Abstract
Gut symbiotic bacteria provide protection and nutrition to the host insect. A high reproductive rate and dispersal ability of the rugose spiralling whitefly help this polyphagous species to develop and thrive on many horticultural crops. In this study, we isolated the cultivable gut bacteria associated with rugose spiralling whitefly and demonstrated their role in the host insect. We also studied the influence of antibiotics on the rugose spiralling whitefly oviposition. A total of 70 gut bacteria were isolated from the second nymphal stage of rugose spiralling whitefly reared on coconut, banana, and sapota using seven growth media. From the 70 isolates, chitinase, siderophore (51), protease (44), and Glutathione-S-Transferase producers (16) were recorded. The activities of chitinase, siderophore, protease, and Glutathione-S-Transferase in the gut bacterial isolates of rugose spiralling whitefly ranged from 0.07 to 3.96 µmol–1 min–1 mL–1, 10.01 to 76.93%, 2.10 to 83.40%, and 5.21 to 24.48 nmol–1 min–1 mL–1 μg–1 protein, respectively. The16S rRNA gene sequence analysis revealed that bacterial genera associated with the gut of rugose spiralling whitefly included Bacillus, Exiguobacterium, Acinetobacter, Lysinibacillus, Arthrobacter, and Pseudomonas. Based on the susceptibility of the gut bacteria to antibiotics, 11antibiotic treatments were administered to the host plant leaves infested with the nymphal stages. The antibiotics were evaluated for their effect on rugose spiralling whitefly oviposition. Among the antibiotic treatments, carbenicillin (100 µg mL–1) + ciprofloxacin (5 µg mL–1) significantly reduced the oviposition (13 eggs spiral–1) and egg hatchability (61.54%) of rugose spiralling whitefly. Disruption of chitinase, siderophore, protease, and detoxification enzyme producers and elimination of these symbionts through antibiotics altered the host insect physiology and indirectly affected whitefly oviposition. In conclusion, gut bacteria-based management strategies might be used as insecticides for the effective control of whiteflies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rugose spiralling whitefly (RSW), Aleurodicus rugioperculatus was originally described as a pest of coconut in Belize and Mexico in 2004 (Martin 2004). In India, RSW was first found in the coconut farms of the Pollachi area of Tamil Nadu and Palakkad area of Kerala during July–August 2016. RSW infestation was recorded in banana, guava, custard apple, sapota, and other ornamental plants such as areca palm/butterfly palm, oleander, and bird of paradise in India (Selvaraj et al. 2017; Stocks and Hodges 2012). Among these, coconut (40–60%) and banana (25–40%) were severely infested, with 18–43 nymphs/sq.cm present at the midrib region of plants (Selvaraj et al. 2017). Presently, RSW is managed by its introduced natural enemy, Encarsia guadeloupae Viggiani, and systemic neonicotinoid chemical pesticides applied to the leaves, soil, and trunk (Taravati et al. 2018). The application of chemical pesticides might cause the development of resistance and negatively affect the natural enemy. Currently, there is no standard management practice available for controlling RSW. Alternatively, a gut bacteria-based management strategy might be utilized for the sustainable management of whiteflies (Indiragandhi et al. 2007).
Insects contain many types of microorganisms in their intestinal tract, and the interaction between the microorganisms and their hosts varies from symbiosis to pathogenesis (Mrázek et al. 2008). Plant phloem sap is rich in carbohydrates but deficient in nitrogen and essential amino acids (Pandey et al. 2013). The gut bacteria of whiteflies provide nutrients and essential amino acids from food (Douglas 1998). They are also a nitrogen source (Gil et al. 2004) and provide resistance to temperature, insecticides, and natural enemies (Montllor et al. 2002; Oliver et al. 2003; Werren 2012), and immunity (Weiss et al. 2011) to their host. Gut bacteria produce iron-binding compounds called siderophores that are used to obtain iron from the host insect for bacterial growth and development (Ciche et al. 2003). Gut bacterial siderophore protects the host insects from entomopathogens (Indiragandhi et al. 2007). A major constraint in the application of chemical pesticides for pest management is insecticide resistance via detoxification enzymes such as glutathione-S-transferase (Kim et al. 2007; Mohan and Gujar 2003). Insecticides ingested by insects can be detoxified by gut bacterial glutathione-S-transferase, which influences the detoxification process in major insects such as Helicoverpa armigera and Plutella xylostella (Genta et al. 2006; Indiragandhi et al. 2007; Lauzon et al. 2003; Xiang et al. 2006).
Gut bacteria play a major role in the synthesis of proteins and enzymes responsible for the transmission of viruses in aphids and honeydew secretion in whiteflies. Disruption of these gut bacteria through antibiotics interferes with the transmission of viruses and honeydew secretion by these insects (Davidson et al. 1994; van den Heuvel et al. 1994). RSW can produce large amounts of honeydew, which completely masks plants with sooty mold growth and disrupts photosynthesis (Kumar et al. 2013). Insect midgut, lined with the peritrophic membrane (PM), consists of chitin microfibrils embedded in a protein-carbohydrate matrix (Merzendorfer and Zimoch 2003), which supplies chitin that acts as a source of nitrogen and carbon for gut bacterial growth and benefits the host insect (Indiragandhi et al. 2007). Using the chitinous morphological framework to disrupt the structural part of the insect using enzymes produced by gut bacteria is an eco-friendly method as it is difficult for insects to develop resistance against microbial enzymes. Gut bacterial chitinases are used for the partial degradation of the old cuticle in insects (Okongo et al.2019).Culture-dependent isolation of gut bacteria provides information about the functional significance of bacterial isolates and their enzymes can be engineered for pest management (Hernández et al. 2015).
Host plants influence the gut microbial diversity in host insects. This observationwas supported by the findings of Jones et al. (2019), who showed that maize and soybean altered the microbial communities in the fall armyworm, Spodoptera frugiperda. The gut bacterial populations of Henosepilachna vigintioctopunctata are influenced by the host plants, Solanum melongena (QZ) and Solanum nigrum (LK). LK isassociated with phylum Cyanobacteria, class Alphaproteobacteria, and genus Ochrobactrum, while QZ supports Bacillus and Lactococcus (Lu et al. 2019).
There are no studies on cultivable gut bacterial diversity in rugose spiraling whiteflies and their relation to the host plants since RSW is a recent invasive species. Understanding the effect of antibiotics on the fitness of host insects is a novel strategy for the sustainable management of whitefly through endosymbionts (Costa et al. 1997). This study was designed to reveal the cultivable gut bacterial diversity of rugose spiraling whitefly reared on three host plants along with their functional significance and also to study the effects of antibiotics on RSW oviposition and offspring development.
