Abstract
2-chloronicotinic acid (2-CA) is a key precursor for the synthesis of a series of pesticides and pharmaceuticals. Nitrilase-catalyzed bioprocess is a promising method for 2-CA production from 2-chloronicotinonitrile (2-CN). In this study, a mutant of nitrilase from Rhodococcus zopfii (RzNIT/W167G) was constitutively overexpressed with Escherichia coli as host, which exhibited a onefold increase in enzymatic activity compared with inducible expression. Biosynthesis of 2-CA using whole cells harboring nitrilase as biocatalysts were investigated and 318.5 mM 2-CA was produced, which was the highest level for 2-CA production catalyzed by nitrilase to date. 2-CA was recovered from the reaction mixture through a simple acidification step with a recovery yield of 90%. This study developed an efficient bioprocess for 2-CA with great potential for industrial application.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
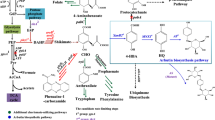
2-Chloronicotinic acid (2-CA) is a key building block for agrochemicals and pharmaceuticals, such as diflufenican, nicosulfuron, mirtazapine, and nevirapine (Jin et al. 2011; Quevedo et al. 2009). Considering the ever-growing demand for 2-CA, numerous efforts have been devoted to developing green and efficient synthetic routes. Currently, 2-CA is produced through chemical methods, including N-oxidation and chlorination of nicotinonitrile or nicotinic acid, oxidation of 2-choloro-3-methylpyridine, and chlorination and Michael addition of ethyl cyanoacetate with acrylaldehyde (Wu et al. 2019). However, these chemical methods suffered from economic and environmental problems due to the harsh reaction conditions, low specificities, and yields. For instance, 2-chloronicotinamide was generated as by-product during the hydrolytic process of 2-chloronicotinonitrile (2-CN) under strong alkaline condition (Scheme 1) (Du et al. 2009). Thus, there is urgent demand to explore and establish efficient synthetic process for 2-CA production.
Nitrilases (EC 3.5.5.1) are a class of industrially important hydrolases that convert nitriles to corresponding carboxylic acids and ammonia in one step (Zhang et al. 2020a, b; Shen et al. 2021; Lauder et al. 2020). Given the prominent regio-, chemo-, and stereo-selectivity under mild conditions, nitrilases are regarded as valuable alternatives to chemical catalysts (Hauer 2020; Wu et al. 2021). Extensive efforts have been devoted to overexpression and efficient production of nitrilase. For instance, increase of gene copy number and co-expressing of ER oxidoreductin 1 were used to enhance the Acidovorax facilis ZJB09122 nitrilase production (Shen et al. 2020). The production of nitrilase from Pseudomonas putida CGMCC3830 was improved using a high density culture strategy by 14.9-fold (Gong et al. 2018). By optimization of culture conditions and glycerol feeding, the production of nitrilase from Alcaligenes sp. ECU0401 was increased by 50-fold (Liu et al. 2011). Nitrilase-mediated bioprocesses have attached a great deal of interests for preparation of high-valued carboxylic acids, such as (S)-3-cyano-5-methylhexanoic acid (Chen et al. 2021; Wu et al. 2020), (R)-3-isobutyl-4-cyanobutanoic acid (Yu et al. 2019, 2021), (R)-o-chloromandelic acid (Xue et al. 2015; Zou et al. 2021) and nicotinic acid (Gong et al. 2017; Teepakorn et al. 2021). However, efficient production of 2-CA by nitrilase has not been reported to date (Zheng et al. 2018; Tang et al. 2018).
In our previous study, a mutant of nitrilase (RzNIT/W167G) from Rhodococcus zopfii was obtained with significantly improved reaction specificity and hydrolysis activity (Zheng et al. 2020). Herein, constitutive expression of the recombinant RzNIT/W167G was investigated and efficient biosynthesis of 2-CA from 2-CN was developed using RzNIT/W167G as biocatalyst (Scheme 1). Characteristics of RzNIT/W167G were investigated, along with its performance evaluation in 2-CN hydrolysis. The results suggested that RzNIT/W167G could serve as a promising biocatalyst for biosynthesis of 2-CA.
