inhabiting hyphomycete from Malaysia. Mycol Res 95:248–250
Kuthubutheen AJ, Nawawi A (1991b) Polytretophora dendroidea sp. nov. and P. calcarata (hyphomycetes) from Malaysia. Mycol Res 95:623–627
Kuthubutheen AJ, Nawawi A (1991c) Dictyochaeta guadalcanalensis comb. nov. and several new records of the genus in Malaysia. Mycol Res 95:1220–1223
Kuthubutheen AJ, Nawawi A (1991d) A key to Dictyochaeta and Codinaea species. Mycol Res 95:1224–1229
Lackner M, de Hoog GS (2011) Parascedosporium and its relatives: phylogeny and ecological trends. IMA Fungus 2(1):39–48
Lantz H, Johnston PR, Park D, Minter DW (2011) Molecular phylogeny reveals a core clade of Rhytismatales. Mycologia 103:57–74
Lechat C, Courtecuisse R (2010) A new species of Ijuhya, I. antillana, from the French West Indies. Mycotaxon 113:443–447
Li JF, Phookamsak R, Jeewon R, Bhat DJ, Mapook A, Camporesi E, Shang QJ, Chukeatirote E, Bahkali AH, Hyde KD (2017) Molecular taxonomy and morphological characterization reveal new species and new host records of Torula species (Torulaceae, Pleosporales). Mycol Prog 16(4):447–461
Li H, Xiao J, Gao YQ, Tang JJ, Zhang AL, Gao JM (2014) Chaetoglobosins from Chaetomium globosum, an endophytic fungus in Ginkgo biloba, and their phytotoxic and cytotoxic activities. J Agric Food Chem 62:3734–3741
Li Q, Wu Y, Lu DD, Xu YF, Lin YR (2015) A new species of Terriera (Rhytismatales, Ascomycota) on Photinia villosa. Mycotaxon 130:27–31
Li Z, Hou C (2016) A new Stictis species from China based on morphological and molecular data. Phytotaxa 286(3):186–192
Lin CG, Dai DQ, Bhat DJ, Hyde KD, Tang LZ, Toanun C (2017) Subsessila turbinata gen. et sp. nov. (Beltraniaceae) a Beltrania-like fungus from Thailand. Mycol Prog 16(4):393–401
Lindau G (1897) Pyrenomycetineae, Laboulbeniineae. In Engler A, Prantl K (eds) Die. Natürlichen Pflanzenfamilien. Teil I (Leipzig) 1. Engelmann, Leipzig, pp 321–505
Liu D (ed) (2011) Molecular detection of human bacterial pathogens. Taylor & Francis/CRC Press, Boca Raton
Liu JK, PhookamsakR DoilomM, Wikee S, Li YM, Ariyawansha H, Boonmee S, Chomnunti P, Dai DQ, Bhat JD, Romero AI, Zhuang WY, Monkai J, Jones EBG, Chukeatirote E, Ko-Ko TW, Zhao YC, Wang Y, Hyde KD (2012) Towards a natural classification of Botryosphaeriales. Fungal Divers 57:149–210
Liu JK, Phookamsak R, Dai DQ, Tanaka K, Jones EBG, Xu JC, Chukeatirote E, Hyde KD (2014) Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov. Roussoella and Roussoellopsis. Phytotaxa 181:1–33
Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura S, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doliom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardena NN, Wanasinghe D, Wisitrassameewong K, Abdel-Aziz FA, Adamcık S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XX, Liu ZY, Matumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E (2015b) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers 72:1–197
Liu JK, Yang J, Maharachchikumbura SS, McKenzie EH, Jones EBG, Hyde KD, Liu ZY (2016) Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycol Prog 15:1157–1167
Liu JK, Hyde KD, Jeewon R, Phillips AJ, Maharachchikumbura SS, Ryberg M, Liu ZY, Zhao Q (2017) Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers 84:75–99
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Locquin M (1984) Mycologie Générale et Structurale. Masson, Paris
Lodge DJ, Hawksworth DL, Ritchie BJ (1996) Microbial diversity and tropical forest functioning. In: Orians G, Dirzo R, Cushman D (eds) Biodiversity and ecosystem processes in tropical forests ecological studies, vol 122. Springer, Berlin, pp 69–100
Lombard L, van der Merwe NA, Groenewald JZ, Crous PW (2015) Generic concepts in Nectriaceae. Stud Mycol 80:189–245
Lombard L, Houbraken J, Decock C, Samson RA, Meijer M, Réblová M, Groenewald JZ, Crous PW (2016) Generic hyper-diversity in Stachybotriaceae. Persoonia 36:156–246
Lu YZ, Liu JK, Hyde KD, Rajesh J, Kang JC, Fan C, Boonmee S, Bhat DJ, Luo ZL, Lin CG, Eungwanichayapant PD (2018) A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers 92 (in press)
