Abstract
The banana bract mosaic virus (BBrMV) is a major virus affecting bananas and plantains. Banana being propagated vegetatively, there arises a high risk of virus transmission through planting materials. Available molecular detection technique like the Reverse Transcriptase Polymerase Chain Reaction needs post-amplification sample handling, predisposing to sample cross contamination. A one-step Reverse Transcription-LoopMediated Isothermal Amplification (RT-LAMP) assay coupled with colorimetric detection was optimised for easy and quick detection of BBrMV in banana. The viral coat protein gene was amplified under isothermal conditions at 65 ºC. The RT-LAMP assay was optimised with respect to concentrations of MgSO4, dNTP, Bst polymerase enzyme and HNB dye. The total RNA purified from symptomatic samples was directly amplified under isothermal conditions by including 100 U M-MLV reverse transcriptase and 20 U RNasin® plus RNase inhibitor in the reaction. With the addition of 120 µM of Hydroxy Naphthol Blue (HNB) dye in the RT-LAMP reaction, the BBrMV-positive samples had a colour change from violet to sky blue after the reaction. The RT-LAMP assay detected BBrMV in 0.1 pg of total RNA isolated from symptomatic plants. Molecular characterisation of RT-LAMP products was done using restriction profiling and sequence analysis. The RT-LAMP assay was validated using field-collected banana leaf samples. The assay successfully detected the virus from symptomatic samples while the healthy samples showed no amplification. Samples sourced from banana plants with symptoms of banana bunchy top virus, banana streak virus and cucumber mosaic virus tested negative in the RT-LAMP assay, thus ensuring the specificity of the assay.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Virus diseases are a cause of concern to banana farmers world over with about 20 different viruses infecting banana, the major ones being the banana bunchy top virus (BBTV), banana bract mosaic virus (BBrMV), banana streak virus (BSV) and cucumber mosaic virus (CMV) (Tripathi et al. 2016). The banana bract mosaic disease, caused by the BBrMV is a major disease in banana and plantain. It was first reported in Philippines in the year 1979 (Magnaye and Espino 1990; Rodoni et al. 1997) and later from Sri Lanka, India, Vietnam, and Thailand (Magnaye and Espino 1990; Bateson and Dale 1995; Rodoni et al. 1997). In India, the bract mosaic disease was first recorded in banana cultivar Nendran in Kerala as Kokkan disease (Samraj et al.1966) with unknown etiology. Later, the causative agent of the Kokkan disease was identified as BBrMV (Rodoni et al. 1997; Singh and Selvarajan 2000). In India, up to 40% yield loss in bananas has been attributed to this disease (Cherian et al. 2002; Thangavelu et al. 2000). The disease significantly reduces the yield of the crop with maximum yield reduction in banana cultivar Robusta, followed by Nendran (Cherian et al. 2002). The bract mosaic disease is characterised by purplish to reddish streaks on bracts, peduncles, pseudostem and fruits (Selvarajan and Jeyabaskaran 2006). During severe infection, there are streaks along the leaf veins and chlorosis of leaves (Selvarajan and Jeyabaskaran 2006). Very severely affected plants often fail to flower and die with stunted growth and pseudostem necrosis. Apart from its spread through vegetative propagules, aphid vectors like Pentalonia nigronervosa, Aphis gossypii and Rhopalosiphum maidis also transmit BBrMV (Magnaye and Espino 1990).
The BBrMV is a single-stranded RNA virus, of the genus Potyvirus (Cherian et al. 2002). Molecular detection methods like RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) and immunological detection based on ELISA(Enzyme-Linked Immuno-Sorbent Assay) are generally used to detect BBrMV (Dassanayake 2001; Bhat 2017). The RT-PCR needs post-PCR sample handling, and hence, there is always a chance of sample cross contamination. Also, it is time consuming, and needs costly equipment for thermal cycling and result analysis. The ELISA is again time and resource consuming. Quick and efficient diagnostics are a pre-requisite to viral disease management and the isothermal amplification platforms are now gaining much importance in plant disease diagnostics (Nair and Manimekalai 2021).