Materials and methods
Bacteria strains, plasmids, and reagents
E. coli BL21 (DE3) and pET-3a were used as the expression host and plasmid, respectively. PrimeSTAR HS DNA polymerase was purchased from Vazyme (Nanjing, China). The restriction enzyme DpnI was purchased from New England biolabs Inc (Ipswich, MA, USA). ClonExpress® II One Step Cloning Kit was purchased from Vazyme biotech Co., Ltd (Nanjing, China). Isopropyl-β-d-thiogalactopyranoside (IPTG), kanamycin, and ampicillin were obtained from Sigma (Shanghai, China). 2-CN and 2-CA were purchased from J&K chemicals (Shanghai, China).
Construction of constitutive expression system
The target gene was amplified using the primer 3a1-F and 3a1-R from the recombinant plasmid previously constructed in our lab (Zheng et al. 2020). The RzNIT/W167G gene was inserted into plasmid pET-3a linearized by 3a2-F and 3a2-R to obtain pET-3a-RzNIT/W167G. The PCR program was 3 min at 95 °C followed by 30 cycles of 15 s at 95 °C, 5 s at 60 °C, and 3.5 min at 72 °C, and a final extension step with 10 min at 72 °C. The amplified products were digested for 3 h at 37 °C using DpnI. The linearized vector and the corresponding amplified fragment were ligated using ClonExpress® II One Step Cloning Kit to obtain the constitutive expression vector. The recombinant plasmids were transformed into E. coli BL21 (DE) cells and then confirmed by sequencing. The primers used in this study are listed in Table S1.
Fermentation of recombinant E. coli overexpressing RzNIT/W167G
E. coli BL21 (DE3)/pET-3a-RzNIT/W167G was grown in a 10 mL LB medium supplemented with ampicillin (100 μg/mL) at 37 °C, 180 rpm for 12 h. Then, the primary seed culture was transferred to 100 mL LB medium with ampicillin (100 μg/mL) at 37 °C, 180 rpm for 12 h. Subsequently, 100 mL of secondary seed culture was transferred to a 5 L fermenter containing 3 L of fermentation medium (pH 7.5), which was consisted of 15 g/L peptone, 10 g/L NaCl, 12 g/L yeast extract, 15 g/L glycerol, 2.28 g/L K2HPO4.3H2O, (NH4)2SO4, 1.36 g/L KH2PO4, 5 g/L (NH4)2SO4, 0.375 g/L MgSO4.7H2O, and 100 μg/mL ampicillin. The fermentation process was carried out at 28 °C with an aeration of 1.3 vvm (air volume/culture volume/minute) and agitation of 500 rpm. The pH of medium was adjusted to 7.5 using ammonia and phosphoric acid, and the fermentation temperature was maintained at 28 °C. The cells were harvested by centrifugation at 10,000 × g for 10 min at 4 °C and washed with sodium phosphate buffer (20 mM, pH 7.5).
The collected cells were resuspended in potassium phosphate buffer (50 mM, pH 7.0), and disrupted by sonication. The supernatant and cell debris were collected as the soluble fraction and insoluble fraction, respectively. Same volume samples of whole-cell fraction, soluble fraction, and insoluble fraction were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The SDS-PAGE was performed using 5% stacking gel and 12% separating gel. Protein bands were stained by Coomassie Brilliant Blue R-250 dye.
Enzyme activity assay
The activity of RzNIT/W167G was assayed in a 1 mL reaction mixture containing an appropriate amount of resting cells or fermentation broth, 50 mM 2-CN, and 200 mM sodium phosphate buffer (pH 7.0). The reactions were carried out at 30 °C with 200 rpm for 20 min, and then terminated by addition of 10 µL HCl (6.0 M). The enzyme activity was determined by measuring the generated 2-CA in reaction mixture using high-performance liquid chromatography (HPLC) equipped with a C18 column (5 μm × 4.6 mm × 250 mm, Welch Materials, Inc., Shanghai, China) at a wavelength of 210 nm. The mobile phase was composed of 0.1% phosphoric acid and acetonitrile (75:25, v/v) with a flow rate of 1 mL/min at 40 °C. One unit of enzyme activity was defined as the amounts of the enzyme required to produce 1 μmol of 2-CA per minute.