Materials and methods
Mass culturing of rugose spiralling whitefly, Aleurodicus rugioperculatus
Coconut leaflets infested with the rugose spiralling whitefly (RSW) were collected from an orchard in Tamil Nadu Agricultural University (TNAU) (11.0123° N, 76.9355° E), Coimbatore, India.The insects were released on mud-potted (pot diameter: 41 cm) plants of coconut, banana, and sapota, which were kept in separate mini net houses (9′ × 5′ × 7′ with nylon net mesh size of 120 µm) and were maintained at an Insectary in the Department of Agricultural Entomology at 31 ± 2 °C, 60–75% RH, and under natural light.
Isolation of cultivable gut bacteria
The second nymphal stage of RSW damages the host plants considerably. For the isolation of gut bacteria, 50 individuals in the second nymphal stage were collected from coconut, banana, and sapota using a hair-brush under the microscope and starved for 24 h to eliminate the transient gut bacteria. Then, the nymphs were surface sterilized with 70% ethanol followed by 5% sodium hypochlorite for 1 min and washed with sterile water 3–5 times to remove adhering contaminants. Next, 0.1 mL of the final wash was added to 5 mL of Tryptic soy broth (TSB) and incubated for 48 h at 28 ± 2 °C to ensure surface sterilization. No growth was noticed in TSB. The second nymphal stage of RSW is very small, and it is difficult to dissect the gut. Hence, the whole RSW nymph was taken for gut bacteria isolation after surface disinfection (Malathi et al. 2017). The nymphs were homogenized with 0.1 M phosphate buffer (pH 7.0). The nymphal homogenates were serially diluted in sterile distilled water and 0.1 mL of was spreaded on seven different bacterial growth media, viz., nutrient agar, Luria Bertani, MacConkey agar, tryptic soy agar, endo agar, Reasoner’s 2A agar, and MRS agar (M/s. HiMedia Laboratories, Mumbai, India) (de Vries et al. 2001). Petri plates containing gut homogenates were incubated for 48 h at 28 ± 2 °C and monitored for the formation of new colonies at an interval of 24 h. The bacterial colonies were selected based on their colony morphology. Selected colonies were subjected to continuous streaking four-six times to obtain a pure culture,which was confirmed through an examination under a microscope. Purified colonies were stored in 60% glycerol at − 80 °C for further experiments (Indiragandhi et al. 2007).
Molecular characterization
A total of 51 chitinase-producing gut bacteria were grown in nutrient broth for genomic DNA isolation using HipurA® Bacterial Genomic DNA Purification Kit (M/s. HiMedia Laboratories, Mumbai, India). Bacterial genomic DNA was then amplified with 16S rRNA gene target universal primers 27F (5′AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) by a thermocycler (Mastercycler® nexus, Eppendorf India Private Limited). The PCR reaction mixture (25 µL) contained 20 ng of template DNA, 0.3 µL of 1 U TaqDNA polymerase (Sigma, India), 2.5 µL of 1 × Taq DNA polymerase buffer (Sigma, India), 2 mM magnesium chloride (Sigma, India), 400 µM dNTPs (Sigma, India), 10 pmol of forward and reverse primer, and the final volume was made up with sterilized deionized water. The thermoprofile condition of PCR consisted of initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 50 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min and hold at 4 °C. Genomic DNA was viewed and documented using a gel documentation and analysis system (Vilber E-box, Germany).
The fluorescent dye terminator method was used for obtaining the nucleotide gene sequences of 16S rRNA, which was then purified by the Millipore-Montage dye removal kit. Sanger sequencing was performed at the M/s. Barcode Biosciences, Bengaluru, India. The 16S rRNA sequences of the isolates obtained from the automatic sequencer were then aligned and identified using the e-server, EzTaxon (http://eztaxon-e.ezbiocloud.net/)(Yoon et al. 2017) to determine their closest relative. The identified sequences were submitted to NCBI (http://www.ncbi.nlm.nih.gov/blast/) under the accession numbers MN782273-MN782284, MN784432-MN784435, MN907648-MN907660, MT027239, and MN907663-MN907689 (Table 2).Based on the molecular confirmation of the bacterial isolates and the representative isolates of each species, 16 bacterial specieswere selected for glutathione-S-transferase and antibiotic susceptibility experiments.
Diversity indices
The Shannon diversity index (H′) and the Margalef index of richness (K) were calculated using the equations \(H^{\prime} = \sum {\left( {Pi} \right)ln\left( {Pi} \right)\,{\text{and }}K = {\text{log }}S{\text{/log }}N{,}}\) respectively. In the equation to calculate the Margalef index, S indicates the number of species,and N indicates the total number of individuals in the sample (Iglesias-Rios and Mazzoni 2014).
Screening of chitinase-producing gut bacteria
Gut bacterial isolates of RSW obtained from the three host plants were screened based on their chitinolytic activity. The bacterial isolates were inoculated in a nutrient broth supplemented with 0.3% colloidal chitin and incubated at 28 ± 2 °C for 24 h. Colloidal chitin was prepared as described by Rodriguez-Kabana et al. (1983). The crude enzyme solution was collected by centrifugation at 4 °C and 5500 ×g for 20 min. The reaction mixture consisted of 0.3% colloidal chitin (0.1 mL), an enzyme solution (0.1 mL), and 0.1 M McIIvaine buffer of pH 6.0 (0.2 mL). The solution was incubated for 25 min at 35 °C. The reaction was terminated by placing the solution in a water bath for 15 min and adding 2.0 mL of 1.5 mmol potassium ferricyanide. The absorbance was measured using a spectrophotometer (®Spectrophotometer 166, Systronics, India) at 420 nm. Known concentrations of N-Acetyl glucosamine (10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 µmol) were used to generate a standard curve for calculating the enzyme activity. One unit of enzyme activity, i.e., the amount of enzyme required to release 1 µmol of N-Acetyl glucosamine min–1 (Wiwat et al. 2000), was calculated. The bacterial isolates were screened based on their chitinolytic activity and subjected to further experiments.