Lumbsch HT, Huhndorf SM (2007) Outline of Ascomycota—2007. Myconet 13:1–58
Lumbsch HT, Huhndorf SM (2010) Outline of Ascomycota—2009. Myconet 14:1–64
Luo J, Zhuang WY (2012) Volutellonectria (Ascomycota, Fungi), a new genus with Volutella anamorphs. Phytotaxa 44:1–10
Luo ZL, Bhat DJ, Jeewon R, Boonmee S, Bao DF, Zhao YC, Chai HM, Su HY, Su XJ, Hyde KD (2017) Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan China. Cryptogam Mycol 38:1–28
Luttrell ES (1951) Taxonomy of the Pyrenomycetes. Curators Univ Mo 24:1–3
Madrid H, da Cunha KC, Gené J, Dijksterhuis J, Cano J, Sutton DA, Guarro J, Crous PW (2014) Novel Curvularia species from clinical specimens. Persoonia 33:48–60
Maharachchikumbura SSN, Guo LD, Chukeatirote E, Bahkali AH, Hyde KD (2011) Pestalotiopsis-morphology, phylogeny, biochemistry and diversity. Fungal Divers 50:167–187
Maharachchikumbura SSN, Guo LD, Cai L, Chukeatirote E, Wu WP, Sun X, Crous PW, Bhat DJ, McKenzie EHC, Bahkali AH, Hyde KD (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers 56:95–129
Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu J, Crous PW (2014) Pestalotiopsis revisited. Stud Mycol 79:121–186
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang SK, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu JK, Liu ZY, Norphanphoun C, Pang KL, Perera RH, Senanayake IC, Shang Q, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers 72:199–301
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat DJ, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, Shang QJ, Xiao YP, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang WY, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang JC, Li QR, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Pang KL, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen TC, Wijayawardene NN (2016) Families of Sordariomycetes. Fungal Divers 79:1–317
Malloch D (1970) New concepts in the Microascaceae illustrated by two new species. Mycologia 62:727–740
Manamgoda DS, Cai L, Bahkali AH, Chukeatirote E, Hyde KD (2011) Cochliobolus: an overview and current status of species. Fungal Divers 51:3–42
Manamgoda DS, Cai L, McKenzie EHC, Crous PW, Madrid H, Chukeatirote E, Shivas RG, Tan YP, Hyde KD (2012a) A phylogenetic and taxonomic re-evaluation of the Bipolaris–Cochliobolus–Curvularia complex. Fungal Divers 56:131–144
Manamgoda DS, Cai L, McKenzie EHC, Chukeatirote E, Hyde KD (2012b) Two new Curvularia species from northern Thailand. Sydowia 64(2):255–266
Marin-Felix Y, Senwanna C, Cheewangkoon R, Crous PW (2017) New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 8(9):1556–1574
Matsushima T (1975) Icones microfungorum a Matsushima lectorum. Nippon Printing, Osaka, pp 1–209
McKenzie EHC (1991a) Dematiaceous hyphomycetes on Freycinetia. 1. Stachybotrys. Mycotaxon 41:179–188
McKenzie EHC (1991b) Dematiaceous hyphomycetes on Freycinetia. 2. Zebrospora gen. nov. Mycotaxon 41:189–193
McKenzie EHC (1991c) Dematiaceous hyphomycetes on Freycinetia. 3. Chalarodes gen. nov. Mycotaxon 42:89–93
McKenzie EHC (1995) Dematiaceous hyphomycetes on Pandanaceae. 5. Sporidesmium sensu lato. Mycotaxon 56:9–29
McKenzie EHC, Kuthubutheen AJ (1993) Dematiaceous hyphomycetes on Freycinetia. 4. Cryptophiale. Mycotaxon 47:87–92
McKenzie EHC, Hyde KD (1996) Index of fungi described from the Pandanaceae. Mycotaxon 57:125–144
McKenzie EHC, Whitton SR, Hyde KD (2002) The Pandanaceae—does it have a diverse and unique fungal biota. Trop Mycol 2:51–61
Mel’nik V, Lee S, Groenewald JE, Crous PW (2004) New hyphomycetes from Restionaceae in fynbos: Parasarcopodium ceratocaryi gen. et sp. nov., and Rhexodenticula elegiae sp. nov. Mycol Res 3:19–28
Mehl JWM, Slippers B, Roux J, Wingfield MJ (2014) Botryosphaeriaceae associated with dieback of Schizolobium parahyba trees in South Africa and Ecuador Forest. Pathology 44:396–408
Mehrotra RS, Aneja KR (1990) An introduction to mycology. New Age International, New Delhi
Miller AN, Huhndorf SM (2004) A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycol Prog 108:26–34
