Abstract
Retinitis pigmentosa (RP) is a rare and heterogeneous group of inherited ocular diseases. However, the relationship between CACNA2D4 mutations and RP is not well understood. In this study, a Chinese autosomal recessive retinitis pigmentosa (arRP) pedigree was enrolled and targeted next-generation sequencing was employed for identifying the causative gene in the proband. These steps were followed by confirmatory Sanger sequencing and segregation analysis. RNA-sequencing (RNA-seq) data and semi-quantitative reverse transcription polymerase chain reaction analysis were then applied to examine the expressions in the human and mouse tissues. Novel compound heterozygous, deleterious missense variants of the CACNA2D4 gene, NM_172364.4: c.G955A (p.D319N) and c.A1822C (p.I608L), were identified in the arRP pedigree, co-segregating with the clinical phenotype in the patient. The CACNA2D4 protein is highly conserved among species. The CACNA2D4 mRNA expression showed the highest expression in the retina of humans and in the later four developmental stages/times of retinal tissues in mice, indicating its role in retina/eye functions and developments. This study is the first to identify novel compound heterozygous mutations c.G955A (p.D319N) and c.A1822C (p.I608L) in the CACNA2D4 gene. These might be disease-causing mutations, thereby extending the mutational spectra. The identification of pathogenic CACNA2D4 variants is expected to enhance our understanding of the genotype–phenotype correlations of arRP for disease diagnosis and genetic counseling. The relationship between the CACNA2D4 variants and diseases/phenotypes other than RP has also been reviewed and discussed in this paper.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinitis pigmentosa (RP, OMIM: 268000) is a rare, heterogeneous group of inherited ocular diseases. RP leads to progressive retinal degeneration, eventually culminating in severe visual impairment or blindness (Fu et al. 2019). The condition affects roughly 1 in 4,000 people in the United States (Ali et al. 2017; Fu et al. 2018). RP is both clinically and genetically heterogeneous and is inherited according to Mendelian inheritance patterns. The most common inheritance pattern of RP is autosomal recessive (50–60%), followed by autosomal dominant (30–40%) and rarely X-linked (5–15%) (Hamel 2014; Hartong et al. 2006). To date, mutations in over 80 genes have been identified to cause RP and result in progressive photoreceptor loss.
The calcium voltage-gated channel auxiliary subunit alpha2delta4 (CACNA2D4) gene (NM_172364.4) (OMIM: 608171), also known as RCD4, is located on chromosome 12p13.33. The gene has 38 exons spanning 128,909 bases in the human genome (GRCh38/hg38) and encodes a putative 1137-amino acid protein with a predicted molecular mass of 127,938 Da (NP_758952.4) (Qin et al. 2002). Mutations in the CACNA2D4 gene have been linked to autosomal recessive cone dystrophy 4 (RCD4, OMIM 610478) or autosomal recessive retinitis pigmentosa (arRP) (Wycisk et al. 2006b; Ba-Abbad et al. 2016). However, the CACNA2D4 gene mutations and diseases/phenotypes, including RP, are not well understood.
The present study aims to decipher the causative gene and its mutations accounting for the occurrence of arRP in a Chinese family. Targeted next-generation sequencing (NGS) (Wang et al. 2014; Zhang et al. 2016; Valencia et al. 2015) was used to identify the possible genetic cause for arRP in this family. The relationship between the CACNA2D4 variants and diseases/phenotypes other than RP has also been reviewed and discussed.
Materials and methods
Pedigree, proband, clinical assessment, and sample collection
Detailed ophthalmic examinations were performed on the recruited pedigree with a proband (M338). Blood samples were collected and gDNA was extracted from the patient (proband) and the parents as well as healthy control volunteers using a standard phenol/chloroform extraction method (Fu et al. 2002, 2020a). Prior approval was obtained from the Ethics Committee of Southwest Medical University. Informed written consent was acquired from all the participating subjects.