Effect of temperature and pH on enzyme activity
The influence of temperature on enzyme activity was examined at various temperatures (20–65 °C) under the standard assay condition. To determine the thermal stability of nitrilase, the resting cells were incubated in sodium phosphate buffer (200 mM, pH 7.0) at 30 and 40 °C, respectively. The influence of pH on enzyme activity was investigated using the following buffers at 50 mM: sodium phosphate buffer (pH 5.5–8.0), Tris–HCl (pH 7.0–9.0), and Gly-NaOH (pH 8.5–10.0). To determine pH stability of the enzyme, the resting cells were incubated at pH 7.0–9.0 at 4 °C. The residual activities were measured after incubation at different conditions.
Effect of cell loading and 2-CA concentration on biosynthesis of 2-CA
To examine the effect of cell loading on biosynthesis of 2-CA, the biotransformation was carried out in a 10 mL reaction mixture containing 1.0–5.0 g dry cell weight (DCW)/L resting cells, 200 mM sodium phosphate buffer (pH 7.0), and 300 mM 2-CN.
In order to investigate the effect of 2-CA on biotransformation, the reaction was carried out in a 10 mL reaction mixture containing 200 mM sodium phosphate buffer (pH 7.0), 4.0 g DCW/L resting cells, 50 mM 2-CN, and different concentrations of 2-CA (0–300 mM). The pH was adjusted to 7.0 with 6.0 M NaOH. The effect of temperature on biotransformation of 2-CN was investigated in 10 mL reaction mixture contained 200 mM phosphate buffer (pH 7.0), 4.0 g DCW/L resting cells, and 100 mM 2-CN at 25 °C and 30 °C. Samples were withdrawn periodically for HPLC analysis as described above.
Purification and recovery of 2-CA
The biotransformation was carried out in a 1 L stirred reactor containing 200 mM sodium phosphate buffer (pH 7.0), 4.0 g dried whole cells, and 350 mM 2-CN. The reaction was performed at 30 °C and 180 rpm. The cells were removed by centrifugation (12,000 × g, 5 min) after the biotransformation and the supernatant was mixed with 6% activated carbon at 180 rpm for 2 h. Then, the activated carbon was removed by filtration. The resulting filtrate was acidified with HCl to pH 1.0–2.0 with stirring. The precipitated 2-CA was filtrated and dried at 50 °C.
Results and discussion
Constitutive expression of RzNIT/W167G
Industrial applications of nitrilases were hindered by several limitations, such as formation of inclusion body, low biomass of cells, and requirement of expensive inducers (Gong et al. 2018; Liu et al. 2011). Constitutive expression for recombinant enzymes without the addition of inducers could simplify the fermentation process and reduce costs. Thus, the constitutive expression for RzNIT/W167G was investigated. E. coli BL21 (DE3)/pET-3a-RzNIT/W167G was cultured in LB medium without induction for constitutive expression, while E. coli BL21 (DE3)/pET-28b-RzNIT/W167G was induced by IPTG.
Expression of RzNIT/W167G in E. coli BL21 (DE3)/pET-3a-RzNIT/W167G and E. coli BL21 (DE3)/pET-28b-RzNIT/W167G was analyzed by SDS-PAGE (Fig. 1A). The molecular mass of RzNIT/W167G was approximately 43 kDa, which was consistent with the molecular mass (41 kDa) calculated from its primary amino acid sequence. The expression level of RzNIT/W167G in E. coli BL21 (DE3)/pET-3a-RzNIT/W167G was higher than that of E. coli BL21 (DE3)/pET-28b-RzNIT/W167G (Fig. 1A, lane 1–2), which was quantified using Image J software and calculated based on calibration with protein standards. As shown in Fig. 1A (lanes 3–6), no precipitation was found in the insoluble fraction, indicating that the inclusion body of nitrilase was not formed by constitutive expression. The enzyme activity of constitutive expression system was determined to be 0.12 U/mL, which was approximately onefold higher than that of inducible expression (0.06 U/mL). Biomass (OD600) of constitutive expression (4.6) was also higher than that of inducible expression (3.5) (Fig. 1B), which may attribute to alleviated toxicity of IPTG to cells in inducible expression (Gong et al. 2018). The results indicated that the constitutive expression system of RzNIT/W167G was successfully constructed and superior to inducible expression system.