The LAMP (Loop Mediated Isothermal Amplification) uses a DNA polymerase having strand displacement property and two pairs of primers recognising six distinct regions on a target gene to amplify a single copy of the target at the pace of 109 copies in an hour (Notomi et al. 2000). It does not require thermal cycling as amplification takes place under isothermal condition, around 60–65 °C which can be provided by a dry bath or a water bath. The RT-LAMP is a highly sensitive, specific and relatively faster diagnostic method for RNA viruses (Chen et al. 2020; Fukuta et al. 2004; Siljo and Bhat 2014). Here, the RNA is converted to cDNA with a reverse transcriptase enzyme and is then amplified using the LAMP routine. In one-step RT-LAMP, the RNA template is directly added to the isothermal reaction mixture along with the reverse transcriptase enzyme, wherein, the reverse transcription and amplification take place simultaneously. Positive LAMP amplicons are detected by looking for the turbidity after amplification or by electrophoresis on an agarose gel. Simple colourimetric identification of LAMP positives is possible with the use of Hydroxy Naphthol Blue (HNB) dye which shows a change in colour from violet to sky blue for positive reactions (Goto et al. 2009). The high rate of target amplification increases the sensitivity of LAMP-based molecular detection. The RT-LAMP was reported to have 100 times more sensitivity than conventional RT-PCR in detecting wheat yellow mosaic virus (Zhang et al. 2011). Development of molecular diagnostics for BBrMV in banana based on RT-LAMP will assist quick screening of banana planting materials and will facilitate the supply of disease-free planting materials for cultivation. Here, we report the optimisation and validation of a one-step RT-LAMP platform with colourimetric detection for rapid and efficient screening of banana for BBrMV.
Materials and methods
Plant materials and RNA isolation
Leaf samples were collected from 12 banana plants with typical symptoms of the BBrMV (Fig. 1) from the Banana Research Station, Thrissur, Kerala. Samples were collected from the banana cultivars Nendran and Kadali. We also sourced leaf samples from symptomless plants in the field and healthy tissue culture plants from Centre for Plant Biotechnology and Molecular Biology, Kerala Agriculture University, Thrissur, Kerala. The samples were stored in RNA Later solution (Invitrogen) till RNA isolation. Leaf samples were also collected from banana plants manifesting symptoms of BSV, CMV and BBTV from Banana Research Station, Kannara. These are other common viruses in banana and the samples were used to test the specificity of the RT-LAMP assay. Total RNA was purified from the banana leaves using RNeasy Plant Mini Kit (Qiagen) following the protocol provided with the kit. The quality of the RNA was checked through electrophoresis on agarose gel and the RNA was quantified using a NanoDrop spectrophotometer.
Designing of RT-LAMP primers
The BBrMV coat protein gene sequence (GenBank Accession no. MK139143.1-BRS3) was used as the template for designing RT-LAMP primers. Six RT-LAMP primers (External primers BrF3 and BrB3, internal primers BrFIP and BrBIP, loop primers BrLF and BrLB) were picked using the software Primer Explorer version 5.0. (http://primerexplorer.jp/lampv5e/index.html); keeping default parameters. First, the external and internal primers were designed using the software and this data was provided to generate suitable loop primers. All six primers were synthesized at Sigma-Aldrich Pvt. Ltd.
Establishment of RT-LAMP assay
The one-step RT-LAMP technique for the detection BBrMV was optimized using an RNA sample from a banana plant showing characteristic symptoms of the bract mosaic disease. Several sets of reactions were carried out to standardize the assay by varying the concentration of MgSO4 (4–8 mM), dNTP (1.4–1.6 mM each), betaine (0.8–1 M) and Bst polymerase enzyme (4–8 U). The HNB dye was used to monitor the positive amplicons and HNB at 120 µM and 150 µM were tried to get the clear colour change. The final optimised reaction cocktail contained 2 µg total RNA, 100 U M-MLV reverse transcriptase (Promega), 20 U RNasin® plus RNase inhibitor (Promega), 1.6 mM each dNTP, 0.2 µM each primer BrF3 and BrB3, 0.8 µM each primer BrFIP and BrBIP and 0.4 µM each primer BrLF and BrLB, 1 M betaine (Sigma), 4 mM MgSO4 (New England BioLabs), 1xThermopol buffer with 2 mM MgSO4, 8 U Bst polymerase enzyme large fragment (New England BioLabs) and 120 µM HNB dye (Sigma). Reaction volume was made up to 25 µL using Molecular biology grade water (Hi-Media). No template control (NTC) was run in all the assays. The reaction was set for 60 min at 65 ºC on a heat block (NeuationiTherm D150-2) and finally enzyme inactivation was done by incubating at 80 °C for 20 min.
