Abstract
The Receptor Binding Domain (RBD) of SARS-CoV-2, located on the S1 subunit, plays a vital role in the virus binding and its entry into the host cell through angiotensin-converting enzyme 2 (ACE2) receptor. Therefore, understanding the dynamic effects of mutants on the SARS-CoV-2 RBD is essential for discovering drugs to inhibit the virus binding and disrupt its entry into the host cells. A recent study reported a double mutant of SARS-CoV-2, L452R-E484Q, located in the RBD region. Thus, this study employed various computational algorithms and methods to understand the structural impact of both individual variants L452R, E484Q, and the double mutant L452R-E484Q on the native RBD of spike glycoprotein. The effects of the mutations on native RBD structure were predicted by in silico algorithms, which predicted changes in the protein structure and function upon the mutations. Subsequently, molecular dynamics (MD) simulations were employed to understand the conformational stability and functional changes on the RBD upon the mutations. The comparative results of MD simulation parameters displayed that the double mutant induces significant conformational changes in the spike glycoprotein RBD, which may alter its biological functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pandemic spread of COronaVIrus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to worldwide devastation on the economies and health systems. WHO reported that the pandemic led to 5,324,969 deaths and 271,376,643 confirmed cases as of December 15th, 2021 (https://covid19.who.int/). In addition to the different host responses mediated by SARS-CoV-2, the pathogenesis of COVID-19 is still not clearly understood (Flerlage et al. 2021; Kang et al. 2021; Prasad et al. 2021; Khatoon et al. 2020). SARS-CoV-2 is an envelope, positive single-strand RNA virus that utilizes its envelope spike protein to enter target host cells. The SARS-CoV-2 genome is the largest among all RNA-viruses, packed with a helical capsid, formed by a nucleocapsid protein and RNA surrounded by an envelope. There are three structural proteins, namely membrane protein (M), an envelope protein (E), and the spike protein (S), associated with the envelope that is mainly involved in replication, assembly and cell fusion (Murugan et al. 2021; Arya et al. 2021). The S protein facilitates the entry of the virus into the host cells with the help of the angiotensin-converting enzyme 2 (ACE2) receptor (Fig. 1). Recently, several SARS-CoV-2 variants have been reported globally that seem to exhibit increased transmissibility and infectivity (Burki 2021; Abdool Karim and de Oliveira 2021). The SARS-CoV-2 variant B.1.1.7 carries 17 mutations in its genome, of which 8 are in the spike protein (Leung et al. 2021). Another variant, B.1.351 (Mwenda et al. 2021) and P.1 variant (Francisco et al. 2021) carries 9 and 11 spike protein mutations, respectively, including 3 mutations in the receptor-binding domain (RBD), K417N/T, E484K, and N501Y. All three variants have the N501Y mutation, and its presence has been associated with increased transmissibility (Leung et al. 2021; Zhao et al. 2021). Very recently, a “double mutant” L452R-E484Q variant of SARS-CoV-2, B.1.617, has been reported (Cherian et al. 2021), which caused a transmission outburst with a vast number of deaths and active cases. These two mutations have been previously described in other SARS-CoV-2 variants and are known to confer immune escape and increase transmissibility and infectivity. The literature strongly supported that the L452R mutation has been reported in B.1.429 variant (Zhang et al. 2021). While the E484Q mutation is found in B.1.1.7, B.1.351, P.1, and B.1.526 variants (Tang et al. 2021; Bhattarai et al. 2021; Antony and Vijayan 2021). Besides these mutations, B.1.617 carries 11 additional mutations, including P681R outside of the RBD, which is located adjacent to the furin cleavage site of the spike, suggesting it might influence the way the spike is processed during infection (Cherian et al. 2021). All these SARS CoV-2 variants carry mutations in the viral RBD region that increases viral transmission, infection rates, disease severity and compromising the immune response, and thus raising concerns about the vaccine effectiveness against the COVID-19 pandemic (Altmann et al. 2021; Kuzmina et al. 2021; Kavitha et al. 2020).