Targeted-NGS sequencing
Targeted-NGS (TGS) analysis was conducted using family M338 according to the instructions for Illumina (Wang et al. 2014; Zhang et al. 2016; Fu et al. 2018; Salvo et al. 2015). The reads were aligned with the hg19 reference genome of humans using Burrow–Wheeler Aligner (BWA) and the public online database (Li and Durbin 2009). Variations in single nucleotide polymorphisms (SNPs) and insertions/deletions were refined using Atlas-SNP2 and Atlas-Indel2 (Challis et al. 2012). Variant frequency data were applied to the CHARGE consortium (Psaty et al. 2009), Human 1000 Genomes (Genomes Project et al. 2010), ANNOVAR (Wang et al. 2010), ESP-6500 (Tennessen et al. 2012), and Exome Aggregation Consortium (ExAC) databases to identify all pathogenic variants in the candidate genes and were assessed by PolyPhen-2, SIFT (Sorting Intolerant From Tolerant), MutationTaster, I-Mutant2.0, and HGMD (Human Gene Mutation Database).
Sanger DNA sequencing and co-segregation analysis
Polymerase chain reaction (PCR) amplification and Sanger DNA sequencing of the variants were applied to the gDNA of the participants for mutation verification and pedigree segregation analysis (Imani et al. 2018a). Locus-specific primer pairs, CACNA2D4-955 and CACNA2D4-1822, were designed using the online Primer3 Program for NM_172364.4: c.G955A or c.A1822C mutations in the CACNA2D4 gene (Table 1).
For sequencing, PCR amplification was performed using a total volume of 20 µL of reaction mixture containing the specific primers CACNA2D4-955L and CACNA2D4-955R for the c.G955A variant or CACNA2D4-1822L and CACNA2D4-1822R for the c.A1822C variant (Table 1), 50 ng of gDNA template, and 10 μL of 2× PCR TaqMaster mix (Tiangen Biothech, Beijing, China) in a Veriti™ 96-Well Thermal Cycler from Applied Biosystems, USA. The amplification conditions were as follows: initial denaturation at 95 ºC for 90 s, followed by 32 cycles at 94 ºC for 40 s, 60 ºC for 60 s, 72 ºC for 40 s, and final extension at 72 ºC for 5 min.
The PCR products were then resolved by electrophoresis on 1.2% agarose gels in 0.5× TAE buffer. The gels were visualized by staining with 0.5 μg/mL ethidium bromide, and specific bands of 563 bp for the c.G955A variant or 339 bp for the c.A1822C variant were gel-extracted and purified. The purified PCR products were then sequenced using the Sanger DNA sequencing method with specific primers, CACNA2D4-955L or CACNA2D4-1822L, as presented in Table 1. All ethnically matched normal controls were amplified and sequenced using the aforementioned primers. Co-segregation analyses for the pedigree were conducted based on Sanger DNA sequencing results.
Protein structure analysis and bioinformatics
A search of the conserved domains of CACNA2D4 was performed using the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) by inputting the amino acid sequences of CACNA2D4 (NP_758952.4) (Marchler-Bauer et al. 2017; Imani et al. 2018b). Homologs of CACNA2D4 were revealed by the NCBI HomoloGene program (https://www.ncbi.nlm.nih.gov/homologene?Db=homologene&Cmd=Retrieve&list_uids=26544).
RNA-seq data analysis for CACNA2D4 expression
To determine the tissue-specificity, the mRNA expression profiles of CACNA2D4 were examined using the RNA-seq data from 27 different tissues (https://www.ncbi.nlm.nih.gov/gene/93589). This project was called HPA RNA-seq normal tissues (Zhou et al. 2019; Cheng et al. 2020; Fu et al. 2020b). The mRNA expression profiles of CACNA2D4 in different human tissues, including the retina, were obtained via the HPA (Human Protein Atlas) (https://www.proteinatlas.org/ENSG00000151062-CACNA2D4/tissue) (Uhlen et al. 2010; Wei et al. 2020; Fu et al. 2020b).
