Abstract
Introduction
Noninvasive prenatal testing (NIPT) has revolutionized prenatal screening for chromosomal aneuploidies in some countries. Its implementation has been sporadic in developing countries. Given the genetic variation of the people in different countries, we evaluated the performance of the SNP-based NIPT in India .
Materials and Methods
The Panorama™ NIPT was performed in 516 pregnancies, which had tested intermediate-to-high risk on conventional first and second trimester screening. Results were confirmed either by invasive diagnostic testing or by clinical evaluation after birth.
Results
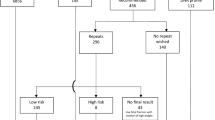
Of 511 samples analyzed, results were obtained in 499 (97.7%). Of these, 480 (98.2%) were low risk and 19 were high risk. A sensitivity of 100% was obtained for detection of trisomies 21, 18, 13 and sex chromosomal abnormalities. The specificity ranged from 99.3 to 100% for abnormalities tested. Taken together, the positive predictive value for trisomies 21, 18, 13 and monosomy X was 85.7%. The average fetal fraction was 8.2%, which is lower than the average observed elsewhere.
Conclusion
This is the first report of detailed experience with NIPT in India and demonstrates comparable performance in all aspects of testing to the results elsewhere.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The incidence of chromosomal disorders in India is 1:166 live births [1]. Given the large population, around 35,000 fetuses with Down syndrome alone are conceived every year [1]. Therefore, screening and diagnosis for chromosomal disorders is important in India, as in other countries. The current prenatal screening for chromosomal abnormalities consists of analyzing blood hormone levels and ultrasonography. However, these procedures are limited by low sensitivity and high false positive rate of 2–7% [2, 3]. Also, conventional screening misses over 10% of affected fetuses [4]. Invasive diagnostic methods, such as amniocentesis or chorionic villus sampling (CVS), are highly sensitive but cannot be offered to all pregnant women as they carry a small but significant risk of miscarriage [5, 6].
NIPT, a recently developed advanced technology provides a significant improvement over conventional testing, with detection rate of over 99% and a false positive rate of less than 0.1% by investigation of cell-free fetal (placental) DNA from maternal blood [7,8,9,10,11]. Furthermore, a significant reduction, 50–70%, of invasive procedures has been observed in setups where NIPT has been implemented [12, 13].
Commercialized NIPT technologies follow either of the two approaches: a counting-based method using massively parallel sequencing (MPSS) or the single nucleotide polymorphism-based approach (SNP), used in the Panorama™ NIPT developed by Natera Inc. (San Carlos, USA). The technology and the performance of the SNP-based method has been described elsewhere and validated both in high and low risk pregnant women [14,15,16,17,18,19,20]. The advantage of this method is that it does not require a reference chromosome, is able to detect vanishing twin, triploidy, maternal mosaicism and is highly accurate [16]. The limitation has been, rarely, the inability to make a call when there is a high genetic homology between parents (consanguinity) [21]. However, improvement of the algorithm has been implemented, such as quantitative multiple model (QMM) that resolves samples with genetic homology [personal communication] and reduction in no-call threshold for sample calling to 2.8% fetal fraction [22]. Several professional societies worldwide have issued guidelines periodically based on available data on appropriate usage of NIPT [13, 23,24,25,26,27].
The Indian population is socioculturally, ethnically and biologically diverse [28] as reflected in the studies of mutations detection that reveal many novel ones. Additionally, the Indian population has a relatively high rate of consanguinity (20–39%) among certain communities [29, 30]. It is not known how these biological factors would influence the SNP-based NIPT test. The collection and handling of samples in a tropical country might also affect the test performance. To study the feasibility of performing NIPT in an Indian population given the above considerations, we conducted an Indian study. The main objective was to evaluate the performance of NIPT for trisomies 21, 18, 13, sex chromosome abnormalities and triploidy in a cohort with intermediate-to-high risk on conventional screening.