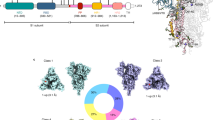
The schematic representation of SARS-CoV-2 envelop protein and receptor binding domain (RBD) region presenting the three mutations, along with the elucidation of ACE2 receptor binding and virus entry into the host cell, followed by step-wise in silico workflow and the tools implemented in the current study
In this regard, a mechanistic study based on structural and molecular dynamics analysis of the RBD variants will be beneficial to understand the molecular mechanism underlying the enhanced transmissibility and infectivity of the RBD variants (Hou et al. 2021; Sixto-López et al. 2021). We have studied the impact of mutations such as L452R, E484Q and L452R-E484Q, which are associated with the host cell attachment and the virus entry into the host cell (Fig. 1). We used various prediction tools to understand the dynamic changes occurring due to the mutations. Further, molecular dynamics (MD) simulations were conducted to understand the potential consequence of the mutations L452R, E484Q and L452R-E484Q. Various parameters like Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of gyration (Rg), Solvent Accessible Surface Area (SASA), Principal Component Analysis (PCA), Free Energy Landscape (FEL) and density distribution analysis were calculated for the MD trajectories to evaluate the structure and dynamics of SARS-CoV-2 RBD (Fig. 1). Substitution of amino acids may alter surface charges that can potentially affect the host receptor binding and significantly impact the structure and function of RBD of spike glycoprotein. Such mutations can even be one of the major causes of resistance in various viral diseases. We believe the study outcomes will help to understand the effect of the emerging SARS-CoV-2 variants on its binding to ACE2 and help correlate the higher transmissibility and infectivity associated with the virus cell entry mechanisms.
Materials and methods
Protein models of WT and mutants
We used the SARS-CoV-2 spike protein RBD crystal structure (PDB ID: 6M0J) (Lan et al. 2020). The structure coordinates were retrieved from the PDB and used as the wild-type (WT) structure for the study. The structure has different chains and RBD chain E was used to create the mutations in the protein. We mapped the three mutations, namely L452R, E484Q, and L452R-E4844Q at the corresponding positions, and mutant structures were constructed using the BuildModel module of FoldX (Schymkowitz et al. 2005).
Structural stability analysis
The online tools Polymorphism Phenotyping (POLYPHEN) (Adzhubei et al. 2010), I-mutant (Capriotti et al. 2005), Protein Variation Effect Analyzer (PROVEAN) (Choi et al. 2012), SNP&GO (Calabrese et al. 2009) and Pmut tool (López-Ferrando et al. 2017) were used to predict the functional impact of selected mutations on the protein. The mutational effect on the structural stability and flexibility were analyzed using DynaMut web server (Rodrigues et al. 2018) through DUET (Pires et al. 2014a), mCSM (Pires et al. 2014b), ENCoM(Frappier et al. 2015) and SDM scores (Worth et al. 2011) to obtain the Gibbs free energy (∆∆G) destabilizing values (Rodrigues et al. 2018). Further, Mutpred (Pejaver et al. 2017), Fathmm (Shihab et al. 2013) and MuPro web servers (Cheng et al. 2006) were also used to understand the threshold scores of the mutants. Consensus predictions from these algorithms indicate a destabilizing role of the mutants on the native structure of the receptor.