Detection of RT-LAMP amplicons
The RT-LAMP products were run on 2 % agarose gel stained with ethidium bromide. Closed tube colourimetric detection of positive samples was enabled by adding 120 µM of HNB dye (Sigma) to the RT-LAMP reaction cocktail and by monitoring the colour change after the reaction.
Molecular characterization of RT-LAMP amplicons
The RT-LAMP products were cut with one selected restriction endonuclease specific for the amplified product to demonstrate the fidelity of the RT-LAMP assay. We initially identified a restriction enzyme with a single internal cut site in the BrF2-BrB2 flanking region based on virtual restriction digestion of the region using NEB cutter software (http://nc2.neb.com/NEBcutter2/). Later, the actual restriction digestion of RT-LAMP product was done using the identified enzyme, Sau3AI (Thermo Fischer Scientific). Restriction fragments were observed on 2.5% agarose gel.
For sequence confirmation, we did RT-PCR using RT-LAMP external primers BrF3 and BrB3. Initially, the total RNA from a symptomatic plant was converted to cDNA using RevertAid first strand cDNA synthesis kit (Thermo Scientific) according to the manufacturer’s protocol. The15 µl PCR reaction mixture contained 40 ng cDNA, 0.6 µM each primer BrF3 and BrB3, 150 µM each dNTPs, 0.5 U Taq DNA polymerase enzyme and 1X PCR buffer with 1.5 mM MgCl2. The PCR program set in the BioRad thermocycler consisted of initial denaturation at 95º C for 1 min and 35 cycles of 30 s denaturation at 95 ºC, 30 s annealing at 55 ºC and 30 s of primer extension at 72 ºC followed by a final extension at 72 ºC for 8 min. The PCR products of expected size were purified from agarose gel using Macherey-NagelTMNucleospin™ gel and PCR clean-up kit. Eluted products were sequenced with BrF3 primer. Sequences were subjected to BLASTN analysis (Altschul et al. 1990).
Sensitivity of one-step RT-LAMP technique
To determine the sensitivity of the one-step RT-LAMP technique, we prepared tenfold serial dilutions of the RNA sample. The RT-LAMP assay was performed with 10 ng, 1 ng, 0.1 ng, 0.01 ng, 1 pg, 0.1 pg, 0.01 pg and 0.001 pg total RNA in the isothermal reaction mixture.
Validation of the RT-LAMP assay
Validation of the RT-LAMP results was done by testing 12 BBrMV symptomatic samples, two healthy samples and samples collected from banana plants showing symptoms BBTV, BSV and CMV.
Results
Establishment of RT-LAMP assay
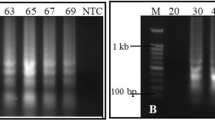
We optimized the RT-LAMP assay for the detection of BBrMV with RNA isolated from one banana plant showing the typical symptoms of BBrMV. Six RT-LAMP primers targeting eight regions of the BBrMV coat protein gene were designed and synthesised (Fig. 2, Table 1). The RT-LAMP reaction containing 120 µl HNB dye in the presence of 1.6 mM each dNTP and 6 mM MgSO4 was violet in colour prior to amplification. The BBrMV positive samples formed ladder-like bands on 2% agarose gel (Fig. 3) representative of stem-loop DNA having inverted repeats of the target. The colour of the reaction mixture changed to sky-blue from violet in case of positive amplification (Fig. 4). Healthy samples and no template control did not show any amplification on the agarose gel and they remained violet at the end of the RT-LAMP reaction. In addition, no amplification or colour change was observed in the case of samples from plants with symptoms of BSV, BBTV and CMV (Fig. 5).
Specificity of one-step RT-LAMP assay. Lanes L: 1 Kb DNA Ladder; 1: No template control; 2: BBrMV symptomatic banana sample; 3: BBTV symptomatic banana sample; 4: BSV symptomatic banana sample; 5: CMV symptomatic banana sample; 6: Healthy banana sample. In agarose gel profile of RT-LAMP products, amplification is observed only for the BBrMV symptomatic banana sample
Molecular typing of RT-LAMP amplicons
For characterizing the amplicons, restriction digestion was done with the in silico-identified enzyme Sau3AI, cutting the RT-LAMP internal primer flanking region. The enzyme has a single cut site generating two fragments of size 100 bp and 45 bp theoretically. The restriction site within the RT-LAMP amplified region has been indicated in Fig. 2. On digesting the RT-LAMP products with Sau3AI, two fragments of predicted sizes (100 bp and 45 bp) were obtained for the diseased samples when run on 2.5% agarose gel (Fig. 6). Thus, the accuracy of RT-LAMP-based detection of BBrMV could be confirmed. The RT-PCR reaction with RT-LAMP external primers produced bands of the expected size, 196 bp, on the agarose gel. Sequencing followed by BLASTN analysis indicated that the sequence corresponds to the BBrMV coat protein gene. It showed 98% identity with BBrMV isolate BRS2 coat protein mRNA, complete CDS (GenBank Acc No: MK139142.1). We deposited a representative sequence in the GenBank database (GenBank Acc No: OL757512).
