Abstract
Trichoderma species are among the most common fungi frequently isolated as saprotrophs from free soil, soil litter, dead wood, and the rhizosphere of different crops. Four sets of species-specific primers were designed from the tef1 and rpb2 genes, in order to identify Trichoderma asperellum (tef1 gene), T. longibrachiatum (tef1 gene), T. virens (tef1 gene), and T. harzianum (rpb2 gene). Here, we report the development of a multiplex PCR assay to detect and distinguish each of these four most common Trichoderma species—viz., T. asperellum, T. harzianum, T. longibrachiatum, and T. virens—simultaneously in a single reaction through their distinct amplicons of 507, 824, 452, and 330 bp, respectively. The developed multiplex PCR technique will provide a rapid, simple, and reliable alternative to conventional methods and a new site for identification of different species of Trichoderma in a single reaction.
Similar content being viewed by others
Introduction
The genus Trichoderma is cosmopolitan and typically soil-borne or wood-decaying fungi (Samuels 1996; Esposito and da Silva 1998). It is very difficult to differentiate Trichoderma species using morphological characteristics due to the increasing number species and the paucity of morphological characteristics (Błaszczyk et al. 2011; Prameeladevi et al. 2012a). In the mid-1990s, DNA-based sequencing methods became popular and provided high divergence through base pairs of particular genes (Samuels 2006). rRNA phylogenetic markers have limited applicability for in situ diversity studies using high-throughput methods (Esposito and da Silva 1998). Molecular markers demonstrate the variation in DNA sequences within and between the species and provide the basis for precise identification. Polymerase chain reaction (PCR) methods have found widespread use for pathogen identification, and a number of PCR-based assays have been developed for use in the diagnosis and characterization of Trichoderma species (Jaklitsch 2009). PCR methods are particularly promising in light of their simplicity, specificity, and sensitivity. Prameeladevi et al. (2011) reported genus-specific primers for detection of Trichoderma based on ITS region. Gene sequences of tef1 are highly informative at the species level, as its small sequence renders easier and cheaper recovery of the sequences, and it also benefits from a paucity of repetitive regions that could produce misleading results owing to comparisons of non-orthologous sequence pairs. For these reasons, tef1 has become the marker of choice for identification of Trichoderma (Prameeladevi et al. 2012b). Sequences of the most variable region of the rpb2 gene (between domains 6 and 7) have proven important in studying the closely related species (Frøslev et al. 2005; Matheny 2005). rpb2 is a single-copy gene of large size with a modest rate of evolutionary change, and provides better phylogenetic resolution in the Ascomycota. PCR priming within these highly conserved regions allows recovery of the rpb2 genes from many different organisms for phylogenetic comparison (Liu et al. 1999). rpb2 and tef1 sequencing shows that the anatomy of the stroma is polyphyletic in nature (Chaverri and Samuels 2003). At present, the PCR method used for the identification of Trichoderma can detect only one species at a time. Hence, the objective of this study was to develop a multiplex PCR assay for simultaneous detection and differentiation of Trichoderma species by combining four species-specific primers in a single PCR reaction that could be used to detect species of Trichoderma under natural conditions.
Materials and methods
Biological material
Species of Trichoderma used in this study were obtained from the Culture Collection Centre (Indian Type Culture Collection [ITCC], New Delhi). They were grown in potato dextrose agar (PDA) slants and maintained at 4 °C for further study.
DNA isolation
A loopful of various species of Trichoderma was introduced separately into flasks containing potato dextrose broth. The cultures were incubated in a stationary state at 25 °C for 5–6 days. The mycelia were harvested after incubation, washed in sterile distilled water, and freeze-dried. The DNA of the individual fungus was extracted according to the method described by Culling (1992). The DNA pellet obtained was rehydrated by the addition of 100 μl TE buffer at 4 °C overnight. The quality and quantity of DNA were estimated using a NanoDrop spectrophotometer.
Design of species-specific primers
Species-specific primers were designed based on the sequence data of the tef1and rpb2 genes (Table 1), downloaded from the NCBI database, to specifically amplify T. asperellum, T. longibrachiatum, T. virens and T. harzianum. Gradient PCR with different annealing temperatures and different concentrations of MgCl2, dNTPs, and Taq DNA polymerase was performed for the development of the individual pairs (Prameeladevi et al. 2012b).