MD simulations
We carried out MD simulations using GROMACS 5.18.3 software package (Pronk et al. 2013; Hess et al. 2008). The WT RBD and all the mutation topology parameter files were generated using the Gromos96a43 force field. The intermolecular (non-bonded) potential, represented as the sum of Lennard–Jones (LJ) force-based switching with a cut-off distance range of 8–10 Å, pairwise Coulomb interaction and the long-range electrostatic force were determined by particle mesh Ewald (PME) approach (Lee et al. 2016). The real-space cutoff was set to 1.2 nm for the PME calculations (Wang et al. 2016). The velocity Verlet algorithm was applied for the numerical integrations and the initial atomic velocities were generated with a Maxwellian distribution at the given absolute temperature. Then the system was immersed with the default SPC/E water model, and protein was placed at the center of the cubic grid box (1.0 nm3). The protein was placed at the center of the cubic grid box (1.0 nm) (Zielkiewicz 2005). The neutralization was performed to make the concentration of the box to 0.15 M. The neutralized system was then subjected to energy minimization using the Steepest Descent (SD) and Conjugate Gradient (CG) algorithms utilizing a convergence criterion of 0.005 kcal mol−1 Å−1. The two-standard equilibration phase was carried out separately NVT (atom, volume, temperature) and NPT (atom, pressure–temperature) ensemble conditions such as constant volume and constant pressure for each protein structure, at similar time scale. We applied the Berendsen thermostat and Parinello–Rahman barostat to maintain the system's temperature and pressure, respectively. The system pressure and temperature were maintained constant at 1 bar and 300 K, with a coupling time of τP = 2 ps and τT = 1 ps. The Periodic Boundary Condition (PBC) was used for integrating the equation of motion by applying the leap-frog algorithm with a 2-fs time step. Finally, to make the system ready for production, the fixing of constraints is achieved by relaxing the grid box with water and counterions. All the simulations, including WT and three mutants, were subjected to 100 ns simulations in the current study. Various parameters were analyzed for the trajectories using GROMACS utilities and Python scripts with MDTraj (McGibbon et al. 2015).
Principal Component Analysis (PCA)
PCA is one of the well-known statistical techniques that helps in the reduction of complexity in analyzing the data and extracts the rigorous motion information in simulations which is essentially correlated for biological function. The eigenvector calculations and eigenvalues identified by diagonalizing the matrix with their projection and the first two principal components were carried out using the Essential Dynamics (ED) method. Then, principal components were selected as reaction coordinates and the free energy of the state, Gα, was calculated using the following equation:
where k is the Boltzmann constant, T is the temperature of simulation. P(qα) corresponds to the estimate of probability density function extracted from the histogram of the MD data constructed using joint probability distribution (PC1 and PC2 as reaction coordinates). Pmax(q) represents the probability of the most probable state. The protein movements and the dynamic motions of WT and L452R, E484Q, double mutant L452R-E484Q throughout the trajectories in the subspace were further identified by Cartesian coordinates projecting the most important eigenvectors from the complete analysis (Ahamad et al. 2021b).
Results
The study was hypothesized to understand the underlying mechanism and molecular basis of SARS-CoV-2 mutations L452R, E484Q, L452R-E484Q on the receptor stability. The various analyses like prediction algorithms and MD simulation results enumerated in this section indicate destabilizing role of the mutants on the protein.
Structural stability analysis
The POLYPHEN scores for L452R and E484Q were high, reflecting deleterious effects on the native protein and a deviation from its WT activity. The results revealed that the mutations L452R and E484Q are predicted to produce functional impact by destabilizing effect on the spike glycoprotein. The deviated role of the mutants were also confirmed from the results of SNP&GO and Pmut tools (Table 1). The impact of the mutations by amino acid substitutions on the biological function of RBD region was predicted by PROVEAN and I-mutant, with high deleterious and destabilizing effects, predicting conformational changes in the protein. Further, the impact on conformational stability upon mutations on the native structure was evaluated using Gibbs free energy (∆∆G) by DynaMut. DUET, mCSM, DynaMut, Mutpred, Fathmm and MuPro scores also predict the impact of amino acid substitutions in the mutants. From the consensus of the results from different methods, it is evident that the selected mutants show deleterious effects upon the amino acid alteration and are found to destabilize the spike glycoprotein RBD, altering its structural stability and function.
Structural dynamics and stability of SARS-CoV-2 variants
The superimposed structure of WT RBD and mutants studied here are shown in Fig. 2A. We studied the effect of mutants on the WT-RBD of SARS-CoV-2 structure in detail with the help of MD simulation allied parameters. The RMSD analysis revealed that the mutations L452R, E484Q and L452R-E484Q were unstable with a RMSD range of ∼0.2 to 0.5 nm compared with WT, which was stable with ∼0.15 nm (Fig. 2B and Table S1). The MD simulation trajectories were further analyzed using Probability Distribution Function (PDF) which revealed that the WT was within the threshold average PDF-RMSD values. Noticeably, the mutant L452R shows a fluctuation of 0.27 nm, and E484Q showed a drift of 0.26 nm, followed by maximum drift in L452R-E484Q with 0.41 nm (Figure S1A). The instability caused by the mutations is confirmed with the RBD deviation analysis, which gives us space to speculate that there is a deformation leading to conformational changes and instability of RBD.