Miller AN, Huhndorf SM, Fournier J (2014) Phylogenetic relationships of five uncommon species of Lasiosphaeria and three new species in the Helminthosphaeriaceae (Sordariomycetes). Mycologia 106:505–524
Miller JH (1949) A revision of the classification of the Ascomycetes with special emphasis on the Pyrenomycetes. Mycologia 41:99–127
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE). Institute of Electrical and Electronics Engineers, New Orleans, pp 1–8
Minter DW (1996) Terriera cladophila. IMI Descriptions of Fungi & Bacteria No. 1296
Mukerji KG (1968) The position of the genus Achaetomium in Pyrenomycetes. Proc Indian Acad Sci 34:288–292
Mukerji KG, Manoharachary C (2010) Taxonomy and ecology of Indian fungi. I.K. International Pvt Ltd, New Delhi, pp 79–80
Müller E, von Arx JA (1962) Die Gattungen der didymosporen Pyrenomyceten. Beitr Kryptogamenflora Schweiz 11:1–922
Müller E, von Arx JA (1973) Pyrenomycetes: meliolales, coronophorales, sphaeriales. In: The fungi: an advanced treatise. Academic Press, New York, pp 87–132
Munk A (1953) The system of the pyrenomycetes. A contribution to a natural classification of the group Sphaeriales sensu Lindau. Dansk Bot Arkiv 15:1–163
Munk A (1957) Danish Pyrenomycetes. A preliminary flora. Dansk Bot Arkiv 17(1):1–491
Nadaf A, Zanan R (2012) World Pandanaceae: an overview. In: Indian Pandanaceae—an overview. Springer, New Delhi
Nag Raj TR (1993) Coelomycetous anamorphs with appendage bearing conidia. Mycologue, Waterloo
Nannizzi A (1934) Repertorio sistematico dei miceti dell’ uomo e degli animali, vol 4. Poligrafia Meimi, Siena, pp 1–557
Nawawi A, Kuthubutheen AJ (1989) Canalisporium, a new genus of lignicolous hyphomycetes from Malaysia. Mycotaxon 34(2):475–487
Nitschke TRJ (1869) Grundlage eines systems der Pyrenomyceten. Verhandlungen des Naturhistorischen Vereins der Preussischen Rheinlande, Westfalens und des Regierungsbezirks Osnabrück 26:70–77
Nylander JAA (2004) MrModeltest 20 Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
Pavlic D, Wingfield MJ, Barber P, Slippers B, Hardy GESJ, Burgess TI (2008) Seven new species of the Botryosphaeriaceae from baobab and other native trees in Western Australia. Mycologia 100(6):851–866
Persoon CH (1795) Observationes mycologicae. Annalen der Botanik (Usteri) 15:1–39
Penzig AGO (1882) Beltrania, un nuovo genere di ifomiceti. Nuovo Giornale Bot Ital Boll Soc Bot Ital 14:72–75
Penzig G, Saccardo PA (1898) Diagnoses fungorum novorum in insula Java collectorum. Ser. II. Malpighia 11:491–530
Petch T (1938) British Hypocreales. Trans Br Mycol Soc 21:243–305
Petrini O (1991) Fungal endophytes of tree leaves. In: Microbial ecology of leaves. Springer, New York, pp 179–197
Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW (2013) The Botryosphaeriaceae: genera and species known from culture. Stud Mycol 76:51–167
Phillips AJL, Alves A, Pennycook SR, Johnston PR, Ramaley A, Akulov A, Crous PW (2008) Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21:29–55
Photita W, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD (2004) Are some endophytes of Musa acuminata latent pathogens? Fungal Divers 16:131–140
Pieckova E (2003) In vitro toxicity of indoor Chaetomium Kunze ex Fr. Ann Agric Environ Med 10:9–14
Pirozynski KA (1963) Beltrania and related genera. Mycol Pap 90:1–37
Pirozynski KA, Hodges CS (1973) New hyphomycetes from South Carolina. Can J Bot 51:157–173
Phookamsak R, Norphanphoun C, Tanaka K, Dai DQ, Luo ZL, Liu JK, Su HY, Bhat DJ, Bahkali AH, Mortimer PE, Xu JC, Hyde KD (2015) Towards a natural classidication of Astrosphaeriella-like species introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellaceaeopsis gen. nov. Fungal Divers 74:143–197
Punithalingam E (1979) Sphaeropsidales in culture from humans. Nova Hedwig 31:119–158
Punithalingam E (1980) Plant diseases attributed to Botryodiplodia theobromae Pat. Cramer, Vaduz
Rajeshkumar KC, Crous PW, Groenewald JZ, Seifert KA (2016) Resolving the phylogenetic placement of Porobeltraniella and allied genera in the Beltraniaceae. Mycol Prog 15:1119–1136