Sensitivity of one-step RT-LAMP technique
In the one-step RT-LAMP assay with tenfold serial dilutions of the RNA sample, we got positive amplification, as indicated by the agarose gel profile, for up to 0.1 pg of the total RNA in the RT-LAMP reaction cocktail (Fig. 7). No amplification was observed with 0.01 pg and 0.001 pg of total RNA.
Sensitivity of the RT-LAMP assay. Lanes L: 1 Kb DNA ladder (GeNei); 1: RT-LAMP with 10 ng total RNA; 2: RT-LAMP with 1 ng total RNA; 3: RT-LAMP with 0.1 ng total RNA; 4: RT-LAMP with 0.01 ng total RNA; 5: RT-LAMP with 1 pg total RNA; 6: RT-LAMP with 0.1 pg total RNA; 7: RT-LAMP with 0.01 pg total RNA; 8: RT-LAMP with 0.001 pg total RNA; 9: No template control. In agarose gel profile of RT-LAMP products, amplification is observed for up to 0.1 pg total RNA in the RT-LAMP reaction
Discussion
Virus diseases distress banana farmers across the world due to their effects on the quality and yield of the crop as well as their spread through planting materials. Bananas are vegetatively propagated through suckers and also through tissue culture plantlets. Hence, an efficient virus indexing platform is essential for making available quality planting materials to farmers. The common detection methods for the BBrMV include ELISA and RT-PCR (Galvez 2020; Balamuralikrishnan et al. 2002). These methods are however restricted to laboratories having specific facilities. Here, we developed a one-step RT-LAMP assay coupled with simple visual detection for providing a quick and sensitive diagnostic method for BBrMV.
The RT-LAMP amplifies an RNA template under isothermal conditions after converting it to cDNA and the method is used to reliably detect RNA viruses. It combines isothermal amplification with reverse transcription, making cDNA from RNA and then running the LAMP reaction. The LAMP does not require thermal cycles (unlike PCR) and is performed at a constant temperature between 60 °C and 65 °C. It allows rapid and specific detection with only one hour of incubation on a dry bath or water bath. The RT-LAMP assay targeted BBrMV coat protein gene and we designed six primers binding to eight regions on the gene, thus ensuring high specificity and sensitivity. Using a heat-stable reverse transcriptase, RT-LAMP can directly detect the pathogen from an RNA sample (Notomi et al. 2000; Przewodowska et al. 2015). In a single incubation at 65 °C, reverse transcription and isothermal amplification take place (Fukuta et al. 2004), and the HNB in the reaction mixture allows closed tube endpoint detection without any additional step. Simple visual detection of positive samples could be made by adding 120 µM of HNB in the RT-LAMP reaction. The HNB is a metal ion indicator that helps to monitor the depletion of Mg2+ ions in the reaction mixture via a change in colour from violet to sky blue in case of positive amplification (Goto et al. 2009); and the HNB-based detection has been successfully used for simple visual identification of LAMP positives (Nahla et al. 2022). Indicators such as HNB, GenefinderTM, or calcein, are often added to the LAMP reaction mixtures before the reaction to identify LAMP amplification products (Tomita et al. 2008).
Diagnostic methods for several plant RNA viruses based on the RT-LAMP routine are available. Koh et al. (2020) reported RT-LAMP technique for the detection of BBrMV in abaca samples. They used GelRed or SYBR Green I dye for the visual evaluation of the LAMP products and the same was added to LAMP products post-amplification. However, in routine detection, closed-tube methods are preferred as they minimise the chances of sample cross contamination; and our HNB-based RT-LAMP has a simple visual detection in a closed-tube system. The RT-LAMP assay has been optimized for detecting citrus tristeza virus from infected citrus plant samples using SYBR Green I-based monitoring (Warghane et al. 2017). Zhang et al. (2016) developed an efficient and robust IC-RT-LAMP assay for field-level monitoring of BBrMV from flowering ginger. Using LAMP, a specific, sensitive, and visible detection technique for barley yellow mosaic virus was developed by Chen et al. (2020). The RT-LAMP was demonstrated to have a detection limit up to 100 times that of conventional RT-PCR and on par with that of real-time RT-PCR in the detection of BBrMV in cardamom (Siljo and Bhat 2014).