MD simulation of SARS-CoV-2 WT RBD and RBD variants. (A) The superimposition of WT and mutant structures of SARS-CoV-2 RBD generated using PyMOL. The protein structure is rendered in cartoon and mutants displayed as sticks (B) C-alpha (C-α) RMSD conformation of WT and the respective mutations, and (C) RMS-Fluctuations depicting the maximum drift in mutations compared to WT noted at 10 ns of simulation time in all the trajectories
We proceeded with further analysis to check the overall flexibility of WT and mutant structures by RMSF to determine the effect of Cα-RMSF for each residue. The results found minimal fluctuations on the WT with a value of ∼0.1 nm and the residual displacement of mutants L452R, E484Q and L452R-E484Q revealed maximum Cα-RMSF fluctuation with ∼0.2 nm, ∼0.4 nm, and ∼0.3 nm, respectively (Fig. 2C). Thus, the above comparative analysis of Cα-RMSF confirmed the loss in the degree of flexibility caused by mutations on SARS-CoV-2 RBD compared to the WT.
The Rg analysis helped in a comparative analysis of the compactness of the protein structures in the WT and the mapped mutant structures during MD simulations. The results revealed that the mutants L452R, E484Q and L452R-E484Q showed a decrease in Rg with a maximum reduction in E484Q compared with WT (Fig. 3A). However, the average PDF-Rg analysis indicates a higher drift on L452R, with 1.77 nm, E484Q with 1.77 nm, L452R-E484Q with 1.78 nm, whereas WT with 1.84 nm (Figure S1B and Table S1). Thus, results from the above analysis suggested that mutants showed a highly destructive impact on the native SARS-CoV-2 structure, inducing structural destabilization, which could be responsible for the loss of compactness, suggesting a cumulative effect on the core structure.
The WT and mutant structures were further subjected to SASA analysis. The system’s core hydrophobic regions were examined, and we found a significant decrease in SASA upon mutations (Fig. 3B). The mutants revealed high SASA fluctuation values, ranging from 109nm2 to 110nm2, in contrast to WT (117nm2). The PDF analysis of SASA on mutants revealed an unsteady change with the values of 117.18nm2 for L452R, 109.98nm2 for E484Q, and 109.43nm2 for L452R-E484Q (Figure S1C and Table S1). The fluctuations from the SASA and PDF-SASA results justified a high loss of hydrophobic contacts between the residues on the SARS-CoV-2 RBD due to the substitution of amino acids.
The change in the number of intramolecular hydrogen bonds formed in the WT and mutant structures was investigated. The overall analysis showed that the L452R consists of 118.84 H-bonds, E484Q with 125.32, L452R-E484Q with 123.93, whereas WT with 119.59 H–bonds (Fig. 3C and Table S2). The investigation revealed the deviations in the hydrogen bonding pattern of the mutants have a significant impact on the structural dynamics and instability of SARS-CoV-2.
PCA results
The analysis further carried out for essential subspace examined where the protein dynamics are noted with eigenvectors associated with the eigenvalues. The dynamic behavior of WT and mutants were compared with the clustering parameters and the results revealed that the clusters are well defined in the WT structures by covering the minimum region, whereas the L452R, E484Q and L452R-E484Q mutants occupied maximum regions with a wider cluster (Fig. 4A). Furthermore, the fluctuations with eigenvector1 and eigenvector2 were analyzed and plotted to understand the atomic fluctuations and intramolecular collective motions. The results revealed that L452R, E484Q, L452R_E484Q mutations showed increased fluctuation than WT SARS-CoV-2 RBD (Fig. 4B-C). The maximum fluctuations were mainly observed in C-terminal ACE2 binding sites present in RBD (Figure S3). The comparative study of WT and mutant SARS-CoV-2 indicates significant conformational changes in mutants leading to consequential disturbances in internal atomic motion causing loss of protein stability.