Multiplex PCR condition
Four species-specific primers (Table 1) designed for identification of T. asperellum, T. harzianum, T. longibrachiatum, and T. virens individually were comixed for a multiplex PCR panel. The different concentrations of four pairs of primers (5, 10, 20, 30, 50, and 100 pmol), genomic DNA template (5, 10, 15, 20, 25 30 ng) and Taq DNA polymerase (0.5, 1, 1.5, 2U; Bangalore Genei, India) were used for optimization of the PCR reaction. The reaction mixture also contained 1.5 mM MgCl2 and 0.2 mM dNTPs in a total volume of 25 μl. The reaction was carried out in a GenePro thermal cycler (Bioer Technology Co., Ltd., China) pre-equilibrated at 96 °C to provide a hot start. The gradient PCR reaction was optimized for the development of multiplex PCR, i.e., 94 °C for 1 min, followed by 30 cycles of 45 s at 94 °C, 45 s at 55–65 °C, and 1 min at 72 °C, with a final extension step for 7 min at 72 °C. PCR products were run on 1.5 % (w/v) agarose gel stained with ethidium bromide. The gel was visualized under UV radiation in a gel documentation system (Bio-Rad Laboratories, CA, USA) and digitally photographed (Canon, Tokyo, Japan).
Cloning and sequencing
To further confirm species-specific markers of each species, the amplified PCR products were eluted from agarose gels and cloned into the pGEM-T Easy Vector (Promega Corporation, WI, USA). Ligations, transformations of Escherichia coli XL blue, and plasmid amplifications were performed following standard procedures (Sambrook and Russell 2001). After cloning, positive colonies were selected, and each colony was cultured overnight in Luria-Bertani liquid medium containing ampicillin 100 mg/L. The size of cloned fragments was verified using the corresponding primers and digested with restriction enzymes in the multiple cloning sites of the vector. The recombinant plasmids were extracted using the alkaline lysis method (Ausubel et al. 2002), and the fragment was sequenced using an automated ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, CA, USA) by Bangalore Genei (Bangalore, India) using M13 universal primers.
Molecular phylogenetic analysis
In order to identify the fungal isolates, tef1and rpb2 gene sequences of four taxa of Trichoderma were downloaded from GenBank and aligned with our sequences for phylogenetic analysis. The nucleotide sequences were aligned using ClustalW multiple sequence alignment (Thompson et al. 1994). Phylogenetic analysis was carried out using the MEGA 5 software program (Tamura et al. 2011), and a neighbor-joining tree was constructed using the Kimura 2-parameter distance model (Kimura 1980). Finally, the CONSENSE program was used to construct the tree.
Results
Development of multiplex PCR
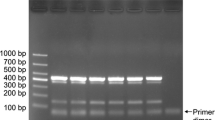
In order to increase the accuracy and efficiency of Trichoderma identification, a multiplex PCR assay was developed that could identify T. asperellum, T. harzianum, T. longibrachiatum, and T. virens based on the banding pattern of specific amplicons. In the multiplex PCR, four sets of primers (T2A F—T2A R, Th1 F—Th1 R, T1 F-T1 R, and T2 F-T2 R) were combined in a single tube for simultaneous identification of four different Trichoderma species. Under optimized conditions, multiplex PCR generated specific amplicons of expected size with their respective DNA templates (Fig. 1). The experiments were repeated a minimum of 10 times to ensure reproducibility, showing that the sensitivity of the multiplex PCR was similar to that of the single-primer-set PCR. Multiplex reactions did not produce any ambiguous or extra amplicons with non-target DNA. These results indicate that four different Trichoderma species can be identified simultaneously in a single PCR. PCR amplification was not significantly affected by changing concentrations of Mgcl2 in PCR reactions. Using optimized reaction parameters, no cross-reactivity with non-target DNA was found. No possible cross-reactions were detected with other Trichoderma species, bacteria, or viral taxa, using BLAST analysis.
Multiplex PCR showing amplified tef1 and rpb2 genes of Trichoderma species. Lanes: M−100 bp ladder, (1) tef1 and rpb2 genes of T. virens, T. asperellum, T. longibrachiatum, and T. harzianum; (2) tef1 gene of T. asperellum, T. longibrachiatum, and T. virens; (3) tef1 gene of T. asperellum and T. longibrachiatum; (4) T. virens; (5) T. asperellum; (6) T. longibrachiatum; (7) T. harzianum; (8) Other Trichoderma species (T. hamatum, T. flavofuscum, and T. fasciculatum); (9) Negative control
Multiplex PCR conditions
The multiplex PCR reaction contained 10 pmol of each species-specific primer and 10 ng of genomic DNA template of each species; 1.5 mM MgCl2, 0.2 mM dNTPs, and 0.5 U of Taq DNA polymerase were added to the reaction in total volume of 25 μl. The PCR conditions were optimized as 94 °C for 1 min, followed by 30 cycles of 45 s at 94 °C, 45 s at 60 °C, and 1 min at 72 °C, with a final extension step for 7 min at 72 °C for amplification of species-specific markers for T. asperellum, T. harzianum, T. longibrachiatum, and T. virens.