We tested the specificity of the RT-LAMP assay by using samples from banana plants showing symptoms of BBTV, CMV and BSV, which are the other major viral pathogens in banana. The absence of positive amplicons in these samples confirmed that our assay specifically detects the presence of BBrMV in banana. In their study, Koh et al. (2020) also determined the specificity of the RT-LAMP assay in abaca by testing samples with symptoms of common viruses in abaca like the BBTV, abaca bunchy top virus and sugarcane mosaic virus. They reported that 35 min of incubation was optimal for reverse transcription coupled with isothermal amplification. In our assay, we gave an incubation time of 60 min so that even the templates with low titre of the pathogen get amplified. However, our incubation on a dry bath coupled with closed-tube detection enables us to observe for positive amplification during the incubation itself.
In our study, the molecular analysis of the RT-LAMP amplicons was done through restriction analysis and sequencing. We digested the amplified RT-LAMP products with Sau3AI enzyme having a single restriction site in the BrFIP/BrBIP amplified region. The BrFIP/BrBIP region of 145 bp had a single recognition site for Sau3AI enzyme producing two fragments of size 100 bp and 45 bp. The fragments of the expected size in the analysis confirmed the fidelity of the amplicons. This restriction analysis of the LAMP products enables simple molecular typing of the amplicons. Restriction digestion has been used to confirm the accuracy of the LAMP assay for detecting phytoplasma 16S rRNA gene (Nair et al. 2016).
In conclusion, we devised a simple, fast, closed tube, colorimetric one-step RT-LAMP for detecting BBrMV in bananas by directly using the total RNA purified from the plant sample. This single-step assay is cost-effective compared to the BBrMV detection methods in bananas already available. It has wide application in the screening of banana planting materials and will enable the distribution of virus-indexed planting materials for cultivation.
Data availability
The data needed to reproduce the findings of this research are provided within the article.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2. (PMID: 2231712)
Balamuralikrishnan M, Doraisamy S, Ganapathy T, Viswanathan R (2002) Serological specificity and titre of Sugarcane mosaic virus polyclonal antisera raised under varying immunization procedures and bleeding time. J Plant Dis Protect 109(6):646–654
Bateson MF, Dale JL (1995) Banana bract mosaic virus: characterisation using potyvirus specific degenerate PCR primers. Arch Virol 140(3):515–527
Bhat AI, Maheshwari Y (2017) Application of immuno-diagnosis for plant viruses occurring in India. A century of plant virology in India. Springer, pp 583–619
Chen Z, Mao S, Zhang W, Fan X, Wu W, Liu C, Zhao K, Lu R (2020) Rapid visual detection method for barley yellow mosaic virus using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Plant Dis 105(9):2658–2663
Cherian KA, Menon R, Suma A, Nair S,Sudeesh MV (2002) Effect of banana bract mosaic disease on yield of commercial varieties in Kerala (Abstr.). In: Global conference on banana and plantain, Bangalore, p 155
Dassanayake EM (2001) Detection of banana bract mosaic potyvirus by immunocapture polymerase chain reaction (IC-PCR). Ann Sri Lanka Dept Agric 3:19–25
Fukuta S, Ohishi K, Yoshida K, Mizukami Y, Ishida A, Kanbe M (2004) Development of immunocapture reverse transcription loop-mediated isothermal amplification for the detection of tomato spotted wilt virus from chrysanthemum. J Virol Meth 121(1):49–55
Galvez LC, Koh RBL, Barbosa CFC, Aquino VM (2020) Multiplex reverse transcription-polymerase chain reaction for simultaneous detection of banana bract mosaic virus (BBrMV) and sugarcane mosaic virus (SCMV) in abaca. Can J Plant Pathol 42(4):572–583
Goto M, Honda E, Ogura A, Nomoto A, Hanaki KI (2009) Colorimetric detection of Loop Mediated Isothermal Amplification reaction by using hydroxy napthol blue. Biotechniques 46:167–172
Koh RBL, Barbosa CFC, Aquino VM, Galvez LC (2020) Rapid, simple detection of banana bract mosaic virus in abaca using a one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay. J Plant Pathol 86:433–441