The MD simulation conformational landscape exhibited by the RBD variants. A 2D Projection plot showing the conformational space of Cα-atom and atomic number monitoring of the RBD WT and mutant structures in the essential subspace along the eigenvector 1 and 2. Average Eigen RMSF values for WT-RBD and the L452R, E484Q and L452R-E484Q mutant systems (B) PC1 and (C) PC2
The WT and mutant structures were further examined for a diagonalized covariance matrix to know the positional fluctuations on the SARS-CoV-2 receptor Cα-atoms. The atomic behavior is identified by the residual motions between WT and mutant structures. The analysis was differentiated with red and blue color representation, where red implies small fluctuations between the atoms and blue denotes large fluctuations (Figure S2). The amplitude and the intensity of WT were magnified with a value of 0.080nm2 (Figure S2A), whereas a high difference is observed in the mutants L452R, E484Q and L452R-E484Q, with a range of 0.258nm2, 0.239nm2 and 0.239nm2, respectively (Figure S2B–D). The comparison of residual displacement in MD trajectories of WT and mutants confirms the atomic deformation of SARS-CoV-2 on the native structure.
FEL results
The free energy changes of the WT and mutants were explored using FEL analysis through eigenvectors plots. The comparison studies indicated stable global free energy minima confined within one basin in the WT (Fig. 5A). However, mutants including L452R, E484Q and L452R-E484Q had more expansive basins and numerous meta-stable conformations associated with multiple energy minima (Fig. 5B–D). From the observations, it is evident that the alterations of amino acids invoke protein structure destabilization of SARS-CoV-2 of RBD.
Moreover, the atomic density distribution analysis also helps understand the changes in atomic orientation, plotted using densmap script. The partial density area of WT was stable with minimum values of 3.95 nm−3 (Fig. 6A), whereas lower density was noticed for the mutations L452R, E484Q and L452R-E484Q, with a range of 3.14 nm−3, 2.74 nm−3 and 2.94 nm−3, respectively (Fig. 6B–D). Thus, the comparison of density distribution affirmed the high impact structural transitions, leading to loss of stability upon amino acid substitution.
Discussion
Various studies and advancements revealed a strong involvement of mutations on the dynamic effect of spike glycoprotein (Ahamad et al. 2020, 2021a; Rezaei et al. 2020). Specifically, many researchers stated that replacing amino acids has resulted in structural changes on the spike glycoprotein of SARS-CoV-2, increasing its susceptibility to proteolysis. However, mechanistic and functional consequences of amino acid substitutions remain unresolved (Mishra et al. 2021; Teng et al. 2021). Khalid et al., 2020 reported an in-depth study on spike glycoprotein-ACE2 interactions exploring both sequence and structural features using a bioinformatics approach. The authors labeled various single nucleotide polymorphism (SNP) as damaging/deleterious/destabilizing that block the interaction, which might halt the virus from entering the host cell (Khalid and Naveed 2020). Ahmed et al. reported mutations occurring in the proteins can affect its conformation, structure folding and stability, which can eventually show effect on protein–protein interactions/thermodynamics. Recent research reported several mutations in the SARS-CoV-2 spike glycoprotein RBD that enhances the infectivity and binding interactions of virus to the host ACE2 receptor. It is necessary to study the conformational, stability and dynamic effect of mutations on native RBD structure. Most of the mutations occur at RBD region that are directly bound to ACE2 which has ability to generate a stable complex with good binding affinity (Alaofi and Shahid 2021).