Comparison of inducible and constitutive expression of nitrilase in E. coli cells. A SDS-PAGE analysis of the distribution of nitrilase. M: protein markers; 1: whole-cell fraction of inducible expression; 2: whole-cell fraction of constitutive expression; 3: soluble fraction of inducible expression; 4: soluble fraction of constitutive expression; 5: insoluble fraction of inducible expression; 6: insoluble fraction of constitutive expression. B Biomass and enzyme activity of inducible and constitutive expression
Constitutive expression of RzNIT/W167G was further performed in a 5 L fermenter. Biomass and enzyme activity gradually increased during the fermentation. As shown in Fig. 2, the highest OD600 was 30.5 and the highest enzyme activity reached 1.03 U/mL after 24 h culture.
Effect of temperature on enzyme activity and thermostability
The nitrilase activity of whole cells was monitored at different temperatures (20–65 °C). The result indicated that the resting cells showed the maximum activity at 45 °C and the activity decreased when the temperature exceeded 50 °C (Fig. 3A). To determine the thermostability, the resting cells were incubated at 30 °C and 40 °C. As shown in Fig. 3B, thermostability of the nitrilase at 30 °C was higher than that at 40 °C, which exhibited a half-life time of 21.8 h. Additionally, 100 mM 2-CN could be completely converted within 20 h at 25 °C and within 16 h at 30 °C (Fig. S1). Thus, 30 °C was chosen as the reaction temperature in the following experiments.
Effect of pH on enzyme activity and stability
The effect of pH on the hydrolysis of 2-CN was investigated. As shown in Fig. 4A, RzNIT/W167G exhibited higher activities at pH between 7.0 and 9.0, and the highest activity was achieved at pH 8.0. However, the nitrilase exhibited better stability at pH 7.0 (Fig. 4B). In addition, it was noteworthy that generation of 2-CA would acidify the reaction mixture, resulting in a decrease in pH value. Thus, 200 mM sodium phosphate buffer (pH 7.0) was selected to maintain pH of the reaction mixture.
Effect of cell loading on biotransformation of 2-CN
The effect of cell loading on enzymatic hydrolysis of 2-CN was investigated at a substrate concentration of 300 mM. As shown in Fig. 5, the concentration of 2-CA increased with the increase of cell loading. High concentration of 2-CN (300 mM) could be completely converted into 2-CA at cell loadings of 4.0 and 5.0 g DCW/L. Insufficient amounts of biocatalysts may lead to long reaction time, while excess biocatalysts were unfavorable for the downstream separation. Thus, 4.0 g DCW/L of resting cells was selected for the biotransformation.
Effect of 2-CA concentration on the biotransformation of 2-CN
Low product tolerance was a common barrier in the enzymatic conversion of nitriles to acids (Sharma et al. 2011; Zhu et al. 2014). For example, 300 mM nicotinic acid caused 44% inhibition of the activity of 3-cyanopyridinase from Nocardia rhodochrous LL100-21 (Vaughan et al. 1989). Thus, the effect of 2-CA concentration on RzNIT/W167G was investigated. Different concentrations of 2-CA were added at the initial of biotransformation. The production of 2-CA was not obviously affected when the added 2-CA concentration increased from 0 to 300 mM, indicating that the product was not an inhibitor for the nitrilase (Fig. S2). These results suggested that RzNIT/W167G is suitable to achieve a high product concentration for efficient industrial production of 2-CA.
Biosynthesis of 2-CA using whole E. coli cells
To further explore the performance of RzNIT/W167G in production of 2-CA, the biotransformation was carried out using whole-cell biocatalyst in a 1 L stirred bioreactor. The biotransformation was performed at 30 °C in the reaction mixture containing 200 mM sodium phosphate buffer (pH 7.0), 4.0 g DCW/L whole cells, and 350 mM 2-CN. The product accumulation reached 318.5 mM with a yield of 91% within 50 h (Fig. 6), which is the highest level for 2-CA production catalyzed by nitrilase to date. The cells were then removed by centrifugation and the impurities were absorbed by activated carbon, in which 2-CA was crystalized by acidification from the supernatant (Fig. 6). The solid 2-CA was obtained by filtration and dried at 50 °C. The yield of 2-CA separated from the reaction mixture was 90% with a purity of 99%.