Materials and Methods
Ten leading institutes collaborated in this study, after review and approval from their respective institutional review boards. The flowchart of the study protocol is shown in Fig. 1. Pregnant women were enrolled based on the specified criteria. Singleton pregnancies, between 11 and 18 weeks gestation (CRL 45–84 mm), intermediate-to-high risk (risk > 1:1000 for trisomy 21 and > 1:400 for trisomy 18, Nuchal Translucency (NT) measure < 95th centile) on biochemical, combined, triple or quadruple screening were included. Score of > 1:250 was defined as high risk and between 1:250 and 1:1000 as intermediate risk. While pregnancies with egg/sperm donor, surrogacy, twin or multiple gestation, with known parental chromosomal abnormalities (including known balanced translocations), where invasive testing was planned, NT > 95th percentile or nuchal fold thickness > 6 mm, ≥ 1 malformation in fetus, bone marrow transplant or malignancy patients were excluded.
Twenty milliliters of peripheral blood was collected in Streck™ tubes after genetic counseling and informed consent. A paternal buccal swab was obtained when available. Samples were transported to Medgenome Laboratory in Bangalore. Laboratory testing was performed using validated protocol from Natera Inc., using SNPs on five chromosomes and analyzed by the cloud-based proprietary NATUS algorithm [10, 14, 15, 22], the enhanced version of which became available during the course of the study. Fetal fraction was reported in each case. In accordance with the legal requirements in India, gender was not revealed to any study personnel [31]. Pertinent details were collected from the pregnant women. For women who underwent invasive test, fluorescence in situ hybridization and karyotyping was done on fetal material. Redraws were requested for these reasons: lysis, low sample volume, fetal fraction below threshold (2.8%), failed quality matrices, and algorithm-based outcomes such as low confidence results. Follow-up information was obtained from the participants till the delivery. Information on false negative outcomes was keenly sought.
Data Analysis
Sensitivity and specificity for each disorder were calculated separately using only confirmed cases after diagnostic tests or clinical evaluation after birth. Samples that received a no-call were not included. Descriptive data analysis was performed. Where applicable, the t test was used for statistical analysis, and p < 0.05 was accepted as significant.
Results
The demographic data did not differ significantly among the various groups, except; in the no-call group, the maternal weight and BMI were higher (though statistically not significant), while the fetal fraction was significantly lower as compared with the other two groups (p < 0.05) (Table 1). The overall results are depicted in Fig. 2, while details of each high-risk result are shown in Table 2. Fourteen (73.7%) of 19 high-risk NIPT calls and 323 (67.3%) of 480 of low-risk NIPT calls had a prior high risk on conventional screening, while 5 of 19 (26.3%) high risk and 157 of 480 (32.7%) low risk were in the intermediate-risk range (p > 0.05). No significant association was found between a high- or low-risk call with advanced maternal age (p > 0.05). Table 3 lists the test performance for each chromosomal abnormality, based on the cytogenetic results; combined positive predictive value (PPV) for all chromosomal abnormalities was 81.25%.
Follow-Up
Follow-up information was received from 477 of 511 (93.3%) cases. The uptake for invasive testing was 16 (84.2%) of 19 women (Table 2). Two women terminated the pregnancy without confirmatory testing (2/19, 10.5%), and one woman (1/19, 5.3%) refused invasive test. These three unconfirmed cases of trisomy 21 were suggestive of abnormality as intrauterine death was reported in one case and ultrasound abnormalities in the other two. Follow-up information was available in 452 (94.2%) of 480 low-risk cases, eleven of which were confirmed normal after invasive test (performed for other reasons) and the rest were clinically normal at birth. Follow-up information was received in 9 (81.8%) of 11 no-call cases (Table 4). No follow-up was obtained in 28 (5.8%) of 480 cases, which were excluded from statistical analysis where applicable. No false negatives were reported till the time of publication.