RNA extraction and semi-quantitative reverse transcription (RT)-PCR
Total RNA was isolated from mice according to our previously reported protocol (Fu et al. 2018). The sequences of the RT-primers, product sizes, and PCR conditions, including the annealing temperature, are listed in Table 1. RT-PCR was conducted using the primer pair RT-cacna2d4 (RT-cacna2d4L and RT-cacna2d4R) targeting the mouse Cacna2d4 gene (NM_001033382.2). The mouse β-actin gene was used as an internal control and was amplified using the primer pair described previously (Cheng et al. 2019) (Table 1). RT-PCR amplification was performed in a total volume of 10 µL using cDNA as a template on a Veriti™ 96-Well Thermal Cycler. The amplification conditions for the mouse Cacna2d4 gene were as follows: initial denaturation at 95 ºC for 90 s, followed by 30 cycles at 94 °C for 40 s, 60 ºC for 60 s, 72 ºC for 40 s, and final extension at 72 ºC for 5 min. The amplified PCR products were then separated on 1.2% agarose gel in 1 × TAE buffer (Liu et al. 2020). The gels were then visualized, and the images were documented as reported previously (Zhou et al. 2019; Cheng et al. 2019).
Results
Pedigree and proband clinical characteristics
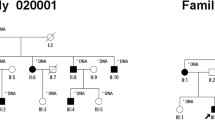
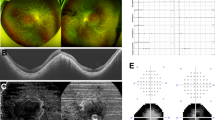
The proband (Fig. 1a, II: 1) was a 62-year-old man belonging to a Chinese family. The patient claimed a reduction in visual acuity and peripheral field loss at the age of 35. His clinical features are summarized in Table 2. Fundus examination revealed pale fundus, optic nerve atrophy, vessel attenuation, osteocyte-like pigmentation, and retinal pigment epithelial degeneration (Fig. 1b). Electroretinogram revealed that the amplitudes of the α and β waves were significantly decreased in the apparent dark vision reaction, and the reaction was extinguished. (Fig. 1c). The proband’s parents had normal eye examinations and showed no retinal disease. Thus, the M338 proband was characterized as arRP.
A M338 pedigree of retinitis pigmentosa. a A pedigree of the M338 family. Normal individual is indicated as a clear circle (female), whereas the affected male patient is indicated as a filled square symbol with an arrow (II: 1) for the compound heterozygous variants of CACNA2D4: NM_172364.4:M1:c.G955A; M2:c.A1822C. b Fundus photography of the right eye of the proband. c Electroretinogram (ERG) of the proband. Upper panels, photopic of 3.0 ERG (right and left eyes); Bottom panels, photopic of 3.0 flicker 30 Hz ERG (right and left eyes). M338 is the molecular number for the proband
NGS analyses and putative pathogenic mutation identification
To assess the disease-causing gene and its mutations, a targeted NGS panel of retinal disease-causing genes was conducted successfully using the gDNA of the M338 patient (Fig. 1, pedigree II: 1). Compound heterozygous, missense variants c.G955A and c.A1822C at exons 8 and 18, respectively, in the CACNA2D4 gene (NM_172364.4) were identified, which led to amino acid (aa) changes (p.D319N and p.I608L) in the CACNA2D4 protein (NP_758952.4) (Fig. 1, II: 1). P olyPhen-2 analysis suggested probable damage (PD) for c.G955A: p.D319N change (score 1) and benign for c.A1822C: p.I608L change (score 0.268); MutationTaster revealed disease-causing (DC) (scores of 1 for c.G955A: p.D319N change and 0.9998 for c.A1822C: p.I608L change); SIFT alluded damaging (scores of 0.02 for c.G955A: p.D319N change and 0.04 for c.A1822C: p.I608L change); and I-Mutant2.0 of the free-energy change values indicated decreased stability (DDG = − 1.00 kcal/mol, < 0) for c.G955A: p.D319N change and increased stability (DDG = 0.75 kcal/mol, > 0) for c.A1822C: p.I608L change. The deleterious and pathogenic aspects of the CACNA2D4 gene: c.G955A and c.A1822C mutations are presented in Table 2. Hence, the compound heterozygous, missense mutations (c.G955A and c.A1822C) in the CACNA2D4 gene are likely to have induced damaged protein function in the patient. These variants were determined to be novel mutations through ExAC or HGMD (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=CACNA2D4) database searches (Table 3).