Conclusions
An efficient bioprocess for 2-CA production was developed using recombinant nitrilase RzNIT/W167G as biocatalyst. The constitutive expression system was established and employed to enhance the biomass and nitrilase production. Under the optimized reaction condition, the final 2-CA concentration reached 318.5 mM using whole cells as biocatalyst. To the best of our knowledge, this is the highest concentration reported for nitrilase-catalyzed synthesis of 2-CA from 2-CN to date. 2-CA was crystalized by a simple acidification step and the recovery yield reached 90%. The obtained results demonstrated the potential of industrial production of 2-CA by the established bioprocess.
References
Chen Z, Wang H, Yang L et al (2021) Significantly enhancing the stereoselectivity of a regioselective nitrilase for the production of (S)-3-cyano-5-methylhexanoic acid using an MM/PBSA method. Chem Commun 57:931–934. https://doi.org/10.1039/d0cc07106d
Du XH, Ge LD, Chen SY, Zhao JB, ZHong ZK, Wang JL, Xu ZY (2009) Patent. CN200910152436.1
Gong JS, Dong TT, Gu BC et al (2017) Semirational engineering accelerates the laboratory evolution of nitrilase catalytic efficiency for nicotinic acid biosynthesis. ChemCatChem 9:3395–3401. https://doi.org/10.1002/cctc.201700665
Gong JS, Zhang Q, Gu BC et al (2018) Efficient biocatalytic synthesis of nicotinic acid by recombinant nitrilase via high density culture. Bioresour Technol 260:427–431. https://doi.org/10.1016/j.biortech.2018.03.109
Hauer B (2020) Embracing nature’s catalysts: a viewpoint on the future of biocatalysis. ACS Catal 10:8418–8427. https://doi.org/10.1021/acscatal.0c01708
Jin LQ, Li YF, Liu ZQ et al (2011) Characterization of a newly isolated strain Rhodococcus erythropolis ZJB-09149 transforming 2-chloro-3-cyanopyridine to 2-chloronicotinic acid. N Biotechnol 28:610–615. https://doi.org/10.1016/j.nbt.2011.04.004
Lauder K, Anselmi S, Finnigan JD et al (2020) Enantioselective synthesis of α-thiocarboxylic acids by nitrilase biocatalysed dynamic kinetic resolution of α-thionitriles. Chem - A Eur J 26:10422–10426. https://doi.org/10.1002/chem.202001108
Liu JF, Zhang ZJ, Li AT et al (2011) Significantly enhanced production of recombinant nitrilase by optimization of culture conditions and glycerol feeding. Appl Microbiol Biotechnol 89:665–672. https://doi.org/10.1007/s00253-010-2866-y
Quevedo CE, Bavetsias V, McDonald E (2009) Microwave-assisted synthesis of 2-aminonicotinic acids by reacting 2-chloronicotinic acid with amines. Tetrahedron Lett 50:2481–2483. https://doi.org/10.1016/j.tetlet.2009.03.034
Sharma NN, Sharma M, Bhalla TC (2011) An improved nitrilase-mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol 38:1235–1243. https://doi.org/10.1007/s10295-010-0902-7
Shen Q, Yu Z, Lv P et al (2020) Engineering a Pichia pastoris nitrilase whole cell catalyst through the increased nitrilase gene copy number and co-expressing of ER oxidoreductin 1. Appl Microbiol Biotechnol 104:2489–2500. https://doi.org/10.1007/s00253-020-10422-4
Shen JD, Cai X, Liu ZQ, Zheng YG (2021) Nitrilase: a promising biocatalyst in industrial applications for green chemistry. Crit Rev Biotechnol 41:72–93. https://doi.org/10.1080/07388551.2020.1827367
Tang XL, Jin JQ, Wu ZM et al (2018) Structure-based engineering of amidase from Pantoea sp. for efficient 2-chloronicotinic acid biosynthesis. Appl Environ Microbiol 85:e02471-e2518. https://doi.org/10.1128/AEM.02471-18