Our study highlighted the significance of amino acid substitutions on the native spike glycoprotein, which presented a significant destabilizing effect. The free energy (∆∆G) scores from the computational prediction algorithms on the RBD upon the mapped mutations also displayed a strong destabilizing impact on the native spike glycoprotein, affecting the receptor binding between the complexes. Herein, we studied three critical RBD mutants L452R, E484Q and L452R-E484Q by implementing the all-atom MD simulations to examine the spike glycoprotein molecular alterations associated with RBD mutations. The calculated RMSD for all the Cα-atoms starting from the central origin showed that the mutant structures projected a diverge manner of deviations throughout the simulations compared to RBD-WT and mutants. RMSF analysis also displayed increased fluctuations on the mutant structures than the WT structures. Rg results portrayed large oscillations and significant fluctuations in mutants compared to WT. The SASA analysis depicted the decreased alterations of mutant in contrary to WT structures for a more extended period enduring a major structural transition. The mutations present in the RBD region showed high flexibility, solvent accessibility and gyration when compared to WT-RBD suggesting the mutation did not alter the conformational compactness. The PCA, FEL and hydrogen bond analysis results also suggest a destabilizing effect for the selected mutations, which have a significant free energy changes when compared to the WT structure. The H-bond analysis comprehended the stable flexibility on WT and mutants demonstrated major changes. Moreover, the energy spectrum, energy wells and atomic motion analysis also aided in detailed stability particulars where significant changes in the energy levels of mutants were investigated and found loss of RBD stability upon mutants compared to WT. To summarize, the study provided a comprehensive view of destabilizing mechanisms on L452R, E484Q and L452R-E484Q of RBD associated with SARS-CoV-2 spike glycoprotein. The prediction tools, MD simulations and the free energy calculations indicate a loss in the stability of the spike glycoprotein due to the RBD mutations.
Conclusions
The RBD mutants, namely L452R, E484Q and L452R-E484Q, are predicted to have a significant deleterious impact on SARS-CoV-2 spike protein. The comparative results of prediction algorithms and MD simulation studies facilitated studying the effect of mutants on the WT-RBD. The MD simulation trajectory investigations, namely, atomic movements, residue flexibility, FEL analysis, hydrogen bond, Rg and SASA, indicate unstable mutant structures. Summarily, the RBD mutants L452R, E484Q and L452R-E484Q displayed a destabilizing and deleterious impact on the RBD native structure.
Data availability
Not applicable.
References
Abdool Karim SS, de Oliveira T (2021) New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N Engl J Med. https://doi.org/10.1056/NEJMc2100362
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
Ahamad S, Hema K, Gupta D (2021a) Structural stability predictions and molecular dynamics simulations of RBD and HR1 mutations associated with SARS-CoV-2 spike glycoprotein. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1889671
Ahamad S, Kanipakam H, Birla S, Ali MS, Gupta D (2021b) Screening malaria-box compounds to identify potential inhibitors against SARS-CoV-2 Mpro, using molecular docking and dynamics simulation studies. Eur J Pharmacol 890:173664
Ahamad S, Kanipakam H, Gupta D (2020) Insights into the structural and dynamical changes of spike glycoprotein mutations associated with SARS-CoV-2 host receptor binding. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1811774
Alaofi AL, Shahid M (2021) Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules 11(9):1273
Altmann DM, Boyton RJ, Beale R (2021) Immunity to SARS-CoV-2 variants of concern. Science 371(6534):1103–1104. https://doi.org/10.1126/science.abg7404371/6534/1103
Antony P, Vijayan R (2021) Molecular dynamics simulation study of the interaction between human angiotensin converting enzyme 2 and spike protein receptor binding domain of the SARS-CoV-2 B. 1.617 variant. Biomolecules 11(8):1244
Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, Prashar V, Gupta GD, Panicker L, Kumar M (2021) Structural insights into SARS-CoV-2 proteins. J Mol Biol 433(2):166725
Bhattarai N, Baral P, Gerstman BS, Chapagain PP (2021) Structural and dynamical differences in the spike protein RBD in the SARS-CoV-2 variants B. 1.1. 7 and B. 1.351. J Phys Chem B 125(26):7101–7107
Burki T (2021) Understanding variants of SARS-CoV-2. Lancet 397(10273):462. https://doi.org/10.1016/S0140-6736(21)00298-1
Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R (2009) Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat 30(8):1237–1244
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33(2):W306–W310
Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S (2021) Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra India. Microorganisms. https://doi.org/10.1101/2021.04.22.440932
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins: Struct, Funct, Bioinform 62(4):1125–1132
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7(10):46688
Flerlage T, Boyd DF, Meliopoulos V, Thomas PG, Schultz-Cherry S (2021) Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat Rev Microbiol. https://doi.org/10.1038/s41579-021-00542-7
Francisco RDS Jr, Benites LF, Lamarca AP, de Almeida LGP, Hansen AW, Gularte JS, Demoliner M, Gerber AL, de Guimaraes CAP, Antunes AKE, Heldt FH, Mallmann L, Hermann B, Ziulkoski AL, Goes V, Schallenberger K, Fillipi M, Pereira F, Weber MN, de Almeida PR, Fleck JD, Vasconcelos ATR, Spilki FR (2021) Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul. Brazil. Virus Res 296:198345. https://doi.org/10.1016/S0168-1702(21)00052-6
Frappier V, Chartier M, Najmanovich RJ (2015) ENCoM server: exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res 43(W1):W395–W400
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447. https://doi.org/10.1021/ct700301q
Hou X, Zhang Z, Gao J, Wang Y (2021) SARS-CoV-2 spike protein N501Y mutation causes differential species transmissibility and antibody sensitivity: a molecular dynamics and alchemical free energy study. Mol Syst Design Eng 6(11):964–974
Kang YW, Park S, Lee KJ, Moon D, Kim YM, Lee SW (2021) Understanding the host innate immune responses against SARS-CoV-2 infection and COVID-19 pathogenesis. Immune Netw 21(1):e1. https://doi.org/10.4110/in.2021.21.e1
Kavitha K, Sivakumar S, Ramesh B (2020) 1, 2, 4 triazolo [1, 5-a] pyrimidin-7-ones as novel SARS-CoV-2 Main protease inhibitors: In silico screening and molecular dynamics simulation of potential COVID-19 drug candidates. Biophysical Chem 267:106478
Khalid Z, Naveed H (2020) Identification of destabilizing SNPs in SARS-CoV2-ACE2 protein and spike glycoprotein: implications for virus entry mechanisms. J Biomol Struct Dyn 40(3):1205–1215
Khatoon F, Prasad K, Kumar V (2020) Neurological manifestations of COVID-19: available evidences and a new paradigm. J Neurovirol 26(5):619–630. https://doi.org/10.1007/s13365-020-00895-4
Kuzmina A, Khalaila Y, Voloshin O, Keren-Naus A, Boehm-Cohen L, Raviv Y, Shemer-Avni Y, Rosenberg E, Taube R (2021) SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe 29(4):522–528. https://doi.org/10.1016/S1931-3128(21)00136-0
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. https://doi.org/10.1038/s41586-020-2180-5
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12(1):405–413
Leung K, Shum MH, Leung GM, Lam TT, Wu JT (2021) Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26(1):2002106. https://doi.org/10.2807/1560-7917.ES.2020.26.1.2002106
López-Ferrando V, Gazzo A, De La Cruz X, Orozco M, Gelpí JL (2017) PMut: a web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res 45(W1):W222–W228
McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, Schwantes CR, Wang LP, Lane TJ, Pande VS (2015) MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J 109(8):1528–1532. https://doi.org/10.1016/j.bpj.2015.08.015
Mishra D, Maurya RR, Kumar K, Munjal NS, Bahadur V, Sharma S, Singh P, Bahadur I (2021) Structurally modified compounds of hydroxychloroquine, remdesivir and tetrahydrocannabinol against main protease of SARS-CoV-2, a possible hope for COVID-19: Docking and molecular dynamics simulation studies. J Mol Liquids 335:116185
Murugan C, Ramamoorthy S, Guruprasad K, Murugan RK, Sivalingam Y, Sundaramurthy A (2021) COVID-19: a review of newly formed viral clades, pathophysiology, therapeutic strategies and current vaccination tasks. Int J Biol Macromol 193:1165–1200
Mwenda M, Saasa N, Sinyange N, Busby G, Chipimo PJ, Hendry J, Kapona O, Yingst S, Hines JZ, Minchella P, Simulundu E, Changula K, Nalubamba KS, Sawa H, Kajihara M, Yamagishi J, Kapin’a M, Kapata N, Fwoloshi S, Zulu P, Mulenga LB, Agolory S, Mukonka V (2020) Bridges DJ (2021) Detection of B.1.351 SARS-CoV-2 Variant Strain—Zambia. MMWR Morb Mortal Wkly Rep 70(8):280–282. https://doi.org/10.15585/mmwr.mm7008e2
Pejaver, V., Urresti, J., Lugo-Martinez, J., Pagel, K.A., Lin, G.N., Nam, H.J., Mort, M., Cooper, D.N., Sebat, J., Iakoucheva, L.M. and Mooney, S.D., 2020. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nature communications, 11(1), 1-13.