Verification of candidate mutations and segregation analysis
Confirmation of the mutations (c.G955A and c.A1822C) and segregation analysis were done by DNA sequencing (Fig. 2). The variants c.G955A and c.A1822C of the CACNA2D4 gene was verified to be compound heterozygous in patient M338 (pedigree II:1; Fig. 2a, b). A heterozygous variant c.G955A in the M338 father and a heterozygous variant c.A1822C in his mother (pedigree I:1, I:2; data not shown) were identified, while the normal males had wild-type forms of the gene (Fig. 2c, d). Thus, we were able to establish that the c.G955A and c.A1822C variants in the CACNA2D4 gene co-segregated with the RP phenotype of the family members. Both variants were absent in the 100 normal controls. These findings indicate the co-segregation of the mutations in the pedigree of the RP family and define their disease-causing roles in pathogenesis.
Pyrogram profiles for variant verification by Sanger DNA sequencing. a, b The sequencing results in II: 1 for CACNA2D4 compound heterozygous mutants for c.G955A and c.A1822C, and c, d The sequencing results in a normal male for wild-type in both the sites c.G955 and c.A1822, respectively. The arrows indicate the mutations at the nucleotide position NM_172364.4: M1:c.G955A; M2: c.A1822 in CACNA2D4
Functional effects of the variants c.G955A (p.D319N) and c.A1822C (p.I608L) in CACNA2D4
Search for conserved domains within the CACNA2D4 protein in the NCBI database revealed that CACNA2D4 had four conserved domains, namely VWA_N (pfam08399) (aa interval: 154–264), vWA_VGCC_like (cd01463) (aa interval: 277–460), dCache_1 (pfam02743) (aa interval: 454–576), and VGCC_alpha2 super family (cl07190) (aa interval: 687–1070) (Fig. 3a). The p.D319N mutation was located in the vWA_VGCC_like domain (Fig. 3c). The CACNA2D4 gene was found to be conserved in the chimpanzee, rhesus monkey, dog, cow, rat, mouse, chicken, zebrafish, mosquito, fruit fly, and frog upon searching for homologs. Orthologous comparisons of the CACNA2D4 protein in Homo sapiens with the abovementioned 12 species indicated that p.D319 and p.I608L of the CACNA2D4 protein were highly conserved (Fig. 3b, c, respectively). In essence, our study has asserted that the pathogenic variants c.G955A (p.D319N) and c.A1822C (p.I608L) of the CACNA2D4 gene with compound heterozygous, missense mutations are responsible for the patient’s arRP disease.
CACNA2D4 protein structure and comparison. a CACNA2D4 domains. Arrows indicate the mutated amino acid positions. Red arrows indicate the missense mutation positions identified in this study, whereas the black arrows indicate the missense/nonsense mutation positions reported earlier. “*” indicates the stop codon. b Orthologous conservation analysis in the CACNA2D4 p.D319 variant in the indicated species. c Orthologous conservation analyses in the CACNA2D4 p.I608 variant in the indicated species
Results of CACNA2D4 and Cacna2d4 mRNA expression profiles
When RNA-seq was performed, CACNA2D4 gene expression in the different human tissues showed that the Reads Per Kilobase of transcript per Million (RPKM) values in the testis and the appendix were the highest, with an approximate score of 1.0 (Fig. 4a), demonstrating that the levels were not quite high in any of the tested tissues. Unfortunately, no eye tissues, including the retina, were available. Thus, the mRNA expression profiles of the CACNA2D4 gene in 61 different tissues and cells, including the retina, were obtained. The results revealed that the CACNA2D4 gene expression was the highest in the human retina, with a consensus normalized expression (NX) value of 47.0. The second highest expression was in the T-cells, but with an NX value of only 4.7 (Fig. 4b). Hence, this finding strongly indicates that CACNA2D4 is highly expressed only in the retinal tissue and plays an important role there.