Ramaley AW (2001) Hyaloseta nolinae its anamorph Monocillium nolinae and Niesslia agavacearum new members of the Niessliaceae, Hypocreales from leaves of Agavaceae. Mycotaxon 79:267–274
Rambaut A (2014) FigTree v14: tree figure drawing tool. http://tree.bio.ed.ac.uk/software/figtree/
Ranghoo VM (1998) Phylogeny of freshwater ascomycetes. Ph.D. thesis. University of Hong Kong: Hong Kong
Rangaswamy BE, Francis F, Prakash KK, Manjunath NS (2013) Variability in airborne bacterial and fungal population in the tertiary health care centre. Aerobiologia 29(4):473–479
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311
Rao V, De Hoog GS (1986) New or critical hyphomycetes from India. Stud Mycol 28:1–84
Reddy KA, Rao V (1984) Bahugada—a new hyphomycete. Curr Sci 53:542–544
Réblová M, Barr ME, Samuels GJ (1999) Chaetosphaeriaceae a new family for Chaetosphaeria and its relatives. Sydowia 51:49–70
Réblová M, Gams W, Seifert KA (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud Mycol 68:163–191
Rehner SA (2001) Primers for elongation factor 1-alpha (EF1-alpha). http://www.aftol.org/pdfs/EF1primer.pdf
Rogerson CT (1970) The Hypocrealean Fungi (Ascomycetes, Hypocreales). Mycologia 62:865–910
Rossman AY (2000) Towards monophyletic genera in the holomorphic Hypocreales. Stud Mycol 45:27–34
Rossman AY, Samuels GJ, Rogerson CT, Lowen R (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol 42:3–238
Rossman AY, McKemy JM, Pardo-Schultheiss RA, Schroers HJ (2001) Molecular studies of the Bionectriaceae using large subunit rDNA sequences. Mycologia 93:100–110
Rossman AY, Seifert KA, Samuels GJ, Minnis AM, Schroers HJ, Lombard L, Crous PW, Põldmaa K, Cannon PF, Summerbell RC, Geiser DM, Zhuang W, Hirooka Y, Herrera C, Salgado-Salazar C, Priscila Chaverri P (2013) Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus 4:41–51
Saccardo PA (1883) Pyrenomyceteae Sylloge Fungorum omnium hucusque cognitorum, vol 2, pp 1–813, Padua
Samuels GJ, Barr ME (1997) Notes on and additions to the Niessliaceae (Hypocreales). Can J Bot 75:2165–2176
Samuels GJ, Candoussau F, Magni F (1997) Fungicolous pyrenomycetes 2. Ascocodinaea gen. nov. and reconsideration of Litschaueria. Mycologia 89:156–162
Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98:1041–1052
Schroers HJ (2001) A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud Mycol 46:214
Schroers HJ, Gräfenhan T, Seifert KA, Nirenberg H (2011) Fusarium buxicola (Cyanonectria) revised and Geejayessia gen. nov. for Nectria desmazierii and related species with Fusarium-like anamorphs. Stud Mycol 68:115–138
Seaver FJ (1909a) The Hypocreales of North America I. Mycologia 1:41–76
Seaver FJ (1909b) The Hypocreales of North America II. Mycologia 1:177–207
Seaver FJ (1910a) The Hypocreales of North America III. Mycologia 2:48–92
Seaver FJ (1910b) Hypocreales. N Am Flora 3:1–88
Seaver FJ (1911) The Hypocreales of North America IV. Mycologia 3:207–230
Seaver FJ, Waterston JM (1941) Contributions to the mycoflora of Bermuda—II. Mycologia 33:310–317
Seifert K, Jones MG, Gams W, Kendrick B (2011) The genera of hyphomycetes. CBS biodiversity series no. 9. CBS-KNAWFungal Biodiversity Centre, Utrecht, pp 1–997
Senanayake IC, Maharachchikumbura SSN, Hyde KD, Bhat JD, Jones EBG, McKenzie EHC, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S, Li WJ, Shang QJ, Stadler M, Wijayawardene NN, Xiao YP, Norphanphoun C, Li Q, Liu XZ, Bahkali AH, Kang JC, Wang Y, Wen TC, Wendt L, Xu JC, Camporesi E (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers 73:73–144
Sherwood MA (1977) The ostropalean fungi. Mycotaxon 5:1–277
Shenoy BD, Jeewon R, Wu WP, Bhat DJ, Hyde KD (2006) Ribosomaland RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol Res 110:916–928
Shenoy BD, Jeewon R, Hyde KD (2007) Impact of DNA sequence-data on the taxonomy of anamorphic fungi. Fungal Divers 26:1–54
Sivanesan A (1984) The bitunicate ascomycetes and their anamorphs. J. Cramer, Vaduz
Sivanesan A (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycol Pap 158:1–261
