Abstract
Bevacizumab is the standard treatment for colorectal cancer (CRC) in the advanced stage. However, poor diagnosis identified due to the bevacizumab resistance in many CRC patients. Previous studies have found that CRC stem cells (CCSCs) and interleukin 22 (IL-22) are involved in the resistance of bevacizumab, however, the mechanism of remains unclear. In this study, we established the bevacizumab drug-resistant cell line HCT-116-R by concentration gradient method, and the cell viability was detected by CCK-8 assay. The resistance of bevacizumab in CRC cell lines HCT-116-R was identified by characterizing epithelial-mesenchymal transition (EMT). Additionally, HCT-116-R cell lines were isolated from CCSCs and their tumorigenicity was validated in nude mice. We observed that that compared with the matched group, the expression of IL-22, IL-22R, STAT3, and GP130 in drug-resistant cells increased distinctly, with blocked IL-22 cells were successfully constructed by lentiviral interference. The level of proteins in stem cell landmarks (EpCAM, CD133), and stem cell landmarks (Oct4, Sox2) was identified by western blotting. Furthermore, the IL-22 role was evaluated by xenograft model. We found that short hairpin RNA (shRNA) suppression of IL-22 expression can restore the sensitivity of drug-resistant CCSCs to bevacizumab, Moreover, xenograft tumor models show that suppression of IL-22 can increase the anti-tumor influence of bevacizumab. In summary, we demonstrated that CCSCs play a major part in bevacizumab-resistant CRC. Inhibiting the signaling pathway of IL-22/STAT3 can improve the anti-tumor influence on bevacizumab in vitro and in vivo. Thus, IL-22 may represent a new anti-bevacizumab target in CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vascular endothelial growth factor receptor (vascular endothelial growth factor, VEGFR) is a tyrosine kinase receptor (Wang et al. 2020). The activated VEGFR-1 is related to the release of growth factors, the migration of macrophages, and the recruitment of vascular endothelial progenitor cells (Benlahfid et al. 2018). The growth and metastasis of tumors require the formation of blood vessels and lymphatic vessels (Zhang et al. 2021). In the absence of vascular support, tumors may become necrosis or even apoptotic (Shah 2022). Therefore, anti-tumor angiogenesis drugs came into being, and bevacizumab, which targets VEGF, first entered people's field of vision.
Bevacizumab is a recombinant humanized monoclonal antibody with the trade name Avastin (Rosenberg et al. 2021). It is the first anti-tumor angiogenesis drug approved for marketing in the United States for the first-line medicine of metastatic colorectal cancer (CRC). At present, the FDA has approved bevacizumab combined with chemotherapy as the first-line medicine for metastatic renal cell carcinoma, breast cancer, metastatic CRC, and advanced non-small cell lung cancer (Chou et al. 2021; Fang et al. 2021; Yoo et al. 2021). Bevacizumab binds to VEGF and prevents it from activating VEGFR-1 and VEGFR-2. Early application of Avastin can make tumor blood vessels shrink rapidly, and at the same time make the surviving tumor vascular structure change from abnormal to normal, reduce intratumor pressure, and help chemotherapy drugs better reach the tumor (Ottaiano et al. 2021).
Although anti-angiogenesis drugs, mainly Avastin, have achieved optimistic therapeutic effects in the clinic, they cannot cure tumors after all. The decline in efficacy is mostly due to drug resistance. Therefore, how to overcome drug resistance has become the key to curing tumors. Hamdollah Zadeh et al. (2015) found that selective cleavage of some RNA-binding proteins in colorectal tumors can regulate the expression of specific VEGF, which makes the mechanism of angiogenesis more complicated. Ishima et al. (2015) studies the effect of nitric oxide on autophagy and found that nitric oxide may inhibit autophagy through the JNK1 or mTOR pathway. In in vivo experiments, the nitric oxide inhibitor SNO-HAS inhibited hypoxia-mediated autophagy. In in vitro tests, SNO-HAS combined with bevacizumab more effectively controlled lung metastases. At the same time, Liang et al. (2015) explored the relationship between Bevac and autophagy. They found that the process of Bevac treatment promotes autophagy and promotes the high expression of the interferon regulatory factor 1 gene, which inhibits autophagy and promotes cell apoptosis. This may be related to the resistance mechanism of bevacizumab treatment. However, the key mechanism of the universality of bevacizumab has not been revealed yet. Here, we found that the bevacizumab resistance of CCSC depends on IL-22 produced by CRC. Bevacizumab resistance is regulated by the IL-22/STAT3 signaling cascade, and these effects are inhibited by IL-22 shRNA. These mechanisms may play a major role in the targeted drug resistance of CRC patients treated with bevacizumab.