The mRNA expression profiles of the genes of human CACNA2D4 and mouse Cacna2d4. The CACNA2D4 expressions in the indicated human tissues (a). The expression for CACNA2D4 in 55 indicated tissues and 6 blood cells (b). The expression profiles of Cacna2d4 in the indicated tissues in mice (c) and the indicated development stages of retina in mice (d). NX, consensus normalized expression. RPKM, Reads Per Kilobase of transcript per Million mapped reads. d indicate day(s); w, week(s); m, month(s); nc, negative control
Meanwhile, the expressions of Cacna2d4 mRNA in the different tissues and developmental stages of the retina were analyzed by quantitative RT-PCR, and the results are shown in Fig. 4c, d. From Fig. 4c, it is evident that the levels of Cacna2d4 mRNA are the highest in the retina, followed by the lens, cornea, and sclera of the eyes. However, no expression was detectable in the uterus, breast, ovary, testis, kidney, spleen, liver, intestine, brain, skeletal muscle, and blood (Fig. 4c). Furthermore, Cacna2d4 mRNA of the retinal tissue was highly expressed in the later four stages of (Fig. 4d). Whole eyeballs of the embryos at 12.5 days and 20.5 days (Fig. 4d) were taken because of technical difficulties in sample collections. The higher expression of Cacna2d4 gene in the retina and other tissues of the eyes further supports the notion that CACNA2D4 plays important roles in the functioning of the retina and the eye as a whole.
Discussion
The CACNA2D4 gene encodes a member of the alpha-2/delta subunit family, a protein with a molecular mass of 127,938 Da in the complex of the voltage-dependent calcium channel. Calcium channels can mediate the influx of calcium ions into the cells when the membranes polarize and consist of a complex in a 1:1:1:1 ratio with alpha-1, alpha-2/delta, beta, and gamma subunits. The abundance of the presynaptic calcium channel and the probability for release are determined by alpha-2-delta expression since the release of the synaptic neurotransmitter is driven by Ca2+ influx through the active zone voltage-gated calcium channels (VGCC) (Hoppa et al. 2012). As a regulatory subunit, CACNA2D4 can alter the properties of the pore-forming alpha-1 subunits of VGCC. CACNA2D4 can also be processed into two peptides (subunits of alpha-2 or delta) and be held together via a disulfide bond. Mutations in the CACNA2D4 gene cause autosomal recessive cone dystrophy, retinal dystrophy, or arRP (Wycisk et al. 2006b; Ba-Abbad et al. 2016; Gustafson et al. 2017; Huang et al. 2015). Interestingly, a homozygous mutation has also been identified in the Cacna2d4 gene of mice, which leads to autosomal recessive cone–rod dysfunction in the visual system (Wycisk et al. 2006a). However, the CACNA2D4 gene mutations and their involvement in RP have not been well studied in the Chinese population (Huang et al. 2015). In this study, we have successfully identified novel compound heterozygous, pathogenic missense variants of the CACNA2D4 gene, namely c.G955A and c.A1822C, by NGS-based genetic diagnosis in a Chinese family, which led to RP. Upon searching the HGMD (access date: December 1, 2020), we found that only seven pathogenic variants of the CACNA2D4 gene have so far been reported, including four missense/nonsense mutations (Fig. 3a), one small deletion, one small insertion, and one gross deletion. The relationship between the CACNA2D4 variants and diseases/phenotypes other than RP have also been discussed, and the relevant literature has been reviewed. Table 4 lists the CACNA2D4 variants and the associated diseases/phenotypes. From the above identified variants, we found that the mutational spectra and disease phenotypes were variable (Goodloe et al. 2014), for example, familial atrial fibrillation was associated with the CACNA2D4 variant: p.S886R (Table 4) (Weeke et al. 2014). To the best of our knowledge, the variants c.G955A and c.A1822C of the CACNA2D4 gene are novel and might cause arRP, thereby extending its mutational spectra.