Cloning and sequencing
The specific amplicons of the species T. asperellum, T. harzianum, T. longibrachiatum, and T. virens were eluted and cloned into the pGEM-T Easy Vector, and sequenced. A BLAST search showed that the amplified products from T. asperellum (507 bp) and T. harzianum (824 bp) were 100 % homologous with the respective sequences in the GenBank database. Homologies of the products derived from T. longibrachiatum (452 bp) and T. virens (330 bp) were 98 % and 99 %, respectively. The sequences were deposited in GenBank, and accession numbers were obtained (Table 2).
Phylogenetic analysis
Nucleotype distribution suggested a close relationship between the same species of Trichoderma and the uniqueness of each of the four species. The phylogenetic tree formed four separate groups of four species, viz., T. asperellum, T. longibrachiatum, T. virens, and T. harzianum (Fig. 2). Phylogenetic analysis of combined tef1 and rpb2 sequences showed the clear separation of each individual species, with 12 sequences with high phylogenetic affinity. Phylogenetic analysis proved the specificity of particular individual species for specific identification.
Phylogenetic relationships of 12 isolates of four different Trichoderma spp. inferred by analysis of tef1 and rpb2 sequences. The neighbor-joining tree was constructed using the Kimura (1980) two-parameter model implemented in the MEGA 5.2 program. * Specific gene of a particular species used to design a specific molecular marker of individual species
Discussion
Trichoderma is one of the most successful biocontrol organisms of the ascomycetes family (Liu et al. 1999; Elad 2000a; 2000b; Consolo et al. 2012; Prameeladevi et al. 2012a; 2012b). Due to the inherent difficulties and inaccuracies associated with distinguishing Trichoderma species based on phenotypic characteristics, molecular markers have been used for authentic identification at the species level. Species-specific primers were developed for the identification of T. asperellum, T. longibrachiatum, and T. virens based on the tef1 gene and for T. harzianum using the rpb2 gene (Prameeladevi et al. 2012b). Identification of unknown Trichoderma biocontrol isolates at the species level may be important, as different species have variable resistance to multiple plant fungal diseases (Elad 2000a: 2000b; Consolo et al. 2012). In earlier studies, the tef1 gene was used for identification of Trichoderma species (Jaklitsch 2009; Jaklitsch 2011; Friedl and Druzhinina 2012; Prameeladevi et al. 2012b), as its capacity for species differentiation is superior to that of the ITS rDNA region due to the variation in sequences (Samuels et al. 1998). In fact, the ITS and tef1 sequences sometimes appeared contradictory, resulting in conflicting identification for many species complexes of fungal genera (Hoyos-Carvajal et al. 2009), and their phylogenetic markers have limited applicability for in situ diversity studies using high-throughput methods (Friedl and Druzhinina 2012). More recently, the applicability of more conserved markers such as rpb2 and chi18-5 are being tested for improved identification of fungal species complexes (Friedl and Druzhinina 2012).
In order to identify the four species of relevant interest for their biocontrol aspects, a multiplex PCR was performed using four pairs of primers. Compatible PCR conditions were standardized for the amplification of primers specific in size to four different biocontrol species, viz., T. asperellum, T. longibrachiatum, T. virens, and T. harzianum. This is the first study to combine four different species-specific primers in order to develop a multiplex PCR panel for rapid and accurate identification of multiple Trichoderma species.
Since the multiplex PCR was developed for only two Trichoderma biocontrol strains (Feng et al. 2011), an attempt was made to develop a multiplex PCR method to identify four different species of Trichoderma, as it was developed for the identification of different fungal species (Luo and Mitchell 2002; Logotheti et al. 2008).
The reconfirmation of specificity of each amplicon in a multiplex PCR is also an important step. In this study, the amplified regions for the identification of the above species were sequenced and reconfirmed. It is evident through these studies that the markers developed using tef1 and rpb2 genes could phylogenetically distinguish all four species, which is consistent with the results demonstrated by Chaverri and Samuels (2003). In the present study, molecular phylogenetic analyses supported the morphology of these four species (Prameeladevi et al. 2012a).
The multiplex PCR technique described here is a precise method of identification in that it has the ability to produce the same result given the same inputs and environmental conditions. This multiplex PCR assay reduces the length of time to obtain results of high quality, and offers high sensitivity and improved detection of the above four biocontrol strains of Trichoderma. The appropriate combination of this new multiplex PCR assay may offer accurate and rapid identification of one or more of these four Trichoderma species in a single reaction. The technique is simple enough to be implemented in any molecular laboratory for detection of these Trichoderma species under natural conditions.