Materials and methods
Reagents and cell lines
The purchased CRC cell line was HCT116 (this cell line is a kind of CRC cell line that can fully simulate the physiology of CRC cells. The cells were cultured in DMEM-F12 medium (1.2 mmol/L thioglycerol, 2 mmol/L glycine, 100 U/mL penicillin, 100 mg/L streptomycin, 10% fetal cattle Serum) continuously for 5 days, then planted in a six-well plate culture flask, and placed in a 37 °C, 5% CO2 incubator. When the cell shape has retracted from the extended state to the oval shape, add DMEM-F12 medium to plant trypsin digestion, and ensure the cells were uniform after repeated pipetting.
Generation of bevacizumab-resistant CRC Cells
DMEM medium containing 10% fetal bovine serum for HCT116 cells, was gradually added with 1, 2, 4, 8, and 16 μmol/L bevacizumab to stimulate, and each concentration lasted for 1 week before replacing the next higher concentration, until the addition of 16 μmol/L bevacizumab for a continuous culture for 1 week. HCT116-R cells were subsequently routinely cultured with a medium containing low concentration (2 μmol/L) of bevacizumab to maintain their resistance to bevacizumab.
Beading and grouping of CCSCs
The immunomagnetic bead method was implemented to separate CD133-positive cells. The logarithmic growth of CRC cells was taken, washed and suspended with PBS to prepare 300 μL cell suspension. 100 μL of FcR blocker was added to the cells, 100 μL of CD133-labeled micromagnetic beads was added to complete the separation of cell subpopulations, mixed well and was let to stand for 30 min at a temperature of 4 °C. 1.0–2.0 mL buffer was added to the cells after mixing, and centrifuged for 10 min at a speed of 5000 r/min, and 500 μL of buffer was added to make the cell suspension; a suitable sorting column was placed in the differential separator, cells were slowly added (to avoid air bubbles) to complete the collection of CD133-positive cells. 1 mL MACS buffer was added to the sorting column and the sorting column was used, equipped with a piston to push out the positive cells labeled with magnetic beads. 100 μL of CD133 positive cells was taken; the cell suspension was adjusted to 2 × 105, and 10 μL of 133/2-PE was added and mixed well. It was then incubated for 10 min in the dark and a buffer was added and centrifuged for 10 min at a speed of 3000 r/min.
Cell proliferation assay
103–104 cells were inoculated in each well; cell suspension after routine digestion of the cells was prepared. The cells were inoculated in a 96-well plate, and were placed in an incubator. After the cells adhered to the wall, a pipette was used to remove the cell culture medium in each well, and continued for 72 h, 5 g/L MTT was added to each well, and the culture was continued for 4 h. 150 μL DMSO was added to each well and after mixing for 15–20 min, the number of cell proliferation was measured and observed under an inverted microscope.
Cell transfection
The isolated and cultured CD133-positive cells were inoculated in a six-well plate at a density of 5 × 106. When the cells grew and converged to 70.0%, DMEM/F12 cell culture medium was used without FBS, and 100 pmol siRNA was added to the cells. FAM siRNA and 50 mL of DMEM-F12 medium without FBS were incubated at a room temperature for 20 min, and then placed in a 37 °C 5% CO2 incubator. The other two purchased CRC cell lines were taken as the normal cell group and the matched group after empty plasmid transfection.
Quantitative real-time reverse transcription PCR
Trizol method was implemented to extract total RNA in the specimen tissue, and a 20 μL reaction system was prepared according to the instructions of the kit (purchased from Beijing Tiangen Biochemical Technology Co., Ltd.), reacted at 37 °C for 60 min, and the mRNA was reverse transcribed into cDNA. The PCR reaction system was confirmed to be 25 μL according to the kit instructions, and the following E-cadherin primer sequences were used: upstream 5′-CCAGCTGTCCACCTCGACCT-3′, downstream 5′-GATGAGAGGCAGCAAGATGGAC-3′; the upstream primer for the amplification of the internal reference GAPDH gene is CCCTCC GCTTCG-CCCTCCGCTTCG-3′, the downstream primer sequence is 5′-GCTGGCGACGCAAAAGA-3′. PCR amplification conditions are 50 °C for 2 min and 95 °C for 10 min. A total of 40 cycles were performed for 15 min and 60 °C for 1 min. After the reaction, the amplification curve and the dissolution curve were recorded. Using β-actin as an internal reference, the relative expression of CD68 was quantified using the 2−ΔΔCT method.