Slippers B, Boissin E, Phillips AJL, Groenewald JZ, Lombard L, Wingfield MJ, Postma A, Burgess T, Crous PW (2013) Phylogenetic lineages in the Botryosphaeriales: a systematic and evolutionary framework. Stud Mycol 76:31–49
Slippers B, Wingfield MJ (2007) Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity ecology and impact. Fung Biol Rev 21:90–106
Soytong K, Kanokmedhakul S, Kukongviriyapa V, Isobe M (2001) Application of Chaetomium species (Ketomium) as a new broad spectrum biological fungicide for plant disease control. Fungal Divers 7:1–15
Sri-Indrasutdhi V, Boonyuen N, Suetrong S, Chuaseeharonnachai C, Sivichai S, Jones EBG (2010a) Wood-inhabiting freshwater fungi from Thailand: Ascothailandia grenadoidia gen. et. sp. Canalisporium grenadoidia sp. nov. with a key to Canalisporium species (Sordariomycetes, Ascomycota). Mycoscience 51(6):411–420
St John H (1962) Revision of the genus Pandanus, Stickman, Part 12. Queensland Pandanus. Pac Sci 16:291–346
St John H (1967) Revision of the genus Pandanus, Stickman, Part 23. Three Australian species of Pandanus. Pac Sci 21:523–530
St John H (1968) Revision of the genus Pandanus, Stickman, Part 28. The Australian species published by Robert Brown. Pac Sci 22:412–421
St John H (1969) Revision of the genus Pandanus, Stickman, Part 33. Further accounts of Australian species and a key to the section Microstigma. Pac Sci 23:89–114
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Spegazzini CL (1911) Mycetes Argentinenses. Series V. Anales Mus Nac Hist Nat Buenos Aires 3(13):446
Spegazzini C (1923) Algunos hongos de Tierra del Fuego. Phys Rev Soc Argent Cienc Nat 7:9–23
Sri-indrasutdhi V, Boonyuen N, Suetrong S, Chuaseeharonnachai C, Sivichai S, Jones EBG (2010b) Wood-inhabiting freshwater fungi from Thailand: Asothailandia grenadoidia gen. et sp. nov., Canalisporium grenadoidia sp. nov. with a key to Canalisporium species (Sordariomycetes, Ascomycota). Mycoscience 51:411–420
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Steyaert RL (1949) Contributions a etude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bull Jardin Bot Brux 19:285–354
Su H, Hyde KD, Maharachchikumbura SS, Ariyawansa HA, Luo Z, Promputtha I, Chai H (2016) The families Distoseptisporaceae fam. nov. Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae with new species from freshwater in Yunnan Province China. Fungal Divers 80:375–409
Su HY, Udayanga D, Luo ZL, Manamgoda DS, Zhao YC, Yang J, Hyde KD (2015) Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226(3):201–216
Su XJ, Luo ZL, Jeewon R, Bhat DJ, Bao DF, Li WL, Hao YE, Su HY, Hyde KD (2018) Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan China. Mycol Prog 16(4):1–15
Subramanian CV (1952) Fungi imperfecti from Madras-III Beltraniella gen. nov. Proc Indian Acad Sci Sect B 36:223–228
Summerbell RC, Gueidan C, Schroers HJ, de Hoog GS, Starink M, Arocha Rosete Y, Guarro J, Scott JA (2011) Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium and Trichothecium. Stud Mycol 68:139–162
Sugiyama K, Sano A, Murakami M, Ogawa T, Mishima H, Otake H, Kamei K, Sugiyama S (2008) Three isolations of Chaetomium globosum from erythematous epilation of canine skin. Sabouraudia 46(5):505–510
Swofford DL (2002) PAUP: Phylogenetic Analysis using Parsimony, version 4.0 b10. Sinauer Associates, Sunderland
Tanaka K, Hirayama K, Yonezawa H, Hatakeyama S, Harada Y, Sano T, Shirouzu T, Hosoya T (2009) Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs, and notes on the phylogeny of selected species from bamboo. Stud Mycol 64:175–209
Tanaka K, Endo M, Hirayama K, Okane I, Hosoya T, Sato T (2011) Phylogeny of Discosia and Seimatosporium and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26:85–98
Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T, Hosoya T (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol 82:75–136
Tanney J, Miller AN (2017) Asexual–sexual morph connection in the type species of Berkleasmium. IMA Fungus 8:99–105
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Tennakoon DS, Hyde KD, Wanasinghe DN, Bahkali AH, Camporesi E, Khan S, Phookamsak R (2016) Taxonomy and phylogenetic appraisal of Montagnula jonesii sp. nov. (Didymosphaeriaceae, Pleosporales). Mycosphere 7(9):1346–1356