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2002) Short protocols in molecular biology, 5th ed. Wiley, New York
Błaszczyk L, Popiel D, Chełkowski J, Koczyk G, Samuels GJ, Sobieralski K, Siwulski M (2011) Species diversity of Trichoderma in Poland. J Appl Genet 522:233–243
Chaverri P, Samuels GJ (2003) Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud Mycol 48:1–36
Consolo VF, Monaco CI, Cordo CA, Salerno GL (2012) Characterization of novel Trichoderma spp. isolates as a search for effective biocontrollers of fungal diseases of economically important crops in Argentina. World J Microb Biot 28:1389–1398
Culling KW (1992) Design and testing of a plant specific PCR primer for ecological evolutionary studies. Mol Ecol 1:233–240
Elad Y (2000a) Trichoderma harzianum T39 preparation for biocontrol of plant diseases – control of Botrytis cinerea, Sclerotinia sclerotiorum and Cladosporium fulvum. Biocontrol Sci Techn 10:499–507
Elad Y (2000b) Biological control of foliar pathogens by means of Trichoderma harzianum and potential modes of action. Crop Prot 19:709–714
Esposito E, da Silva M (1998) Systematic and environmental application of the genus Trichoderma. Crit Rev Microbiol 24:89–98
Feng XM, Holmberg AIJ, Sundh I, Ricard T, Melin P (2011) Specific SCAR Markers and Multiplex Real-Time PCR for Quantification of Two Trichoderma Biocontrol Strains in Environmental Samples. BioControl 56:903–913
Friedl MA, Druzhinina IS (2012) Taxon-specific metagenomics of Trichoderma reveals a narrow community of opportunistic species that regulate each other’s development. Microbiology 158:69–83
Frøslev TG, Matheny PB, Hibbett DS (2005) Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): a comparison of RPB1, RPB2, and ITS phylogenies. Mol Phylogenet Evol 37:602–618
Hoyos-Carvajal L, Orduz S, Bissett J (2009) Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet Biol 46:615–631
Jaklitsch WM (2009) European species of Hypocrea Part I. The green-spored species. Stud Mycol 63:1–91
Jaklitsch WM (2011) European species of Hypocrea. Part II: species with hyaline ascospores. Fungal Divers 48:1–250
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotides sequences. J Mol Evol 2:87–90
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 16(12):1799–1808
Logotheti M, Kotsovili-Tseleni A, Arsenis G, Legakis NI (2008) Multiplex PCR for the discrimination of A. fumigatus, A. flavus, A. niger and A. terreus. J Microb Meth 76:209–211
Luo G, Mitchell TG (2002) Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol 40:2860–2865
Matheny PB (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Mol Phylogenet Evol 35:1–20
Prameeladevi T, Kamil D, Prabhakaran N, Pandey P (2011) Development of genus specific rDNA based marker for detection of Trichoderma species. J Mycol Plant Pathol 41:600–604
Prameeladevi T, Prabhakaran N, Kamil D, Borah JL, Pandey P (2012a) Development of species specific markers for detection of Trichoderma species. Vegetos 2502:207–217
Prameeladevi T, Prabhakaran N, Kamil D, Pandey P, Borah JL (2012b) Characterization of Indian native isolates of Trichoderma spp. and assessment of their bio-control efficiency against plant pathogens. Afr J Biotechnol 1185:15150–15160
Sambrook J, Russell DW (2001) Molecular cloning a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Samuels GJ (1996) Trichoderma: a review of biology and systematics of the genus. Mycol Res 100:923–935
Samuels GJ (2006) Trichoderma: systematics, the sexual state, and ecology. Phytopathology 96:195–206
Samuels GJ, Petrini O, Kuhls K, Lieckfeldt E, Kubicek CP (1998) The Hypocrea schweinitzii complex and Trichoderma sect. Longibrachiatum. Stud Mycol 41:1–54
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Acknowledgments
The authors thank the Head of the Division of Plant Pathology, Indian Agricultural Research Institute, New Delhi, for help in various aspects of this study. Financial support from the Department of Biotechnology of the Government Of India, New Delhi, is gratefully acknowledged.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prabhakaran, N., Prameeladevi, T., Sathiyabama, M. et al. Multiplex PCR for detection and differentiation of diverse Trichoderma species. Ann Microbiol 65, 1591–1595 (2015). https://doi.org/10.1007/s13213-014-0998-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0998-5