Siderophore production assay
Siderophore-producing bacterial isolates were identified using the ChromAzurol S (CAS) agar plate method as per Milagres et al. (1999). The autoclaved basal agar medium and CAS indicator solution were allowed to cool to 50 °C. The glucose solution (50%) was prepared and autoclaved. Once cooled, 2 mL of the 50% glucose solution was added to the autoclaved basal agar medium. Then, 10 mL of the CAS indicator solution was added carefully along the walls of the flask with constant stirring. Once mixed thoroughly, the resulting CAS agar medium (100 mL) was plated in sterile plates. Next, 10 µL of culture (1 × 107 cfu mL–1, 24 h old) was spotted on CAS agar plates and incubated for 48 h at 28 ± 2 °C. The production of siderophore was indicated by the appearance of a zone with an orange halo around the spotted colonies. The production of siderophore was calculated by the following formula,
Detection of protease activity
The protease enzyme activity of the bacterial isolates was determined using a skim milk agar medium (Cattelan et al. 1999). The medium consisted of peptone (0.1%), NaCl (0.5%), and skim milk (10%). Bacterial culture (1 × 107 cfu mL–1, 10 µL, 24 h old) was spotted on skim milk agar plates and incubated for 24 h at 28 ± 2 °C. The diameter of the clearing zone around the spot was measured. The protease activity was calculated by the following formula,
Estimation of Glutathione-S-Transferase (GST) in gut bacterial isolates of RSW
Gut bacterial isolates of RSW were inoculated in nutrient broth and incubated for 24 hat 28 ± 2 °C. After incubation, the bacterial cells were harvested by centrifugation (8000 ×g for 1 min) and washed twice with buffer K. Then, the cell pellets were suspended in 3–4 volumes of buffer K and subjected to sonication for cell disruption in three 20-s bursts at 50% power and 0 °C. The disrupted cell homogenates were centrifuged for 5 min at 14,000 ×g and 10 °C. Bacterial GST was estimated through the CDNB (1-chloro-2, 4-dinitrobenzene) substrate method as per Lau et al. (1980) and Zablotowicz et al. (1995). The reaction volume (2 mL) contained 1.0 mmol of CDNB and 1.0 mmol of reduced glutathione. The reaction mixture was vortexed and observed spectrophotometrically (®Spectrophotometer 166, Systronics, India) at 340 nm for 5.0 min. This experiment was replicated thrice and control reactions (without enzyme) were maintained conjugation. A unit of enzyme activity (µmol of CDNB/DCNB conjugated min–1 mg of protein –1) was calculated using the extinction coefficient of 9.6 mM–1 cm–1. Enzyme activity was correlated with the amount of protein in each sample, and protein concentration was measured by the standard method of Bradford (1976).
Antibiotic susceptibility test for gut bacterial isolates of RSW
Gut bacteria associated with RSW were subjected to sensitivity tests by the Kirby-Bauer disk diffusion method against different antibiotics (Erythromycin E15, Streptomycin S10, Rifampicin RIF5, Polymyxin-B PB300, Vancomycin VA30, Cefotaxime CTX30, Doxycycline DO20, Trimethoprim TR5, Ciprofloxacin CIP5, Colistin CL10, Ampicillin AMP10, Nalidixin NA30, Bacitracin B8, Tetracycline TE30, Carbenicillin CB100, Kanamycin K30, Spectinomycin SPT100, Chloramphenicol C30, and Novobiocin NV30) (M/s. HiMedia Laboratories, Mumbai, India). Gut bacterial isolates of RSW were inoculated in nutrient broth and incubated for 24 h at 28 ± 2 °C. After incubation, the bacterial isolates were spread on nutrient agar plates and allowed to dry for 5 min and each antibiotic disc (HiMedia, India) was placed on the surface of the agar using sterilized forceps and incubated for 24 h at 28 ± 2 °C. Then, the diameter of the inhibition zone was measured and compared to the diameter of the inhibition zone published by the Clinical Laboratory Standards Institute (CLSI) to interpret the sensitivity of the antibiotics.
Effect of antibiotics on RSW oviposition
Effect of the antibiotic treatment (T1-Carbenicillin 100 µgmL–1; T2-Ciprofloxacin 5 µgmL–1; T3-Erythromycin 15 µg mL–1; T4-Cefotaxime 30 µg mL–1;T5-Carbenicillin 100 µg mL–1 + Ciprofloxacin 5 µg mL–1;T6-Carbenicillin 100 µg mL–1 + Erythromycin 15 µg mL–1; T7-Carbenicillin 100 µg mL–1 + Cefotaxime 30 µg mL–1; T8-Ciprofloxacin 5 µg mL–1 + Erythromycin 15 µg mL–1; T9-Ciprofloxacin 5 µg mL–1 + Cefotaxime 30 µg mL–1; T10-Erythromycin 15 µg mL–1 + Cefotaxime 30 µg mL–1; T11-control) on the oviposition of RSW was evaluated through a bioassay as per the protocol of the Insecticide Resistance Action Committee susceptibility test 016 (IRAC 2009). The above-mentioned antibiotics were applied on coconut leaves infested with the RSW nymphal stages. Clip cages were placed on antibiotic-treated leaves with RSW in the second nymphal stage to feed and oviposit. The oviposition parameters of RSW, including the number of eggs laid/spiral and the percentage of eggs that hatched into immature stages were recorded.
Scanning electron microscopy
Fresh, hydrated progeny of antibiotic-treated and control population of whitefly eggs and nymphal stages were placed on sticky carbon tabs on aluminum stubs and sputter-coated with gold and palladium in a ratio of 1:1. The coated specimens were visualized at 10 kV using a scanning-transmission electron microscope (Quanta 250, FEI, Netherland) in the SEM mode at magnifications of 100 × to 1000 × and documented as microphotographs.
Statistical analysis
Data were analyzed by performing analysis of variance (ANOVA), and the means were compared using generalized linear models (GLMs) with Tukey’s HSD test. All the analyses were performed using IBM SPSS Statistics 22 (Spss 2013).
Results
Gut bacterial isolation
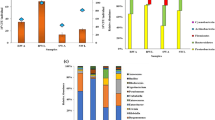
A total of 70 gut bacterial isolates were obtained from the nymphal stages of rugose spiralling whitefly (RSW) cultured on three different host plants, coconut, banana, and sapota, using seven different growth media. No bacterial growth was observed in the MacConkey, MRS, and Endo agar media. From the culture-dependent isolation, we recorded 17 gut bacterial isolates of RSW from coconut, 32 isolates from banana, and 20 isolates from sapota (Table S1). The highest bacterial diversity and species richness were observed in TSA (2.30: 3.90), followed by those in NA (2.08: 3.36), R2A (1.95: 3.08), and LB (1.61: 2.48), respectively. The maximum bacterial diversity and species richness were observed for the isolates of RSW from coconut (2.20: 3.64), followed by those in the isolates of RSW from banana (2.20: 3.64) and sapota (1.61: 2.50), as indicated by the Shannon and Margalef diversity indices (Table 1).
Bacillus (80–100%) was noted the most in the RSW reared on all the tested host plants. The genera Bacillus (30%), Acinetobacter (10%), and Exiguobacterium (10%) were observed in the RSW reared on coconut plants. Bacillus (81%), Lysinibacillus (11%), Arthrobacter (4%), and Pseudomonas (4%) were recorded in the RSW from banana plants. However, only Bacillus (100%) was present in the RSW reared on sapota plants. The NA medium supported the isolation of Bacillus (87%) and Exiguobacterium (13%), whereas, in the LB medium, Bacillus (64%), Lysinibacillus (27%), and Acinetobacter (9%) were dominant. Bacillus (88%), Arthrobacter (6%), and Pseudomonas (6%) were isolated in TSA; however, in the R2A growth medium, only Bacillus species were reported.