Redraws
Of 511 cases, 30 (5.9%) redraws were requested due to a combination of pre-analytic and post-analytic factors. The causes of redraw were: low sample volume/moderate to severe lysis (7), low confidence result (10), fetal fraction below threshold (5), failed libraries (4), failed quality control (2), ambient genotype contamination and an uninformative DNA pattern one each. Pre-analytical factors accounted for 23% of the redraws, while the laboratory processing and algorithm-based redraws were 76.7%. Redraws were received in 22 (73.3%) of 30 samples, and a definitive result was obtained in 19 (86.3%) of 22 redraws.
Turnaround Time
Most samples (97.2%) were received within 48 h. Most (88.8%) samples were reported within 12 days (Fig. 3). The average turnaround time for reporting was 7 calendar days.
Fetal Fraction (FF)
The average FF in the no-call cases (3.9%) was significantly lower than in samples that received a call (8.7%) (Welch two-sample t test, p < 0.001) (Table 1). The lowest FF for which a call was made was 3%. Five (1%) of 511 cases were found to have a FF below threshold (2.8%). Redraws were received in 3 of 5 cases with low FF; however, a call on redraw could be made only in one (Table 4). The FF was directly proportional to gestational age and inversely proportional to maternal weight (Fig. 4).
Comparison of fetal fraction with gestational age and maternal weight. Fetal fraction of cfDNA is plotted against gestational age (weeks) and maternal weight (n = 445) in kilograms A: Thick line with in box plot represents mean fetal fraction of each bin. Box plot representing quartile 2 and quartile 4, i.e., with in 25th and 75th percentile and outer hash marks representing lower and upper data. Total number of NIPT study subjects, 511. a Small dip was observed at weeks 19–20 which could be explained by the smaller sample size in this cohort. b Percentile of subjects with respect to each bin mentioned above the outer hash mark. Solid black line in each box is the average fetal fraction of that group. Outliers are represented as dark color dots next to hash marks of each bin
No-call
The overall no-call rate was 2.3% (12/511). This was defined as no result on the original sample (when no redraw is received) or on a redraw. Redraws were received in only 3 of the 11 samples, which were no-calls again (Table 4).
Ethnicity
Information was available in 453 (88.6%) of 511 samples confirming that the sample cohort consisted of pregnant women of Indian origin.
Consanguinity
Of 511 samples, consanguinity information was available in 479 (93.7%). Of these 17 (3.5%) were consanguineous, and all 17 samples received a call either the regular algorithm or QMM. Only 3 (17.6%) of 17 pregnant women who reported consanguinity yielded an uninformative DNA pattern on initial analysis. All three samples received a definite call by QMM algorithm. No-call due to uninformative DNA pattern was observed in 7 (1.3%) of 511 (all chromosome no-call—5 cases, and no-call on chromosome X—2 cases) samples. Of these, only one (partial no-call) had history of consanguinity and interestingly also screened high risk of trisomy 21. Of the samples with total uninformative DNA pattern, 80% (4/5) could be resolved on QMM algorithm.
Obesity
Obesity (defined as BMI > 30) was observed in 39 (13.9%) of 281 pregnant women. Calls were available in 38 (97.4%) cases on the first draw, of which one was high risk of trisomy 21 (confirmed on invasive testing). Only 2.7% (1/37) did not receive a call on the first run, and a redraw was requested but was not received.
Discussion
Overall, 97.9% samples received a result within 12 days. The good performance in this study is consistent with the overall performance of NIPT [8, 19, 20, 32]. The overall positive predictive value of trisomy 21, trisomy 18, trisomy 13 and monosomy X was 85.7%, which was comparable to previous publication—82.9% for the four aneuploidies [23]. However, the PPV for trisomy 21 was 80%, which was slightly lower than 90.9% in the clinical validation study [20]. If the three unconfirmed high-risk trisomy 21 cases were true positives, the PPV would increase to 84.6%. Nevertheless, this PPV is much higher than the conventional screening methods [8, 33]. The PPV for sex chromosome abnormalities including monosomy X was 75%, which is higher than that published previously—48.4% [34]. It is important to note here that PPV is not intrinsic to the test but depends on the prevalence of the condition in the tested population [35]. An overwhelming number of women (96.6%), intermediate-to-high risk on conventional screening, were low risk on NIPT. Therefore, potentially, these women avoided invasive procedures for aneuploidy confirmation [13]. The high negative predictive value is a significant benefit in terms of reassurance to pregnant women.