Magnaye LV, Espino RRC (1990) Banana bract mosaic, a new disease of banana. Symptomatology. Philipp Agric 73:55–59
Nahla Binth T, Nair S, Loius V (2022) Colorimetric detection platform for banana bunchy top virus (BBTV) based on closed-tube loop mediated isothermal amplification (LAMP) assay. Virus Dis 33(3):303–308
Nair S, Manimekalai R (2021) Phytoplasma diseases of plants: molecular diagnostics and way forward. World J Microbiol Biotechnol 37:102. https://doi.org/10.1007/s11274-021-03061-y
Nair S, Manimekalai R, Gangaraj P, Hegde V (2016) Loop mediated isothermal amplification (LAMP) assay for detection of coconut root wilt disease and arecanut yellow leaf disease phytoplasma. World J Microbiol Biotechnol 32:108
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63
Przewodowska A, Zacharzewska B, Chołuj J et al (2015) A one-step, real-time reverse transcription loop mediated isothermal amplification assay to detect potato virus Y. Am J Potato Res 92:303–311. https://doi.org/10.1007/s12230-015-9430-3
Rodoni BC, Ahlawat YS, Varma A, Dale JL, Harding RM (1997) Identification and characterization of banana bract mosaic virus in India. Plant Dis 81(6):669–672
Samraj J, Menon MR, Christudas SP, Satyarajan PK (1966) Kokkan a new disease of banana (Musa paradisiaca Linn). Agric Res J Kerala 4(1):116
Selvarajan R, Jeyabaskaran KJ (2006) Effect of banana bract mosaic virus (BBrMV) on growth and yield of cultivar nendran (plantain, AAB). Indian Phytopathol 59:496–500
Siljo A, Bhat AI (2014) Reverse-transcription loop-mediated isothermal amplification assay for rapid and sensitive detection of banana bract mosaic virus in Cardamom (Elettaria cardamomun). Eur J Plant Pathol 138:209–214
Singh SJ, Selvarajan R, Singh HP (2000) In Banana-improvement, production and utilization, ed. by H.P. Singh, K.L. Chadha. pp 381–383
Thangavelu R, Selvarajan R, Singh HP (2000) Status of banana streak virus and banana bract mosaic virus diseases in India. In Singh HP, Chadha KL (eds) Banana: improvement, production and utilization. In: Proceedings of the conference on challenges for banana production and utilization in 21st century. Trichy, India: AIPUB, NRCB, pp 364–376
Tomita N, Mori Y, Kanda H, Notomi T (2008) Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3(5):877–882
Tripathi S, Patil BL, Verma R (2016) Viral diseases of banana and their management. In: Gaur R, Petrov N, Patil B, Stoyanova M (eds) Plant viruses: evolution and management. Springer, Singapore. https://doi.org/10.1007/978-981-10-1406-2_17
Warghane A, Misra P, Bhose S, Biswas KK, Sharma AK, Reddy M, Ghosh DK (2017) Development of a simple and rapid reverse transcription loop mediated isothermal amplification (RT-LAMP) assay for sensitive detection of Citrus tristeza virus. J Virol Methods 250:6–10
Zhang Z, Liu X, Li D, Yu J, Han C (2011) Rapiddetection of Wheat yellow mosaic virus by reverse transcription Loop Mediated Isothermal Amplification. Virol J 8:550
Zhang J, Borth WB, Lin B, Dey KK, Melzer MJ, Shen H, Pu X, Sun D, Hu JS (2016) Deep sequencing of banana bract mosaic virus from flowering ginger (Alpinia purpurata) and development of an immunocapture RT-LAMP detection assay. Arch Virol 161(7):1783–1795
Acknowledgements
Accession number: GenBank database Accession number—OL757512.
Author information
Authors and Affiliations
Contributions
SN conceptualized the idea, designed the experiments and edited the MS. MM conducted the experiments and drafted the manuscript. VL identified the diseased samples and arranged the samples for the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Research involving human participants and/or animals
There is no involvement of human participants or animals in the research.
Informed consent
All authors are informed about the paper and have given their consent for the publication of the paper.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Madhu Kovileri, M., Nair, S. & Loius, V. One-step Reverse Transcription-LoopMediated Isothermal Amplification (RT-LAMP) for closed-tube colorimetric detection of banana bract mosaic virus in Banana (Musa spp.). 3 Biotech 13, 131 (2023). https://doi.org/10.1007/s13205-023-03550-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03550-x