Functional significance of cultivable gut bacteria isolated from rugose spiralling whitefly
Out of the 70 isolates, 51 gut bacteria were found to be positive for both chitinase and siderophore production and 44 isolates were positive for protease activity. The chitinolytic activity was the highest in Bacillus siamensis SBRSW9 (3.96 µmol–1 min–1 mL–1) followed by that in Bacillus altitudinis SBRSW2 (3.01 µmol–1 min–1 mL–1), which were isolated from the RSW of banana plants; the lowest chitinolytic activity was recorded in Bacillus altitudinis SSRSW18 (0.07 µmol–1 min–1 mL–1) from the RSW of sapota plants. The maximum proteolytic activity was recorded in Bacillus albus SCRSW11 (76.93%), followed by that in Bacillus subtilis subsp. Stercoris SBRSW19 (71.43%).The minimum proteolytic activity was noted in Bacillus altitudinis SSRSW13 (10.01%) from RSW collected from coconut, banana, and sapota plants, respectively. The maximum siderophore production was recorded in Bacillus subtilis subsp. Stercoris SBRSW19 (83.40%) from banana and the minimum siderophore production was in Bacillus zanthoxyli SCRSW13 (2.10%) from coconut (Table 2).
The Glutathione-S-Transferase activity (GST) for the gut bacterial isolates of RSW ranged from 5.21 to 24.48 nmol–1 min–1 mL–1 μg protein–1 (Fig. 1). The maximum activity of GST was recorded in Bacillus altitudinis SCRSW14 (23.42 nmol–1 min–1 mL–1of 42.9 μg protein–1 min–1), Lysinibacillus xylanilyticus SBRSW13 (24.38 nmol–1 min–1 mL–1of 52.5 μg protein–1 min–1), and Pseudomonas stutzeri SBRSW22 (23.44 nmol–1 min–1 mL–1of 49.00 μg protein–1 min–1), while the minimum enzyme activity was observed in Acinetobacter refrigeratoris SCRSW5 (5.21 nmol–1 min–1 mL–1of 20.3 μg protein–1 min–1) (F = 486.199, P < 0.001).
Glutathione-S-Transferase (GST) activity of gut bacterial isolates from rugose spiralling whitefly (RSW). GST enzyme activity was estimated using CDNB substrate. Values in each column are mean of 3 replications of ± Standard Error (SE). Means in a column followed by a different letter(s) are significantly different (F = 486.199, P < 0.001; Tukey’s HSD test)
Antibiotic susceptibility test
Sixteen gut bacterial isolates of RSW were tested to assess their susceptibility toward19 antibiotics. The majority of the gut bacterial isolates were susceptible to most of the tested antibiotics, except Ampicillin AMP10, Colistin CL10, Kanamycin K30, and Polymyxin-B PB300. Among the 16 gut bacterial isolates, seven bacterial isolates were susceptible to Erythromycin E15 and Ciprofloxacin CIP5, and six bacterial isolates were susceptible to Carbenicillin CB100 and Cefotaxime CTX 30. Hence, Erythromycin E15, Ciprofloxacin CIP5, Carbenicillin CB100, and Cefotaxime CTX 30 were selected based on the maximum zone of inhibition toward bacterial isolates and were usedfor the antibiotic bioassay (Table S2).
Antibiotic bioassay
Eleven antibiotic treatments were evaluated for RSW oviposition and egg hatchability (%). The number of eggs laid per spiral (Fig. 3a, b) and the egg hatchability of RSW were significantly reduced by the three antibiotic treatments, viz., CB100 + CIP5 (13 eggs spiral–1 and 61.54%) followed by CIP5 + CTX30 (15 eggs spiral–1 and 80.00%), and CB100 (16 eggs spiral–1and 81.25%) compared to the values of the parameters in the RSW of the control population (29 eggs spiral–1and 100%) (Fig. 2).
Effect of antibiotics on rugose spiralling whitefly oviposition. CB100- Carbenicillin 100 µg mL−1, CIP5- Ciprofloxacin 5 µg mL−1, CTX30- Cefotaxime 30 µg mL−1, E15- Erythromycin 15 µg mL−1 sprayed on coconut leaves infested with the RSW nymphal stages and clip cages were placed on antibiotic-treated leaves to observe the effects of antibiotics on RSW oviposition. Values in each column are mean of 3 replications of ± Standard Error (SE). Means in a column followed by a different letter(s) are significantly different (P = 0.05; Tukey’s HSD test)
Scanning electron microscopy
The progeny of the antibiotic-treated RSW showed variation in the morphometry of the egg and nymphal stages. The SEM micrograph showed longer and narrower eggs and nymphs after antibiotic treatment of whiteflies compared to the eggs and nymphs of control whiteflies. The antibiotic-treated eggs and nymphs were longer (320.2 μm and 722.9 μm) than those in the control population (311.6 μm and 675.3 μm). Similarly, eggs (121.7 μm) and nymphs (431.3 μm) were narrower in the antibiotic-treated samples than the eggs (137.1 μm) and nymphs (473.3 μm) in the samples of the control population (Fig. 3c–f). Additionally, lesser wax threads were observed on the nymphal progeny of the antibiotic-treated whiteflies than those on the nymphal progeny of the whiteflies in the control population (Fig. 3e, f).
Effect of antibiotics on RSW oviposition and progeny development. (a) image of egg spirals laid by control (b) antibiotic-treated RSW adults; (c) egg laid by control RSW adult (d) egg laid by antibiotic-treated RSW adult; (e) nymphal progeny from control adult (f) nymphal progeny from antibiotic-treated adult. Eggs laid by RSW in antibiotic treatment, carbenicillin 100 µg mL−1 + ciprofloxacin 5 µg mL−1 treated on coconut leaves showed the least no of eggs per spiral in treated than control viewed in image analyser (Leica M205C) using LAS X software at 40 × magnification (3a and 3b) and egg size, shape changes observed under scanning electron microscope at 500 × magnification (c and d) in antibiotic-treated population. Nymphal progeny of antibiotic-treated RSW adults were observed the wax threads present in the treated population over control (e and f) in scanning electron microscope at 250 × magnification
Discussion
This study revealed the cultivable gut bacteria of rugose spiralling whitefly (RSW) nymphs reared on three different host plants. Based on the molecular confirmation of bacterial isolates, 16 bacterial species were identified, which included Acinetobacter, Arthrobacter, Exiguobacterium, Bacillus, Lysinibacillus, and Pseudomonas. Bacillus species were obtained from all selected host plants and isolation media. Similarly, 11bacterial genera were isolated from sweet potato whitefly, Bemisia tabaci, which included Pseudomonas, Deinococcus, Sphingomonas, Acinetobacter, Staphylococcus, Modestobacter, Micrococcus, Bacillus, Kocuria, Microbacterium, Erwinia, Brevibacterium, Exiguobacterium, and Moraxella (Ateyyat et al. 2010; Indiragandhi et al. 2010; Visôtto et al. 2009).