The average fetal fraction in Indian pregnant women was 8.3%, which is lower than the average fetal fraction range published in the Western population (10.69%) [11, 19]. These results are, however, consistent with lower fetal fractions in women of South Asian origin between 11 and 14 weeks [36]. This study did not observe negative impact of fetal fractions on NIPT calls in obese women, probably because the degree of obesity observed among Indian women is lower than in the women in other studied population, and mean BMI observed in the no-call cohort was only 27.3 [37]. The no-call rate observed was identical to the no-call rate in the validation study [20]. The no-call rates in other studies ranged between 2.9 and 8.1% [8, 19, 35]. Previous publications and guidelines have indicated that no-calls are likely to be aneuploid [11, 19]. From our no-call cases, a suspected T18 could not be confirmed. In practice, therefore, the decision to resample or proceed to invasive testing, given a no-call result, should be made based on reason for the no-call, the original fetal fraction, the probability of receiving a result on a redraw, established guidelines and the applicable regulations of prenatal testing.
Genetic counseling is mandatory, as recommended by several professional societies [27]. In the present study, a fetus with confirmed 47, XXX, was continued to term after counseling, and the baby is doing well postnatally. All other confirmed high-risk pregnancies were terminated. Unfortunately, two cases high risk on NIPT were terminated without confirmation. This undesirable outcome observed in other studies too remains a challenge [20].
Although consanguinity is cited as one of the reasons for inability to make a call in the SNP-based NIPT [21], it did not impact the current study. Limitations of the study were the small sample size, a biased cohort, as samples were preselected and a lack of complete follow-up (follow-up information was received in 93.3% subjects) and the inability to determine the cause of false positives.
Indian Council of Medical Research recommends genetic screening services to all pregnant women in the National and Family Welfare Program [38]. NIPT, in view of its strong performance in the Indian scenario, safety, accuracy and its easy extension to peripheral areas would be a valuable addition to the prenatal screening program, once the cost comes down. The experience gained in this study has enabled us to consider the implications and suggest modifications to international guidelines before applying these to India [39]. Currently, NIPT may be used in cases with high-risk results on conventional screening to avoid unnecessary invasive tests. Further reduction in cost and greater awareness would provide the benefits of this remarkable technology more widely.
References
Verma IC. Noninvasive prenatal testing: the Indian perspective. J Fetal Med. 2014;1:113–8.
Guanciali-Franchi P, Iezzi I, Palka C, et al. Comparison of combined, stepwise sequential, contingent, and integrated screening in 7292 high-risk pregnant women. Prenat Diagn. 2011;31(11):1077–81.
Russo ML, Blakemore KJ. A historical and practical review of first trimester aneuploidy screening. Semin Fetal Neonatal Med. 2014;19(3):183–7.
Padula F, Cignini P, Giannarelli D, et al. Retrospective study evaluating the performance of a first-trimester combined screening for trisomy 21 in an Italian unselected population. J Prenat Med. 2014;8(3–4):50–6.
Norton ME, Rink BD. Changing indications for invasive testing in an era of improved screening. Semin Perinatol. 2016;40(1):56–66. https://doi.org/10.1053/j.semperi.2015.11.008 (Epub 2015 Dec 24. Review).
Akolekar R, Beta J, Picciarelli G, et al. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015;45(1):16–26.
Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401.