Theissen F, Sydow H (1917) Synoptische Tafeln. Ann Mycol 15:389–491
Theissen F, Sydow H (1918) Vorentwürfe zu den Pseudosphaeriales. Ann Mycol 16:1–34
Thambugala KM, Wanasinghe DN, Phillips AJL, Camporesi E, Bulgakov TS, Phukhamsakda C, Ariyawansa HA, Goonasekara ID, Phookamsak R, Dissanayake A, Tennakoon DS, Tibpromma S, Chen YY, Liu ZY, Hyde KD (2017) Mycosphere notes 1–50: grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8(4):697–796
Than PP, Prihastuti H, Phoulivong S, Taylor PW, Hyde KD (2008) Chilli anthracnose disease caused by Colletotrichum species. J Zhejiang Univ Sci B 9(10):764
Thongkantha S, Lumyong S, McKenzie EHC, Hyde KD (2008) Fungal saprobes and pathogens occurring on tissues of Dracaena lourieri and Pandanus spp. in Thailand. Fungal Divers 30:149–169
Tian Q, Liu JK, Hyde KD, Wanasinghe DN, Boonmee S, Jayasiri SC, Luo ZL, Taylor JE, Phillips AJ, Bhat DJ, Li WJ (2015) Phylogenetic relationships and morphological reappraisal of Melanommataceae (Pleosporales). Fungal Divers 74:267–324
Tibpromma S, Boonmee S, Wijayawardene NN, Maharachchikumbura SSN, McKenzie EHC, Bahkali AH, Jones EBG, Hyde KD, Itthayakorn P (2016a) The holomorph of Parasarcopodium (Stachybotryaceae) introducing P. pandanicola sp. nov. on Pandanus sp. Phytotaxa 266:250–260
Tibpromma S, Bhat J, Doilom M, Lumyong S, Nontachaiyapoom S, Yang JB, Hyde KD (2016b) Three new Hermatomyces species (Lophiotremataceae) on Pandanus odorifer from Southern Thailand. Phytotaxa 275:127–139
Tibpromma S, McKenzie EHC, Karunarathna SC, Mortimer PE, Xu J, Hyde KD, Hu DM (2016c) Muyocopron garethjonesii sp. nov. (Muyocopronales, Dothideomycetes) on Pandanus sp. Mycosphere 7:1480–1489
Tibpromma S, Daranagama DA, Boonmee S, Promputtha I, Nontachaiyapoom S, Hyde KD (2017a) Anthostomelloides krabiensis gen. et. sp. nov. (Xylariaceae) from Pandanus odorifer. Turk J Bot 41:107–116
Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK, Bhat DJ, Jones EBG, McKenzie EHC, Camporesi E, Bulgakov TS, Doilom M, de Azevedo Santiago ALCM, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang JB, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KNA, Latha KPD, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon DS, Li JF, Dayarathne MC, Perera RH, Xiao Y, Wanasinghe DN, Senanayake IC, Goonasekara ID, de Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KWT, Luo ZL, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Meŝič A, Dutta AK, de Souza CAF, Richter C, Lin CG, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, da Silva GA, Plautz HL Jr, Ariyawansa HA, Lee H, Kuśan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, Mikśìk M, Samarakoon MC, Wijayawardene NN, Kim NK, Matocčec N, Singh PN, Tian Q, Bhatt RP, de Oliveira RJV, Tulloss RE, Aamir S, Kaewchai S, Svetasheva STY, Nguyen TTT, Antonìn V, Li WJ, Wang Y, Indoliya Y, Tkalčec Z, Elgorban AM, Bahkali AH, Tang AMC, Su HY, Zhang H, Promputtha I, Luangsa-ard J, Xu JC, Yan J, Chuan KJ, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJL, Nontachaiyapoom S, Wen TC, Karunarathna SC (2017b) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261
Tibpromma S, Hyde K, Bhat J, Mortimer P, Xu J, Promputtha I, Doilom M, Yang J, Tang A, Karunarathna S (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33:25–67
Tsui CKM, Sivichai S, Berbee ML (2006) Molecular systematics of Helicoma, Helicomyces and Helicosporium and their teleomorphs inferred from rDNA sequences. Mycologia 98:4–104
Tulasne LR, Tulasne C (1863) Selecta Fungorum Carpologia: Xylariei-Valsei-Spaeriei 2
Tulasne LR, Tulasne C (1865) Selecta Fungorum Carpologia: Nectriei-Phacidiei-Pezizei 3
Uecker FA (1994) Ontogeny of the ascoma of Glomerella cingulata. Mycologia 86(1):82–88
Varghese KIM, Rao VG (1978) Two new setose hyphomycetes from India. Bot Notiser 131:215–217
Verma P, Singh S, Singh R (2013) Seven species of Curvularia isolated from three lakes of Bhopal. Adv Life Sci Technol 8:13–15