Pires DE, Ascher DB, Blundell TL (2014a) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 42(W1):W314–W319
Pires DE, Ascher DB, Blundell TL (2014b) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30(3):335–342
Prasad K, AlOmar SY, Alqahtani SAM, Malik MZ, Kumar V (2021) Brain disease network analysis to elucidate the neurological manifestations of COVID-19. Mol Neurobiol. https://doi.org/10.1007/s12035-020-02266-w
Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D (2013) GROMACS 45: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854
Rezaei S, Sefidbakht Y, Uskokovic V (2020) Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS-CoV and the effects of mutations on the binding affinity. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1860829
Rodrigues CH, Pires DE, Ascher DB (2018) DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res 46(W1):W350–W355
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L (2005) The FoldX web server: an online force field. Nucleic Acids Res 33:W382-388
Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 34(1):57–65
Sixto-López Y, Correa-Basurto J, Bello M, Landeros-Rivera B, Garzón-Tiznado JA, Montaño S (2021) Structural insights into SARS-CoV-2 spike protein and its natural mutants found in Mexican population. Sci Rep 11(1):1–16
Tang JW, Toovey OT, Harvey KN, Hui DD (2021) Introduction of the South African SARS-CoV-2 variant 501Y. V2 into the UK. J Infection 82(4):e8–e10
Teng S, Sobitan A, Rhoades R, Liu D, Tang Q (2021) Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief Bioinform 22(2):1239–1253
Wang H, Nakamura H, Fukuda I (2016) A critical appraisal of the zero-multipole method: Structural, thermodynamic, dielectric, and dynamical properties of a water system. J Chem Phys 144(11):114503
Worth CL, Preissner R, Blundell TL (2011) SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 39(2):W215–W222
Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E (2021) Emergence of a Novel SARS-CoV-2 variant in southern California. JAMA 325(13):1324–1326. https://doi.org/10.1001/jama.2021.16122776543
Zhao S, Lou J, Cao L, Zheng H, Chong MKC, Chen Z, Chan RWY, Zee BCY, Chan PKS, Wang MH (2021) Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the UK: an early data-driven analysis. J Travel Med. https://doi.org/10.1093/jtm/taab011
Zielkiewicz J (2005) Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J Chem Phys 123(10):104501. https://doi.org/10.1063/1.2018637
Acknowledgements
We acknowledge the Bioinformatics infrastructure grant to ICGEB by the Department of Biotechnology, Government of India (no. BT/PR40151/BTIS/137/5/2021). SA is a recipient of a Research Associate fellowship from the Indian Council of Medical Research (ICMR), India (2019–6039 File No. ISRM/11(83)/2019). KH is a recipient of the DBT-BioCARe-Women Scientist fellowship awarded by the Department of Biotechnology, India (BT/PR31715/BIC/101/1233/2019). VK sincerely thanks Amity University, Noida, for providing facilities.
Author information
Authors and Affiliations
Contributions
SA: contributed to literature mining, mutation modeling, MD simulations, and write-up in the manuscript. KH, SA: contributed to the write-up and corrections in the manuscript. VK: contributed to design the hypothesis, major inputs, co-correspondence corrections in the manuscript. DG: contributed to the designing the hypothesis, major inputs, correspondence and corrections in the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics statement
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahamad, S., Hema, K., Ahmad, S. et al. Insights into the structure and dynamics of SARS-CoV-2 spike glycoprotein double mutant L452R-E484Q. 3 Biotech 12, 87 (2022). https://doi.org/10.1007/s13205-022-03151-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03151-0