Western blotting
The concentration gel and separation gel required for electrophoresis were prepared according to the concentration of 10% and 5%, and the total protein loading amount was 30 μg per well. The extraction method of total cell protein, then the appropriate amount of protein lysate was added to the collected cell pellet by centrifugation, and shaken on ice for 10 min (after shaking for 30 s, place on ice for 15 s, repeat), at 12,000 r/min at 4 °C. The supernatant was collected in the centrifuge tube and mixed with sample buffer evenly. After boiling for 5 min, the prepared total cell protein sample was stored at − 80 °C. After electrophoresis at 120 V constant voltage for 60 min, it was electrophoresed at 350 mA constant current for 60 min. The primary antibody was implemented at a dilution ratio (ABCG2, abcam, 11,000; E-cadherin, abcam, 11,000; Vimentin, abcam, 11,000; β-actin, abcam, 11,0000; STAT3, abcam, 11,000; IL-22R, abcam, 11,000; CD133, abcam, 11,000; Oct4, abcam, 11,000; Sox2, abcam, 11,000; AFP, abcam, 11,000; G-6-P, abcam, 11,000).
The primary antibody was incubated overnight at 4 °C in a shaker; the secondary antibody was implemented at a dilution ratio of 15,000 and incubated for 1 h in the dark. β-Actin was implemented as the internal reference protein. ImageJ software was implemented to investigate the relative grayscale of western blotting exposure bands.
IHC
Immunohistochemical kit, DAB enzyme substrate chromogenic agent, mouse anti-human Ki67 monoclonal antibody, and rabbit anti-human CD33 monoclonal antibody were purchased from AbCAM (1:400). The steps were performed according to the instructions. Paraffin sections were routinely dewaxed, hydrated, and microwave antigenic repair, followed by the addition of primary antibody, secondary antibody, DAB color rendering, and hematoxylin restain. The radiographs were reviewed by two pathologists independently. According to the staining results of tumor and normal cell nuclei, the expression of > was negative in 50% of tumor cells without staining, and the expression of > was positive in 50% of tumor cells and normal cells.
Subcutaneous xenograft tumor model
Nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The cells were centrifuged and the cells were collected. The PBS buffer was used to make the cells into a single-cell suspension with a specific concentration, the single-cell suspension was mixed with Matrigel ׃1 in equal proportions and was placed on ice. When the nude mouse was injected, the mouse was fixed with the left hand, and the syringe was held with the right hand. The back of the thigh was gently pierced on the left and right sides of the nude mouse under the skin. 100 μL of Matrigel-containing cell suspension was injected on each side. The needle gently was gently pulled to avoid leakage of liquid. After 1 week, the tumor formation of nude mice was observed every day, and the nude mice were dissected when the average subcutaneous tumor volume reached 1 cm3. Before the observation, the mice were anesthetized by intraperitoneal injection of pentobarbital sodium at 35 mg/kg, and the corresponding amount of 15 mg/mL luciferin potassium solution was injected into each mouse intraperitoneally at the concentration of 10 μL/g body weight. After 5 min, the tumor growth was observed by in vivo imaging, and the tumor fluorescence value was quantitatively analyzed. This study was approved by the animal ethics committee of the second hospital of Hebei Medical University.
Statistical analysis
Statistical software SPSS 17.0 was used to perform data analysis, and the data were expressed as mean ± SD. Multiple groups were compared by analysis of variance (ANOVA), two groups were compared by independent sample t-test, and the inhibition rate curve of cell activity was performed using Graph Prism software, A p value less than 0.05 was considered significant.