Varied cultivable gut bacterial populations were present in the nymphal stage of RSW, which might directly or indirectly influence the nutritional role of the host insect. In this study, a high density of Bacillus was found in the RSW nymphal stage. Bacillus and Staphylococcus in the whitefly Bemisia argentifolii produce medium-length sugars from derived sucrose and increase the stickiness of honeydew secreted by homopteran insects (Davidson et al. 2000). Bacillus species present in the nymphal stage of RSW might be responsible for the production of honeydew, which increases the incidence of sooty mold. RSW produces copious amounts of honeydew, which leads to the growth of the sooty mold fungus, Capnodium spp., that disrupts the photosynthesis of the plant, and thus, indirectly affects the quality of coconuts (Mannion 2010; Sundararaj and Selvaraj 2017).
The results suggested that bacterial isolation media and host insect plants influence the cultivable gut bacterial diversity of homopteran insects. The selectionof bacterial growth media affects the number and diversity of bacterial phylotypes in the isolation of cultivable bacterial populations from host insects (Davidson et al. 2000). In the present study,various species of cultivable bacteria were retrieved from TSA, NA, R2A, and LB. Cultivable bacterial diversity was the highest in coconut, followed by that in banana and sapota. This was probably because RSW is primarily a pest of coconut.
Similar studies suggested that host plants influence the number and diversity of cultivable gut bacteriain the host insect (Broderick et al. 2004). Host plants have a positive impact on the shaping of microbial communities associated with Spodoptera littoralis (Tang et al. 2012), Helicoverpa spp. (Priya et al. 2012; Tang et al. 2012; Xiang et al. 2006), Lymantria dispar (Broderick et al. 2004; Mason and Raffa 2014), and Leptinotarsa decemlineata (Chung et al. 2017). Plant characters such as leaf surface, wax composition, and the availability of sugars in plants might influence bacterial community composition in the host insect (Lindow and Brandl 2003).
Potential bacterial isolates were screened based on their chitinase-producing nature, and the screened isolates were studied to understand their functional significance. Seventy gut bacterial isolates were found in RSW, of which, 51 isolates produced chitinase and siderophore and 44 isolates exhibited protease activity. Insect midgut lined with the peritrophic membrane (PM) supports digestion and nutrient absorption. Gut bacteria produce chitinase, protease, and siderophores, which provide protection and enhance the growth of host insects (Indiragandhi et al. 2007).
Gut bacteria conserve the thickness of the PM, which affects the diffusion of nutrients across the insect gut (Shen and Jacobs-Lorena 1997). Gut bacteria produce enzymes that can inactivate the immune system of the host insect (Marokházi et al. 2004). In pest management strategy, chitinase and protease-producing gut bacteria were reported to enter the host gut through feeding and disrupt the thickness of the peritrophic membrane; thus, causing a nutrient imbalance in the host insect leading to insect mortality (Krishnamoorthy et al. 2020; Okongo et al. 2019).
Symbiotic bacteria associated with the host insect produce detoxifying enzymes like Glutathione-S-transferase, which provide resistance against insecticides (Baek et al. 2005; Mohan and Gujar 2003). In the present study, glutathione-S-transferase activity in the gut bacterial isolates of RSW ranged from 5.21 to 24.48 nmol–1 min–1 mL–1 μg protein–1. Xenobiotic and endogenous compounds that are detoxified by the glutathione-S-transferase enzyme include DDT, abamectin, and organophosphate insecticides (Hayes and Wolf1988; Pavlidi et al. 2015). Higher GST activity was reported in B. tabaci Q biotype than in B biotype and showed less susceptibility to insecticides (Kim et al. 2007; Seo et al. 2007). Yang et al. (2016) reported a higher GST activity in the thiamethoxam-resistant B. tabaci THQR strain compared to the GST activity in the susceptible strain of B. tabaci THQS. Similarly, Singh and Walker (2006) reported that Bacillus sp. in Nilaparvata lugens populations were able to degrade the environmental pollutants, including organophosphorus compounds. Furthermore, Krishnamoorthy et al. (2020) found that Bacillus sp.in papaya mealybug might have arole in the detoxification of profenophos and chlorpyrifos OP compounds that were used for the management of the mealybug complex.
The water-soluble antibiotic fumagillin was used for treating Nosema infections in Bombyx occidentalis (Whittington and Winston 2003) and Apis mellifera (Webster 1994), but it showed a negative impact on the developmental stages of Bombyx occidentalis and A. mellifera. Antibiotic-treated individuals of wood tiger moth, Parasemia plantaginis, laid a lesser number of eggs and showed lower immune competence. Antibiotic treatment had deleterious effects on insect populations and reproductive success (Dickel et al. 2016). Similarly, the results of this study revealed that the number of eggs laid per spiral and egg hatchability were affected in the populations treated with Carbenicillin 100 µgmL–1 + Ciprofloxacin 5 µgmL–1 relative to the values in the control population (Fig. 3a, b).
Antibiotics with a different mode of action alter the endosymbiont population of the whiteflies (Ahmed et al. 2010; Costa et al. 1997). Carbenicillin disrupted the components required for the synthesis of peptidoglycans in the bacterial cell wall by inactivating the transpeptidase enzyme (Butler et al. 1970). Ciprofloxacin impaired the secretion of DNA gyrase, which is responsible for DNA synthesis (Zweerink and Edison 1986). Costa et al. (1997) reported that carbenicillin disrupted the bacterial cell wall and negatively affected the developmental time and offspring emergence in whiteflies. Thetreatment of secondary symbionts through antibiotics in B. tabaci probably negatively affects the host insect (Ridley et al. 2013; Ruan et al. 2006; Shan et al. 2016). Rifampicin and oxytetracycline treatment negatively affected the growth and development of the offspring of B. tabaci (Costa et al. 1993, 1997; Ruan et al. 2006; Xue et al. 2012).