Teepakorn C, Zajkoska P, Cwicklinski G et al (2021) Nitrilase immobilization and transposition from a micro-scale batch to a continuous process increase the nicotinic acid productivity. Biotechnol J 16:e2100010. https://doi.org/10.1002/biot.202100010
Vaughan PA, Knowles CJ, Cheetham PSJ (1989) Conversion of 3-cyanopyridine to nicotinic acid by Nocardia rhodochrous LL100-21. Enzyme Microb Technol 11:815–823. https://doi.org/10.1016/0141-0229(89)90055-0
Wu Y, Wu C, Yan S, Hu B (2019) Solubility determination of 2-chloronicotinic acid and analysis of solvent effect. J Chem Eng Data 64:5578–5583. https://doi.org/10.1021/acs.jced.9b00661
Wu ZM, Hao CL, Tong T et al (2020) Purification of (S)-3-cyano-5-methylhexanoic acid from bioconversion broth using an acetone/ammonium sulfate aqueous two-phase system. Process Biochem 89:186–192. https://doi.org/10.1016/j.procbio.2019.10.023
Wu S, Snajdrova R, Moore JC et al (2021) Biocatalysis: enzymatic synthesis for industrial applications. Angew Chemie - Int Ed 60:88–119
Xue YP, Shi CC, Xu Z et al (2015) Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering. Adv Synth Catal 357:1741–1750. https://doi.org/10.1002/adsc.201500039
Yu S, Yao P, Li J et al (2019) Improving the catalytic efficiency and stereoselectivity of a nitrilase from: Synechocystis sp. PCC6803 by semi-rational engineering en route to chiral γ-amino acids. Catal Sci Technol 9:1504–1510. https://doi.org/10.1039/c8cy02455c
Yu S, Li J, Yao P et al (2021) Inverting the enantiopreference of nitrilase-catalyzed desymmetric hydrolysis of prochiral dinitriles by reshaping the binding pocket with a mirror-image strategy. Angew Chemie - Int Ed 60:3679–3684. https://doi.org/10.1002/anie.202012243
Zhang Q, Lu XF, Zhang Y et al (2020a) Development of a robust nitrilase by fragment swapping and semi-rational design for efficient biosynthesis of pregabalin precursor. Biotechnol Bioeng 117:318–329. https://doi.org/10.1002/bit.27203
Zhang XH, Wang CY, Cai X et al (2020b) Upscale production of (R)-mandelic acid with a stereospecific nitrilase in an aqueous system. Bioprocess Biosyst Eng 43:1299–1307. https://doi.org/10.1007/s00449-020-02326-4
Zheng RC, Jin JQ, Wu ZM et al (2018) Biocatalytic hydrolysis of chlorinated nicotinamides by a superior AS family amidase and its application in enzymatic production of 2-chloronicotinic acid. Bioorg Chem 76:81–87. https://doi.org/10.1016/j.bioorg.2017.11.001
Zheng RC, Dai AD, Zheng, YG (2020) Patent CN112063607-A
Zhu XY, Gong JS, Li H et al (2014) Bench-scale biosynthesis of isonicotinic acid from 4-cyanopyridine by Pseudomonas putida. Chem Pap 68:739–744. https://doi.org/10.2478/s11696-013-0521-7
Zou S, Hua D, Jiang Z et al (2021) A integrated process for nitrilase-catalyzed asymmetric hydrolysis and easy biocatalyst recycling by introducing biocompatible biphasic system. Bioresour Technol. https://doi.org/10.1016/j.biortech.2020.124392
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFE0129400) and Natural Science Foundation of Zhejiang Province (No. LR19B060001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dai, AD., Wu, ZM., Zheng, RC. et al. Constitutive expression of nitrilase from Rhodococcus zopfii for efficient biosynthesis of 2-chloronicotinic acid. 3 Biotech 12, 50 (2022). https://doi.org/10.1007/s13205-022-03119-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03119-0