Results
Established bevacizumab-resistant colorectal cancer cell lines HCT-116-R
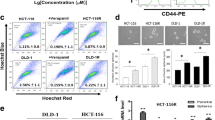
To study the mechanism of bevacizumab resistance, we implemented HCT-116 cells to generate bevacizumab-resistant CRC cells. HCT-116-R were fusiform, with weak contact between cells and easy to remove from the surface of the culture flask. The IC50 of the parental HCT-116 cell line is 6.235 μM, which was consistent and the drug-resistant cell line, the IC50 (10.588 μM) displayed a higher concentration of metastasis than the parent (Fig. 1A). In addition, compared with HCT-116 and HT-29 cells, HCT-116-R cells displayed typical characteristics of EMT, restrained expression of E-cadherin, multidrug resistance gene (ABCG2), and Vimentin expression was up-regulated (Fig. 1B–D). These outcomes manifest that HCT-116 cells resistant to bevacizumab have been successfully established.
Isolation and identification of anti-HCT-116-RCSCs
EpCAM, and CD133 were common CCSC landmarks; the results of the qRT-PCR experiment showed that the mRNA level of stem cell landmarks (EpCAM+, CD133+) in positive cells was higher than in the negative cells (Fig. 2A). Flow cytometry was performed to sort cells expressing CD133 and EpCAM negative or positive cells from HCT-116-R, subsequently, apoptosis cell proliferation tests were performed, but there was no notable difference between the positive and negative cells (Fig. 2B, C). To further study the tumorigenicity of stem cells in vivo, CCSCs, HCT-116-R cells were inoculated into nude mice for xenograft tumors. The outcomes displayed that CCSC-derived tumor volume was more massive than that of HCT-116-R-derived tumors (Fig. 2D). CCSC-derived tumors have more expressions of CD133 and Ki67 (Fig. 2E), indicating that CCSC has more aggressive growth of xenograft tumor.
Separation and identification of CCSC-R. A qPCR to detect the sorted negative and positive. B, C Apoptosis and cell proliferation were used to compare the negative and positive cells. D Nude mouse xenotransplantation experiment. E IHC of the mouse-derived tumor. R+: positive (EpCAM+, CD133+) drug-resistant CCSCs, R−: negative (EpCAM−, CD133−) drug-resistant CCSCs, B+: positive (EpCAM+, CD133+) CCSCs, B−: negative (EpCAM−, CD133−) CCSCs (***P < 0.005, ****P < 0.0001)
High expression of IL-22/STAT3 signaling pathway protein in drug-resistant CCSCs
The expressions of GP130, IL-22, IL-22R, and STAT3 in drug-resistant CCSCs were higher than in the control cells (CCSCs) (Fig. 3A). Compared with CCSCs, the STAT3 protein level in drug-resistant CCSCs was triggered (Fig. 3B, C). The outcomes displayed that in drug-resistant CCSCs, the IL-22/STAT3 signaling pathway was distinctly activated. Blocking IL-22 by shRNA can restrain IL-22R (Fig. 3D–F), key genes related to pluripotency (Oct4, Sox2), epithelial cell adhesion molecule (EpCAM), and stem cells (CD133). It also observes upregulated landmarks of hepatocyte differentiation (AFP, glucose-6-phosphate) (Fig. 4A–C). These outcomes manifest that the suppression of IL-22/STAT3 restrains the gene expression related to drug-resistant CCSCs stem cells.
Blocking the signaling pathway of IL-22/STAT3 in drug-resistant CCSCs. A The levels of GP130 mRNA, IL-22R, IL-22, and STAT3 in CCSCs and drug-resistant CCSCs. B, C The STAT3 protein expression in CCSCs and drug-resistant CCSCs. D–F It was confirmed by qPCR and WB that the IL-22 signal was blocked in drug-resistant CCSCs
Blockade of bevacizumab combined with IL-22 triggers the anti-tumor influence on xenograft tumors
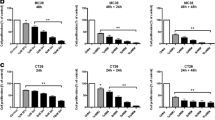
To clarify the role of IL-22 in CCSCs, the exploration of tumorigenicity was conducted with a tumor xenograft model in nude mice. The outcomes displayed that the tumors in the shRNA IL-22 group grew more slowly and were smaller than the matched group (Fig. 5A, B). The tumor volume in the bevacizumab combined shRNA-IL-22 group was smaller than that in the shRNA-IL-22 group (Fig. 5B). The levels of IL-22 and STAT3 in tumor tissues were determined by Western blotting (Fig. 5C, D). CCSC landmarks (EpCAM, CD133), stem cell landmarks (Oct4, Sox2) and angiogenic factors (VEGF, VEGFR) were explored in tumor tissues of nude mice. The outcomes displayed that the expression of CCSC landmarks, stem cell landmarks and angiogenesis factors in the shRNA IL-22 group was restrained (Fig. 6A, B). The above outcomes illustrate that IL-22 gene knockdown can restrain the expression of stem cell-related genes, inhibit the survival of stem cells and tumorigenesis, and trigger the sensitivity of HCT-116-R cells to bevacizumab. It plays an important role in maintaining the stem cell function of stem cells.