SEM micrographs showed that theeggs and nymphs of antibiotic-treated whiteflies were longer and narrower, respectively,compared to the eggs and nymphs of the whiteflies in the control population. Buchnera elimination with rifampicin, negatively affected the body size, body mass, length,width, fertility, and nutritional requirement of A.pisum (Lv et al. 2018). Our results were similar to those of Ruan et al. (2006), who reported that the offspring of B. tabaci were longer in the population treated with rifampicin and tetracycline thanthe offspring of B. tabaci in the control population.
Presently, the invasive rugose spiraling whitefly is managed by its natural enemy, the Encarsia guadeloupae wasp. The practical utility of this natural enemy in the field was difficult to test due to the lack of knowledge on standard mass-production procedures. Chitinase, siderophore-, and protease-producing gut bacteria and the elimination of symbionts through antibiotics negatively affected the host insect physiology. Rapid and widespread infestation of RSW occurs on host plants due to its high fecundity. Antibiotics negatively affect the development of the progeny of whiteflies. Gut bacteria might be considered as insecticides for the sustainable management of RSW. However, the bio-control efficacy of these isolates is yet to be tested by contact leaf assay or pot culture under field conditions.
References
Ahmed MZ, Ren SX, Xue X, Li XX, Jin GH, Qiu BL (2010) Prevalence of endosymbionts in Bemisia tabaci populations and their in vivo sensitivity to antibiotics. Curr Microbiol 61(4):322–328. https://doi.org/10.1007/s00284-010-9614-5
Ateyyat MA, Shatnawi M, Al-Mazra’awi M (2010) Isolation and identification of culturable forms of bacteria from the sweet potato whitefly Bemesia tabaci Genn. (Homoptera: Aleyrodidae) in Jordan. Turk J Agric for 34(3):225–234. https://doi.org/10.3906/tar-0902-35
Baek JH, Kim JI, Lee DW, Chung BK, Miyata T, Lee SH (2005) Identification and characterization of ace1-type acetylcholinesterase likely associated with organophosphate resistance in Plutella xylostella. Pestic Biochem Phys 81(3):164–175. https://doi.org/10.1016/j.pestbp.2004.12.003
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70(1):293–330. https://doi.org/10.1128/AEM.70.1.293-300.2004
Butler K, English AR, Ray VA, Timreck AE (1970) Carbenicillin: chemistry and mode of action. J Infecdis. https://doi.org/10.1093/infdis/122.supplement_1.s1
Cattelan AJ, Hartel PG, Fuhrmann JJ (1999) Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Am J 63(6):1670–1680. https://doi.org/10.2136/sssaj1999.6361670x
Chung SH, Scully ED, Peiffer M, Geib SM, Rosa C, Hoover K, Felton GW (2017) Host plant species determines symbiotic bacterial community mediating suppression of plant defenses. Sci Rep 7(1):1–13. https://doi.org/10.1038/srep39690
Ciche TA, Blackburn M, Carney JR, Ensign JC (2003) Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol 69(8):4706–4713. https://doi.org/10.1128/AEM.69.8.4706-4713.2003
Costa HS, Johnson MW, Ullmann DE, Tabashnik BE (1993) The antibiotic, oxytetracycline interferes with Bemisia tabaci oviposition, development, and ability to induce squash silverleaf. Ann Entomol Soc Am 86(6):740–748. https://doi.org/10.1093/aesa/86.6.740
Costa HS, Enneberry TJ, Toscano NC (1997) Effects of antibacterial materials on Bemisia argentifolii (Homoptera: Aleyrodidae) oviposition, growth, survival, and sex ratio. J Econ Entomol 90(2):333–339. https://doi.org/10.1093/jee/90.2.333
Davidson EW, Segura BJ, Steele T, Hendrix DL (1994) Microorganisms influence the composition of honeydew produced by the silverleaf whitefly, Bemisia argentifolii. J Insect Physiol 40(12):1069–1076. https://doi.org/10.1016/0022-1910(94)90060-4
Davidson EW, Rosell RC, Hendrix DL (2000) Culturable bacteria associated with the whitefly, Bemisia argentifolii (Homoptera: Aleyrodidae). Fla Entomol. https://doi.org/10.2307/3496151
de Vries EJ, Breeuwer JAJ, Jacobs G, Mollema C (2001) The association of western flower thrips, Frankliniella occidentalis, with a near Erwinia species gut bacteria: transient or permanent? J Invertebr Pathol 77(2):120–128. https://doi.org/10.1006/jipa.2001.5009
Dickel F, Freitak D, Mappes J (2016) Long-term prophylactic antibiotic treatment: effects on survival, immunocompetence and reproduction success of Parasemia plantaginis (Lepidoptera: Erebidae). J Insect Sci 16(1):46. https://doi.org/10.1093/jisesa/iew035
Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43(1):17–37. https://doi.org/10.1146/annurev.ento.43.1.17
Genta FA, Dillon RJ, Terra WR, Ferreira C (2006) Potential role for gut microbiota in cell wall digestion and glucoside detoxification in Tenebrio molitor larvae. J Insect Physiol 52(6):593–601. https://doi.org/10.1016/j.jinsphys.2006.02.007
Gil R, Latorre A, Moya A (2004) Bacterial endosymbionts of insects: insights from comparative genomics. Environ Microbiol 6(11):1109–1122. https://doi.org/10.1111/j.1462-2920.2004.00691.x
Hayes JD, Wolf CR (1988) Role of glutathione transferase in drug resistance. Academic Press, London
Hernández N, Escudero JA, Millán ÁS, González Zorn B, Lobo JM, Verdú JR, Suárez M (2015) Culturable aerobic and facultative bacteria from the gut of the polyphagic dung beetle Thorectes lusitanicus. Insect Sci 22(2):178–190. https://doi.org/10.1111/1744-7917.12094
Iglesias-Rios R, Mazzoni R (2014) Measuring diversity: looking for processes that generate diversity. Nat Conserv 12(2):156–161. https://doi.org/10.1016/j.ncon.2014.04.001
Indiragandhi P, Anandham R, Madhaiyan M, Poonguzhali S, Kim GH, Saravanan VS, Sa T (2007) Cultivable bacteria associated with larval gut of prothiofos-resistant, prothiofos-susceptible and field-caught populations of diamondback moth, Plutella xylostella and their potential for, antagonism towards entomopathogenic fungi and host insect nutrition. J Appl Microbiol 103(6):2664–2675. https://doi.org/10.1111/j.1365-2672.2007.03506.x