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Von Arx JA, Müller E (1954) Die Gattungen der amerosporen Pyrenomyceten. Beitr Kryptogamenfl Schweiz 11(1):1–434
Von Arx JA, Guarro J, Figueras MJ (1986) The Ascomycete genus Chaetomium. Beihefte zur Nova Hedwigia 84:1–162
Wang XW, Lombard L, Groenewald JZ, Li J, Videira SIR, Samson RA, Crous PW (2016) Phylogenetic reassessment of the Chaetomium globosum species complex. Persoonia 36:83–133
Wang Y, Hyde KD, McKenzie EH, Jiang YL, Li DW, Zhao DG (2015) Overview of Stachybotrys (Memnoniella) and current species status. Fungal Divers 71(1):17–83
Wang Y, Xu L, Ren W, Zhao D, Zhu Y, Wu X (2012) Bioactive metabolites from Chaetomium globosum L18, an endophytic fungus in the medicinal plant Curcuma wenyujin. Phytomedicine 19:364–368
Wardah, Setyowati FM (2009) Ethnobotanical study on the Genus Pandanus L. f. in certain areas in Java, Indonesia. Biodiversitas 10(3):146–150
Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Stud Mycol 73:115–180
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Whitton SR, Hyde KD, McKenzie EHC (1998) Microfungi on the Pandanaceae a new species of Stictis (Ostropales). Fungal Divers 2:169–174
Whitton SR, McKenzie EHC, Hyde KD (1999) Microfungi on the Pandanaceae: Troposporopsis gen. nov. Fungal Divers 3:173–177
Whitton SR, McKenzie EHC, Hyde KD (2000a) Dictyochaeta and Dictyochaetopsis species from the Pandanaceae. Fungal Divers 4:133–158
Whitton SR, McKenzie EHC, Hyde KD (2001a) Microfungi on the Pandanaceae: Paraceratocladium seychellarum sp. nov. and a review of the genus. Fungal Divers 7:175–180
Whitton SR, McKenzie EHC, Hyde KD (2001b) Microfungi on the Pandanaceae: Nakatopsis gen. nov., a new hyphomycete genus from Malaysia. Fungal Divers 8:163–171
Whitton SR, McKenzie EHC, Hyde KD (2001c) Microfungi on the Pandanaceae: Stachybotrys with three new species. N Z J Bot J 39:489–499
Whitton SR, McKenzie EHC, Hyde KD (2002a) Microfungi on the Pandanaceae: a revision of the hyphomycete genus Balaniopsis with two new species. Mycoscience 43:67–72
Whitton SR, McKenzie EHC, Hyde KD (2002b) Microfungi on the Pandanaceae: two new species of Camposporium and key to the genus. Fungal Divers 11:177–187
Whitton SR, McKenzie EHC, Hyde KD (2003) Microfungi on the Pandanaceae: Zygosporium a review of the genus and two new species. Fungal Divers 12:207–222
Whitton SR, McKenzie EHC, Hyde KD (2012) Fungi associated with Pandanaceae. Springer, Dordecht. ISBN 978-94-007-4446-2
Whitton SR, McKenzie EHC, Hyde KD (2000b) Microfungi on the Pandanaceae: Acrodictys with two new species. Fungal Divers 4:159–169
Whitton SR, McKenzie EHC, Hyde KD, Fröhlich J (2001d) Microfungi on the Pandanaceae: Polytretophora macrospora sp. nov. Mycoscience 42:555–558
Wijayawardene NN, Mckenzie EHC, Hyde KD (2012) Towards incorporating anamorphic fungi in a natural classification–checklist and notes for 2011. Mycosphere. 3(2):157–228
Wongsawas M, Wang HK, Hyde KD, Lin FC (2009) Dictyosporium zhejiangensis sp. nov., a new freshwater anamorphic fungus from China. Cryptogam Mycol 30(4):355–362
Wijayawardene NN, Hyde KD, Tibpromma S, Wanasinghe DN, Thambugala KM, Tian Q, Wang Y, Fu L (2017a) Towards incorporating asexual fungi in a natural classification: checklist and notes 2012–2016. Mycosphere 8:1457–1554
Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang K-L, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro JS, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel-Wahab MA, Takamatsu S, Bensch K, de Silva NI, De Kese A, Karunarathna A, Boonmee S, Pfister DH, Lu Y-Z, Luo Z-L, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng X-Y, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera RH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma X, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC (2017b) Notes for genera-Ascomycota. Fungal Divers 86:1–594
Wijayawardene NN, Hyde KD, Lumbsch T, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R (2018) Outline of Ascomycota—2017. Fungal Divers 88:167–263