Developing subcutaneous xenografts in nude mice by drug-resistant CCSCs blocks the signaling pathway of IL-22/STAT3. A Tumor images of nude mice by subcutaneous xenografts. There were 5 mouse tumors in each treatment group. B Nude mouse xenotransplantation experiment. C Growth Transplanted tumor growth cure from day 1 to11 in each treatment group. D, E Analyzing the expression of STAT3 protein, and IL-22R in transplanted tumors, confirms the blocking of IL-22/STAT3 signaling pathway
Discussion
IL-22 was named IL-10-related T cell-derived inducible factor (IL-TIF) when it was discovered. Due to its unique function, it has become the most in-depth study of IL-10 family members (Koltsova and Grivennikov 2014). IL-22 is mainly secreted by adaptive immune cells (CD4 positive T cells, CD8 positive T cells, etc.) and innate immune cells (LTi cells, NK cells, etc.) (Mossner et al. 2020). IL-22 also plays an important role in the occurrence and development of tumors. IL-22 through STAT3-mediated activation of histone 3 lysine 79 (H3K79) methylase destroys the telomere silencing factor DOT1L and enhances the tumorigenicity of colon cancer (Kryczek et al. 2014). The main sources of IL-22 in colon cancer are Th22 and ILC3 cells. Ablative treatment of ILCs in colon cancer mouse models can significantly reduce tumor and adenoma formation, and IL-22 antibody treatment can improve colitis symptoms and reduce tumor burden (Yanagisawa et al. 2021).
As a monoclonal immunoglobulin G antibody, bevacizumab mainly binds to VEGF and prevents it from activating VEGFR-1 and VEGFR-2. Early application of bevacizumab can rapidly shrink tumor blood vessels, and at the same time turn the surviving tumor vascular structure from abnormal to normal, reduce intratumor pressure, and help chemotherapeutics to better reach the tumor to play a role; its continuous application can also inhibit tumors (Yan et al. 2021). We found that the bevacizumab resistance of CCSC depends on IL-22 produced by CRC. Bevacizumab resistance is regulated by the IL-22/STAT3 signaling cascade, and these effects are inhibited by IL-22 shRNA.
Studies have found that there is a part of endothelial cells in tumors, which not only have the functional characteristics of normal endothelial cells, but also have specific gene mutations from the tumor. Recent research results suggest that this endothelial cell with the phenotypic characteristics of tumor cells can be transformed from tumor cells or tumor stem cells (Bruno et al. 2021). Wei et al. (2019) found that glioblastoma stem cells contain a small group of endothelial precursor cells, which can be further differentiated into mature endothelial cells, and the endothelial cells have the same genetic characteristics as the tumor cells from which they are derived. Both internal and external experiments have confirmed that glioblastoma stem cells can differentiate into tumor endothelial cells. Zamani et al. (2020) found that the blood vessels of mouse glioblastoma express both endothelial cell antigens and tumor cell markers. The results suggest that glioblastoma can differentiate into tumor endothelial cells. In addition, there are many cases of ovarian cancer, lung cancer, melanoma, etc. Differentiation of tumor endothelial cells has also been observed in various tumors. However, the differentiation mechanism of tumor endothelial cells is still unclear. It has been reported that inhibition of signals or administration of secretion inhibitors can block the differentiation of glioma stem cells into endothelial precursor cells. The differentiation of mature endothelial cells has no effect; while inhibiting the signal or giving bevacizumab treatment interferes with the differentiation process of endothelial precursor cells into mature endothelial cells but has no effect on the differentiation of glioma stem cells into endothelial precursor cells (Rocha et al. 2018). Cancer stem cells acquired endothelial cell phenotype under the induction of endothelial culture medium, and human endothelial cells were also detected in the center of the transplanted tumor, suggesting that the transformation of endothelial cells requires stimulation. However, the study found that ovarian cancer stem cells differentiated tumor-forming endothelial cells do not depend on it but require activation of signaling pathways (Cui et al. 2018). In addition, studies have also found that tumor endothelial cells often appear in hypoxic areas deep in tumors, and hypoxia can promote tumor cell formation and express endothelial cell-related markers (Wang et al. 2017). It is found that the center of the tumor is richer in endothelial cells derived from tumor stem cells than on the surface of the tumor. The above results indicate that hypoxia plays an important role in the transformation of tumor endothelial cells (Mendonça et al. 2019). Our research shows that by blocking IL-22 signal transduction, CCSCs are weakened, and animal experiments have also proved this expression.