Indiragandhi P, Yoon C, YangJO CS, Sa TM, Kim GH (2010) Microbial communities in the developmental stages of B and Q biotypes of sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). J ApplBiol Chem 53(5):605–617. https://doi.org/10.3839/jksabc.2010.093
IRAC (2009) IRAC susceptibility test method. Insecticide Resistance Action Committee. https://irac-online.org/content/uploads/2009/09/Method_016_v3_june09.pdf. Accessed Jun 2021
Jones AG, Mason CJ, Felton GW, Hoover K (2019) Host plant and population source drive diversity ofmicrobial gut communities in two polyphagous insects. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-39163-9
Kim EH, Sung JW, Yang JO, Ahn HG, Yoon CM, Seo MJ, Kim GH (2007) Comparison of insecticide susceptibility and enzyme activities of biotype B and Q of Bemisia tabaci. Korean J Pesti Sci 11(4):320–330
Krishnamoorthy R, Arul Jose P, Janahiraman V, Indira Gandhi P, Gandhi Gracy R, Jalali SK, Senthil Kumar M, Malathi V, Anandham R (2020) Function and insecticidal activity of bacteria associated with papaya mealybug, Paracoccus marginatus Williams & Granara de Willink (Hemiptera: Pseudococcidae). Biocontrol Sci Technol 30(8):762–778. https://doi.org/10.1080/09583157.2020.1765983
Kumar V, Mckenzie CL, Mannion C, Stocks I, Smith T, Osborne LS (2013) Rugose Spiralling whitefly Aleurodicus rugioperculatus Martin (Hemiptera: Aleyrodidae). J Edis. https://doi.org/10.32473/edis-in1015-2013
Lau EP, Niswander L, Watson D, Fall RR (1980) Glutathione-Stransferase is present in avariety of microrganisms. Chemosphere 9(9):565–569. https://doi.org/10.1016/0045-6535(80)90074-0
Lauzon CR, Potter SE, Prokopy RJ (2003) Degradation and detoxification of the dihydrochalcone phloridzin by Enterobacter agglomerans, a bacterium associated with the apple pest, Rhagoletis pomonella (Walsh) (Diptera: Tephritidae). Environ Entomol 32(5):953–962. https://doi.org/10.1603/0046-225X-32.5.953
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69(4):1875–1883. https://doi.org/10.1128/AEM.69.4.1875-1883.2003
Lu J, Guo W, Chen S, Guo M, Qiu B, Yang C, Lian T, Pan H (2019) Host plants influence the composition of the gut bacteria in Henosepilachna vigintioctopunctata. PLoS ONE 14(10):e0224213. https://doi.org/10.1371/journal.pone.0224213
Lv N, Wang L, Sang W, Liu CZ, Qiu BL (2018) Effects of endosymbiont disruption on the nutritional dynamics of the Pea Aphid Acyrthosiphon pisum. Insects 9(4):161. https://doi.org/10.3390/insects9040161
Malathi VM, Jalali SK, Lyju VJ, Gracy RG, More RP, Anandham R, Thulasi A, Venkatesan T (2017) Associated bacterial diversity of insecticide-susceptible and-resistant brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae) analyzed by culture-dependent and-independent methods. Phytoparasitica 45(5):683–693. https://doi.org/10.1007/s12600-017-0629-3
Mannion C (2010) Rugose Spiralling whitefly, a new whitefly in South Florida. Tropical Research and Education Center, University of Florida 5: 17
Marokházi J, Lengyel K, Pekár S, Felföldi G, Patthy A, Gráf L, Fodor A, Venekei I (2004) Comparison of proteolytic activities produced by entomopathogenic Photorhabdus bacteria: strain-and phase-dependent heterogeneity in composition and activity of four enzymes. Appl Environ Microbiol 70(12):7311–7320. https://doi.org/10.1128/AEM.70.12.7311-7320.2004
Martin JH (2004) Whiteflies of Belize (Hemiptera: Aleyrodidae). Part 1-introduction and account of the subfamily Aleurodicinae Quaintance & Baker. Moscas blancas de Belice (Hemiptera: Aleyrodidae). Parte 1-introduccion y descripcion de la subfamilia Aleurodicinae Quaintance & Baker. Zootaxa 681(1): 1–119. https://doi.org/10.11646/zootaxa.681.1.1
Mason CJ, Raffa KF (2014) Acquisition and structuring of midgut bacterial communities in gypsy moth (Lepidoptera: Erebidae) larvae. Environ Entomol 43(3):595–604. https://doi.org/10.1603/EN14031
Merzendorfer H, Zimoch L (2003) Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206(24):4393–4412. https://doi.org/10.1242/jeb.00709
Milagres AMF, Machuca A, Napoleao D (1999) Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J Microbiol Methods 37(1):1–6. https://doi.org/10.1016/s0167-7012(99)00028-7
Mohan M, Gujar GT (2003) Local variation in susceptibility of the diamondback moth, Plutella xylostella (Linnaeus) to insecticides and role of detoxification enzymes. Crop Prot 22(3):495–504. https://doi.org/10.1016/S0261-2194(02)00201-6
Montllor CB, Maxmen A, Purcell AH (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27(2):189–195. https://doi.org/10.1046/j.1365-2311.2002.00393.x
Mrázek J, Štrosová L, Fliegerova K, Kott T, Kopečný J (2008) Diversity of insect intestinal microflora. Folia Microbiol 53(3):229–233. https://doi.org/10.1007/s12223-008-0032-z
Okongo RN, Puri AK, Wang Z, Singh S, Permaul K (2019) Comparative biocontrol ability of chitinases from bacteria and recombinant chitinases from the thermophilic fungus Thermomyces lanuginosus. J Biosci Bioeng 127(6):663–671. https://doi.org/10.1016/j.jbiosc.2018.11.007
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci 100(4):1803–1807. https://doi.org/10.1073/pnas.0335320100
Pandey N, Singh A, Rana VS, Rajagopal R (2013) Molecular characterization and analysis of bacterial diversity in Aleurocanthus woglumi (Hemiptera: Aleyrodidae). Environ Entomol 42(6):1257–1264. https://doi.org/10.1603/EN13110
Pavlidi N, Tseliou V, Riga M, Nauen R, Van Leeuwen T, Labrou NE, Vontas J (2015) Functional characterization of glutathione S-transferases associated with insecticide resistance in Tetranychus urticae. PesticBiochem Physiol 121:53–60. https://doi.org/10.1016/j.pestbp.2015.01.009
Priya NG, Ojha A, Kajla MK, Raj A, Rajagopal R (2012) Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS ONE 7(1):e30768. https://doi.org/10.1371/journal.pone.0030768
Ridley EV, Wong ACN, Douglas AE (2013) Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster. Appl Environ Microbiol 79(10):3209–3214. https://doi.org/10.1128/AEM.00206-13
Rodriguez-Kabana R, Godoy G, Morgan-Jones G, Shelby RA (1983) The determination of soil chitinase activity: conditions for assay and ecological studies. Plant Soil 75(1):95–106. https://doi.org/10.1007/BF02178617