Winter G (1885) Ascomyceten: Gymnoasceen und Pyrenomyceten. In: Rabenhorst L (ed) Kryptogamen-Flora von Deutschland Oesterreich und der Schweiz 1(2)
Winter G (1885) Pilze-Ascomyceten. In: Rabenhorst GL (ed) Kryptogamen-Flora von Deutschland Oesterreich und der Schweiz, vol 1, pp 65–528
Yang J, Maharachchikumbura SSN, Liu JK, Hyde KD, Jones EBG, Al-Sadi AM, Liu ZY (2018) Pseudostanjehughesia aquitropica gen. et sp. nov. and Sporidesmium sensu lato species from freshwater habitats. Mycol Prog 17(5):591–616
Yang T, Groenewald JZ, Cheewangkoon R, Jami F, Abdollahzadeh J, Lombard L, Crous PW (2017) Families, genera, and species of Botryosphaeriales. Fungal Biol 121(4):322–346
Zare R, Gams W, Starink-Willemse M, Summerbell RC (2007) Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwig 85:463–489
Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH (2006) An overview of the systematics of the Sordariomycetes based on four-gene phylogeny. Mycologia 98:1076–1087
Zhang SN, Hyde KD, Jones GEB, Cheewangkoon R, Boonmee S, Doilom M, Mapook A, Liu JK (2017) Novomicrothelia pandanicola sp. nov., a non-lichenized, Trypetheliaceae species from Pandanus. Phytotaxa 321(3):254–264
Zhang T, Deng X, Yu Y, Zhang M, Zhang Y (2016) Pseudochaetosphaeronema ginkgonis sp. nov., an endophyte isolated from Ginkgo biloba. Int J Syst Evol Microbiol 66(11):4377–4381
Zhang Y, Crous PW, Schoch CL, Bahkar AH, Guo LD, Hyde KD (2011) A molecular, morphological and ecological re-appraisal of Venturiales—a new order of Dothideomycetes. Fungal Divers 51:249–277
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales. Fungal Divers 53:1–221
Zhang Y, Wang HK, Fournier J, Crous PW, Jeewon R, Pointing SB, Hyde KD (2009) Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers 38:225–251
Zoller S, Scheidegger C, Sperisen C (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31(5):511–516
Acknowledgements
We would like to thank Molecular Biology Experimental Center at Kunming Institute of Botany for their help with sequencing work. Saowaluck Tibpromma thanks the Mushroom Research Foundation (MRF), Chiang Rai, Thailand, Mae Fah Luang University for financial support and Dr. Shaun Pennycook for nomenclature correctness. Kevin D. Hyde thanks the Thailand Research Fund grants entitled Biodiversity, phylogeny and role of fungal endophytes on above parts of Rhizophora apiculata and Nypa fruticans (Grant No. RSA5980068), the future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (Grant No. DBG6080013), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (Grant No. RDG6130001), Mae Fah Luang University for the grant “Biodiversity, phylogeny and role of fungal endophytes of Pandanaceae” (Grant No. 592010200112), “Diseases of mangrove trees and maintenance of good forestry practice’’ (Grant No. 60201000201) and Taxonomy diversity, phylogeny and evolution of fungi in Capnodiales (Grant No. 666713), and the Chinese Academy of Sciences (Project Number 2013T2S0030), for the award of Visiting Professorship for Senior International Scientists, at Kunming Institute of Botany, and Chiang Mai University. Ausana Mapook, Benjarong Thongbai, Chada Norphanphoun, Chayanard Phukhamsakda, Monika Dayarathne, Nimali Indeewari de Silva, Qiuju Shang, Saranyaphat Boonmee, Sirinapa Konta, Tharanga Aluthwaththa, Watcharachai Jaidee and Yuanpin Xiao are thanked for their help. Samantha C. Karunarathna thanks CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2018PC0006), and the National Science Foundation of China (NSFC) for funding this research work under the project code 31750110478. Peter E. Mortimer thanks the National Science Foundation of China (NSFC) project codes 41761144055 and 41771063. Alan JL Phillips acknowledges the support from Biosystems and Integrative Sciences Institute (BioISI, FCT/UID/Multi/04046/2013).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tibpromma, S., Hyde, K.D., McKenzie, E.H.C. et al. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93, 1–160 (2018). https://doi.org/10.1007/s13225-018-0408-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-018-0408-6