Some limitations exist in this study. First, we did not select more cell lines for validation, and this is an aspect for improvement in future work. Additionally, we have only discovered the preliminary mechanism of IL-22, ongoing studies analyzing shifts more comprehensive exploration of its mechanism at the proteomic level. In summary, our study preliminarily proved that the tumor angiogenesis-related factors that block IL-22 signal transduction are significantly reduced, and tumor growth is inhibited. Therefore, in the bevacizumab-resistant node, targeting CSCs to inhibit the IL-22/STAT3 signaling pathway in rectal cancer may be a promising treatment method and a personalized treatment method for patients.
Data availability
Available upon request.
References
Benlahfid M, Traboulsi W, Sergent F et al (2018) Endocrine gland-derived vascular endothelial growth factor (EG-VEGF) and its receptor PROKR2 are associated to human colorectal cancer progression and peritoneal carcinomatosis. Cancer Biomark 21:345–354. https://doi.org/10.3233/CBM-170499
Bruno S, Herrera Sanchez MB, Chiabotto G et al (2021) Human liver stem cells: a liver-derived mesenchymal stromal cell-like population with pro-regenerative properties. Front Cell Dev Biol 9:644088
Chou HH, Chen WC, Yang LY et al (2021) Postoperative adjuvant dose-dense chemotherapy with bevacizumab and maintenance bevacizumab after neoadjuvant chemotherapy for advanced ovarian cancer: a phase II AGOG/TGOG trial. Eur J Obstet Gynecol Reprod Biol 262:13–20. https://doi.org/10.1016/j.ejogrb.2021.04.017
Cui C, Chen X, Liu Y et al (2018) Galactosyltransferase v activates notch1 signaling in glioma stem-like cells and promotes their transdifferentiation into endothelial cells. J Biol Chem 293:2219–2230. https://doi.org/10.1074/jbc.RA117.000682
Fang C, Lin J, Zhang T et al (2021) Metastatic colorectal cancer patient with microsatellite stability and BRAFV600E mutation showed a complete metabolic response to PD-1 blockade and bevacizumab: a case report. Front Oncol 11:652394. https://doi.org/10.3389/fonc.2021.652394
Hamdollah Zadeh MA, Amin EM, Hoareau-Aveilla C et al (2015) Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol Oncol 9:167–178. https://doi.org/10.1016/j.molonc.2014.07.017
Ishima Y, Inoue A, Fang J et al (2015) Poly-S-nitrosated human albumin enhances the antitumor and antimetastasis effect of bevacizumab, partly by inhibiting autophagy through the generation of nitric oxide. Cancer Sci 106:194–200. https://doi.org/10.1111/cas.12577
Koltsova EK, Grivennikov SI (2014) IL-22 gets to the stem of colorectal cancer. Immunity 40:639–641
Kryczek I, Lin Y, Nagarsheth N et al (2014) IL-22+CD4+ T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 40:772–784. https://doi.org/10.1016/j.immuni.2014.03.010
Liang J, Piao Y, Henry V et al (2015) Interferon-regulatory factor-1 (IRF1) regulates bevacizumab induced autophagy. Oncotarget 6:31479–31492. https://doi.org/10.18632/oncotarget.5491
Mendonça L, Trindade A, Carvalho C et al (2019) Metastasis is impaired by endothelial-specific Dll4 loss-of-function through inhibition of epithelial-to-mesenchymal transition and reduction of cancer stem cells and circulating tumor cells. Clin Exp Metastasis 36:365–380. https://doi.org/10.1007/s10585-019-09973-2
Mossner S, Kuchner M, Modares NF et al (2020) Synthetic interleukin 22 (IL-22) signaling reveals biological activity of homodimeric IL-10 receptor 2 and functional cross-talk with the IL-6 receptor gp130. J Biol Chem 295:12378–12397. https://doi.org/10.1074/jbc.ra120.013927
Ottaiano A, Scala S, Santorsola M et al (2021) Aflibercept or bevacizumab in combination with FOLFIRI as second-line treatment of mRAS metastatic colorectal cancer patients: the ARBITRATION study protocol. Ther Adv Med Oncol 13:1758835921989223. https://doi.org/10.1177/1758835921989223