Ruan YM, Xu J, Liu SS (2006) Effects of antibiotics on fitness of the B biotype and a non-B biotype of the whitefly Bemisia tabaci. Entomol Exp Appl 121(2):159–166. https://doi.org/10.1111/j.1570-8703.2006.00466.x
Selvaraj K, Gupta A, Venkatesan T, Jalali SK, Sundararaj R, Ballal CR (2017) First record of invasive rugose spiralling whitefly Aleurodicus rugioperculatus Martin (Hemiptera: Aleyrodidae) along with parasitoids in Karnataka. J Biol Control 31(2):74–78. https://doi.org/10.18311/jbc/2017/16015
Seo MJ, Yang JO, Yoon CM, Youn YN, Kim GH (2007) Differentiation in feeding behaviour of biotypes B and Q of Bemisia tabaci (Homoptera: Aleyrodidae) against three insecticides. Korean J Appl Entomol 46(3):401–408. https://doi.org/10.5656/KSAE.2007.46.3.401
Shan HW, Zhang CR, Yan TT, Tang HQ, Wang XW, Liu SS, Liu YQ (2016) Temporal changes of symbiont density and host fitness after rifampicin treatment in a whitefly of the Bemisia tabaci species complex. Insect Sci 23(2):200–214. https://doi.org/10.1111/1744-7917.12276
Shen Z, Jacobs-Lorena M (1997) Characterization of a novel gut-specific chitinase gene from the human malaria vector Anopheles gambiae. J Biol Chem 272(46):28895–28900. https://doi.org/10.1074/jbc.272.46.28895
Singh BK, Walker A (2006) Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30(3):428–471. https://doi.org/10.1111/j.1574-6976.2006.00018.x
SPSS (2013) IBM SPSS statistics 22. IBM Corp, New York
Stocks IC, Hodges G (2012) The rugose spiralling whitefly, Aleurodicus rugioperculatus Martin, a new exotic whitefly in South Florida (Hemiptera: Aleyrodidae). Gainesville (FL): Florida Department of Agriculture and Consumer Services, Division of Plant IndustryAleurodicus rugioperculatus pest-alert. DACS-P-01745.pdf. Accessed 7 Mar 2017
Sundararaj R, Selvaraj K (2017) Invasion of rugose spiralling whitefly, Aleurodicus rugioperculatus Martin (Hemiptera: Aleyrodidae): a potential threat to coconut in India. Phytoparasitica 45(1):71–74. https://doi.org/10.1007/s12600-017-0567-0
Tang X, Freitak D, Vogel H, Ping L, Shao Y, Cordero EA, Andersen G, Westermann M, Heckel DG, Boland W (2012) Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLoS ONE 7(7):e36978. https://doi.org/10.1371/journal.pone.0036978
Taravati S, Mannion C, McKenzie C, Osborne L (2018) Oviposition preference of rugose spiraling whitefly (Hemiptera: Aleyrodidae) on five host plant species. Fla Entomol 101(4):611–616
van den Heuvel JFJM, Verbeek M, van der Wilk F (1994) Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J Gen Virol 75(10):2559–2565. https://doi.org/10.1099/0022-1317-75-10-2559
Visotto LE, Oliveira MGA, Ribon AOB, Mares-Guia TR, Guedes RNC (2009) Characterization and identification of proteolytic bacteria from the gut of the velvetbean caterpillar (Lepidoptera: Noctuidae). Environ Entomol 38(4):1078–1085. https://doi.org/10.1603/022.038.0415
Webster TC (1994) Fumagillin affects Nosema apis and honey bees (Hymonopterai Apidae). J Econ Entomol 87(3):601–604. https://doi.org/10.1093/jee/87.3.601
Weiss BL, Wang J, Aksoy S (2011) Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol 9(5):e1000619. https://doi.org/10.1371/journal.pbio.1000619
Werren JH (2012) Symbionts provide pesticide detoxification. Proc Natl Acad Sci 109(22):8364–8365. https://doi.org/10.1073/pnas.1206194109
Whittington R, Winston ML (2003) Effects of Nosema bombi and its treatment fumagillin on bumble bee (Bombus occidentalis) colonies. J Invertebr Pathol 84(1):54–58. https://doi.org/10.1016/S0022-2011(03)00123-X
Wiwat C, Thaithanun S, Pantuwatana S, Bhumiratana A (2000) Toxicity of chitinase-producing Bacillus thuringiensisssp. kurstaki HD-1 (G) toward Plutella xylostella. J Invertebr Pathol 76(4):270–277. https://doi.org/10.1006/jipa.2000.4976
Xiang H, Wei GF, Jia S, Huang J, Miao XX, Zhou Z, Zhao LP, Huang YP (2006) Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can J Microbiol 52(11):1085–1092. https://doi.org/10.1139/w06-064
Xue X, Li SJ, Ahmed MZ, De Barro PJ, Ren SX, Qiu BL (2012) Inactivation of Wolbachia reveals its biological roles in whitefly host. PLoS ONE 7(10):e48148. https://doi.org/10.1371/journal.pone.0048148
Yang X, He C, Xie W, Liu Y, Xia J, Yang Z, Guo L, Wen Y, Wang S, Wu Q (2016) Glutathione S-transferases are involved in thiamethoxam resistance in the field whitefly Bemisia tabaci Q (Hemiptera: Aleyrodidae). Pestic Biochem Physiol 134:73–78. https://doi.org/10.1016/j.pestbp.2016.04.003
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613. https://doi.org/10.1099/ijsem.0.001755
Zablotowicz RM, Hoagland RE, Locke MA, Hickey WJ (1995) Glutathione-S-transferase activity and metabolism of glutathione conjugates by rhizosphere bacteria. Appl Environ Microbiol 61(3):1054–1060. https://doi.org/10.1128/aem.61.3.1054-1060.1995
Zweerink MM, Edison A (1986) Inhibition of Micrococcus luteus DNA gyrase by norfloxacin and 10 other quinolone carboxylic acids. Antimicrob Agents Chemothe 29(4):598–601. https://doi.org/10.1128/aac.29.4.598
Acknowledgements
This work was supported by Department of Science and Technology, Government of India-New Delhi, under grant GOI- DST (SERB) /EMR/2016/005815.
Author information
Authors and Affiliations
Contributions
MS Conducted all the laboratory experiments and manuscript drafting. JSK Conceived the hypothesis designed the experiment and corrected the manuscript. RA Assisted in laboratory experiments and drafted the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest, no animals or humans are used in this study and all authors accepted the final version submitted to the journal.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saranya, M., Kennedy, J.S. & Anandham, R. Functional characterization of cultivable gut bacterial communities associated with rugose spiralling whitefly, Aleurodicus rugioperculatus Martin. 3 Biotech 12, 14 (2022). https://doi.org/10.1007/s13205-021-03081-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-03081-3