Norton ME, Brar H, Weiss J, et al. Non-invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207(2):137.e1–8.
Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901.
Nicolaides KH, Syngelaki A, Ashoor G, et al. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. YMOB. 2012;207(374):e1–6.
Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–97.
Larion S, Warsof SL, Romary L, et al. Association of combined first-trimester screen and noninvasive prenatal testing on diagnostic procedures. Obstet Gynecol. 2014;123(6):1303–10.
Dondorp W, De Wert G, Bombard Y, et al. Non-invasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. Eur J Hum Genet. 2015;23(11):1438–50.
Zimmermann B, Hill M, Gemelos G, et al. Non-invasive prenatal aneuploidy testing at chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn. 2012;32(13):1233–41.
Samango-Sprouse C, Banjevic M, Ryan A, et al. SNP-based non- invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Prenat Diagn. 2013;33(7):643–9.
Curnow KJ, Wilkins-Haug L, Ryan A, et al. Detection of triploid, molar, and vanishing twin pregnancies by a single-nucleotide polymorphism-based noninvasive prenatal test. Am J Obstet Gynecol. 2015;212(1):79e1–9.
Wapner RJ, Babiarz JE, Levy B, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015;212(3):332.e1–9.
Gross SJ, Stosic M, McDonald-McGinn DM, et al. Clinical experience with a SNP-based noninvasive prenatal test for 22q11.2 deletion syndrome. Ultrasound Obstet Gynecol. 2016;47:177–83.
Pergament E, Cuckle H, Zimmermann B, et al. Single-nucleotidepolymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol. 2014;124(21):210–8.
Dar P, Curnow KJ, Gross SJ, et al. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am J Obstet Gynecol. 2014;211(5):527.e1–17.
Dar P, Shani H, Evans MI. Cell-f ree DNA comparison of technologies. 2016;1–13.
Ryan A, Hunkapiller N, Banjevic M, et al. Validation of an enhanced version of a single -nucleotide polymorphism-based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagn Ther. 2016;40(3):219–23.
Gregg AR, Gross SJ, Best RG, et al. Noninvasive Prenatal Screening Work Group of the American College of Medical Genetics and Genomics: ACMG statement on noninvasive pre-natal screening for fetal aneuploidy. Genet Med. 2013;15(5):395–8.
Benn P, Borrell A, Crossley J, et al. Aneuploidy screening: a position statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis, January 2011. Prenat Diagn. 2011;31(6):519–22.
Benn P, Borell A, Chiu R, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn. 2013;33(7):622–9.
Benn P, Borrell A, Chiu RW, et al. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn. 2015;35(8):725–34.
Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive pre-natal screening for fetal aneuploidy, update: a position statement of the American College. Genet Med. 2016;2016:1–10.
Tamang R, Thangaraj K. Genomic view on the peopling of India. Investig Genet. 2012;3(20):1–9.
Hamamy H. Consanguineous marriages preconception consultation in primary health care settings. J Community Genet. 2012;3(3):185–92.
Gupta V. Genomic efficiency of endogamy in India. Int J Hum Genet. 2011;11(3):199–201.
Ministry of Health and Family Welfare, Government of India: The Pre-natal Diagnostic Techniques (Regulation and Prevention of Misuse Act. 1994;57 and The Pre-natal Diagnostic Techniques (Regulation and Prevention of Misuse) Amendment Act, 2002 (No. 14 of 2003).
Ashoor G, Syngelaki A, Wagener M, et al. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206(4):322.e1–5.
Stokowski R, Wang E, White K, et al. Clinical performance of non-invasive prenatal testing (NIPT) using targeted cell-free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenat Diagn. 2015;35(12):1243–6.
Porreco RP, Garite TJ, Maurel K, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. Am J Obstet Gynecol. 2014;211(4):365.e1–12.
Lutgendorf MA, Stoll KA, Knutzen DM, et al. Noninvasive prenatal testing: limitations and unanswered questions. Genet Med. 2014;16(4):281–5.