Rocha R, Torres Á, Ojeda K et al (2018) The adenosine A3 receptor regulates differentiation of glioblastoma stem-like cells to endothelial cells under hypoxia. Int J Mol Sci 19:1228. https://doi.org/10.3390/ijms19041228
Rosenberg JE, Ballman KA, Halabi S et al (2021) Randomized Phase III Trial of gemcitabine and cisplatin with bevacizumab or placebo in patients with advanced urothelial carcinoma: results of CALGB 90601 (Alliance). J Clin Oncol 39:2486–2496. https://doi.org/10.1200/JCO.21.00286
Shah AA (2022) Tumor Angiogenesis and VEGFR-2: mechanism, pathways and current biological therapeutic interventions. Curr Drug Metab 22:50–59. https://doi.org/10.2174/18755453mtewxnzq0x
Wang R, Bhattacharya R, Ye X et al (2017) Endothelial cells activate the cancer stem cell-associated NANOGP8 pathway in colorectal cancer cells in a paracrine fashion. Mol Oncol 11:1023–1034. https://doi.org/10.1002/1878-0261.12071
Wang JX, Wu HL, Zhu M, Zhou R (2020) Role of anti-epidermal growth factor receptor therapy compared with anti-vascular endothelial growth factor therapy for metastatic colorectal cancer: an update meta-analysis of randomized clinical trials. Pathol Oncol Res 26:159–166. https://doi.org/10.1007/s12253-017-0365-5
Wei Y, Shi D, Liang Z et al (2019) IL-17A secreted from lymphatic endothelial cells promotes tumorigenesis by upregulation of PD-L1 in hepatoma stem cells. J Hepatol 71:1206–1215. https://doi.org/10.1016/j.jhep.2019.08.034
Yan J, Yu J, Yuan S et al (2021) Musculin is highly enriched in Th17 and IL-22-producing ILC3s and restrains pro-inflammatory cytokines in murine colitis. Eur J Immunol 51:995–998
Yanagisawa H, Sugimoto M, Miyashita T (2021) Mathematical simulation of tumour angiogenesis: angiopoietin balance is a key factor in vessel growth and regression. Sci Rep 11:419. https://doi.org/10.1038/s41598-020-79824-8
Yoo C, Kim JH, Ryu MH et al (2021) Clinical outcomes with multikinase inhibitors after progression on first-line atezolizumab plus bevacizumab in patients with advanced hepatocellular carcinoma: a multinational multicenter retrospective study. Liver Cancer 10:107–114. https://doi.org/10.1159/000512781
Zamani ARN, Avci ÇB, Ahmadi M et al (2020) Estradiol modulated colorectal cancer stem cells bioactivity and interaction with endothelial cells. Life Sci 257:118078. https://doi.org/10.1016/j.lfs.2020.118078
Zhang Y, Liu J, Zou T et al (2021) DPSCs treated by TGF-β1 regulate angiogenic sprouting of three-dimensionally co-cultured HUVECs and DPSCs through VEGF-Ang-Tie2 signaling. Stem Cell Res Ther 12:281. https://doi.org/10.1186/s13287-021-02349-y
Funding
This project was supported by the Research project plan of Medical Science in Hebei Province. No. 20210875.
Author information
Authors and Affiliations
Contributions
XQ: conceptualization; investigation; methodology; writing a original draft. HR: data curation; analysis; review and editing. LY: analysis; review and editing. LL: visualization; supervision; resources review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Research involving human participants and/or animals
This study was approved by the animal ethics committee of the second hospital of Hebei Medical University.
Informed consent
Not applicable.
Ethics approval and consent to participate
This study was approved by the animal ethics committee of the second hospital of Hebei Medical University.
Consent for publication
All authors agree for publication.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qin, X., Ruan, H., Yuan, L. et al. Colorectal cancer tumor stem cells mediate bevacizumab resistance through the signal IL-22-STAT3 signaling pathway. 3 Biotech 13, 327 (2023). https://doi.org/10.1007/s13205-023-03742-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03742-5