Revello R, Sarno L, Ispas A, et al. Screening for trisomies by cell-free DNA testing of maternal blood: consequences of a failed result. Ultrasound Obstet Gynecol. 2016;47(6):698–704.
Ng M, Fleming T, Robinson M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384(9945):766–81.
Rasaily R, Mathur A, Singh MP. Feasibility of introducing genetic services in the National Family Welfare Programme in India. Indian J Med Res. 2009;130(4):404–12.
Verma IC, Dua-Puri R, Bijarnia-Mahay S. ACMG 2016 update on noninvasive prenatal testing for fetal aneuploidy: implications for India. J Fetal Med. 2017;4:1–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors Dr. Ramprasad VL, Dr. Priya Kadam, Dr. Venkataswamy E, Shruti Lingaiah, Riyaz Akhtar, Francis Kidangan, Chandran R., Kiran C., and Ravi Kumar G. R. were/are employed with Medgenome Laboratories Private Limited during the course of the project.
Ethical approval
The study was reviewed and approved by ethical review board of the institutions that participated in the study. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Prof. I.C. Verma is the HOD in the Department of Centre of Medical Genetics Sir Ganga Ram Hospital, New Delhi, India; Ratna Puri is Professor and Chairperson, Institute of Medical Genetic and Genomics, Sir Ganga Ram Hospital, New Delhi, India; Eswarachary Venkataswamy is Associate Director Q&A, Medgenome Labs Pvt. Ltd, Bengaluru, India; Tulika Tayal is Maternal and Fetal Medical Consultant, Rainbow Hospital, Hyderabad, India; Sheela Nampoorthiri is Clinical Professor, Department of Paediatric Genetics, Amrita Institute of Medical Sciences and Research Center, Kochi, India; Chitra Andrew is Senior Consultant, Obstetrics and Gynecology, Sri Ramachandra Medical College, Chennai, India; Madhulika Kabra is Additional Professor and Officer-in-Charge, Genetic Unit, All India Institute of Medical Sciences, New Delhi, India; Rashmi Bagga is Professor, Obstetrics and Gynecology, Postgraduate Institute and Medical Research Center, Chandigarh, India; Mamatha Gowda is Assistant Professor, Obstetrics and Gynecology, Jawaharlal Nehru Institute of Postgraduate Medical Education and Research, Pondicherry, India; Meenu Batra is Consultant Radiologist, Feto-maternal Medicine, CIMAR Fertility Center, Kochi, India; Sridevi Hegde is Head, Department of Medical Genetics, Manipal Hospital, Bengaluru, India; Anita Kaul is Senior Consultant and Coordinator, Indraprastha Apollo Hospital, New Delhi, India; Neerja Gupta is Assistant Professor, Division of Genetics, All India Institute of Medical Sciences, New Delhi, India; Pallavi Mishra is Biochemist, All India Institute of Medical Sciences, New Delhi, India; Jayshree Ganapathi Subramanian is NIPT Project Coordinator, Sir Ganga Ram Hospital, New Delhi, India; Shruti Lingaiah is Senior Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Riyaz Akhtar is Senior Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Francis Kidangan is Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Chandran R. is Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Kiran C is Senior Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Ravi Kumar GR is Research Associate, Medgenome Labs Pvt. Ltd, Bengaluru, India; Ramprasad V.L. is the Chief Operating Officer, Medgenome Labs Pvt. Ltd, Bengaluru, India; Priya Kadam is NIPT Project Director, Medgenome Labs Pvt. Ltd, Bengaluru, India.
Rights and permissions
About this article
Cite this article
Verma, I.C., Puri, R., Venkataswamy, E. et al. Single Nucleotide Polymorphism-Based Noninvasive Prenatal Testing: Experience in India. J Obstet Gynecol India 68, 462–470 (2018). https://doi.org/10.1007/s13224-017-1061-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13224-017-1061-9