Orthologous comparison of the CACNA2D4 protein of H. sapiens with 12 other species indicated that these proteins are higher conserved. The CACNA2D protein has four conserved domains, namely VWA_N (pfam08399), vWA_VGCC_like (cd01463), dCache_1 (pfam02743), and VGCC_alpha2 super family (cl07190). The p.D319N mutation is located in the vWA_VGCC_like domain. The exact biochemical function of this domain is not clear; however, the alpha 2 delta complex has been reported to regulate various functional properties of the channel complex, suggesting its role in complex stability. Four other studies have documented that missense/nonsense variants are located in the VGCC_alpha2 super family domain of the CACNA2D protein (Fig. 3), but the clinical phenotypes of the patients are entirely different (Table 4).
Human CACNA2D4 gene expression based on the RNA-seq of 61 different tissues and cells revealed highest expression in the retina and only very low expression in others. Our results from mouse tissues showed that Cacna2d4 mRNA is expressed only in the retina, lens, sclera, and cornea of the eye, suggesting that the Cacna2d4 protein plays important roles in the development and functioning of the retina/eye. Using KO-mice model, Kerov et al. established that Cacna2d4 is essential for maintaining the structural and functional integrity of the rod and cone synapses. Thus, their disruption by CACNA2D4 gene mutations might contribute to visual impairment (Kerov et al. 2018). To sum up the findings, our study has revealed that the CACNA2D4 compound heterozygous, missense mutations c.G955A and c.A1822C are likely to lead to vision impairment and arRP. However, further studies are needed to determine whether these variants are truly the causative factors for different clinically variable diseases.
Conclusion
In conclusion, our study is the first to identify two novel compound heterozygous missense variants, c.G955A and c.A1822C, in CACNA2D4, which may be the disease-causing mutations for RP in the present Chinese pedigree, thereby expanding its mutation spectrums. Targeted-NGS thus provides an effective approach for genetic diagnosis (Fu et al. 2018; Adams and Eng 2018; Rahbaran et al. 2019). The identification of pathogenic CACNA2D4 variants also enhances our understanding of the genotype/phenotype correlations in RP for gene diagnosis as well as for genetic counseling.
Abbreviations
- RP:
-
Retinitis pigmentosa
- arRP:
-
Autosomal recessive retinitis pigmentosa
- CACNA2D4:
-
Homo sapiens calcium voltage-gated channel auxiliary subunit alpha2delta 4 gene
- NGS:
-
Next-generation sequencing
- TGS:
-
Targeted next-generation sequencing
- RNA-seq:
-
RNA-sequencing
- RT-PCR:
-
Revere transcriptional-polymerase chain reaction
- ExAC:
-
The Exome Aggregation Consortium database
- HGMD:
-
The Human Gene Mutation Database
- RPKM:
-
Reads Per Kilobase of transcript per Million
- OMIM:
-
Online Mendelian Inheritance in Man
- NX:
-
Consensus normalized expression
- VGCC:
-
Voltage-gated calcium channels
- BWA:
-
Burrow–Wheeler Aligner
- ERG:
-
Electroretinogram
References
Adams DR, Eng CM (2018) Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med 379(14):1353–1362. https://doi.org/10.1056/NEJMra1711801
Ali MU, Rahman MSU, Cao J, Yuan PX (2017) Genetic characterization and disease mechanism of retinitis pigmentosa; current scenario. 3 Biotech 7(4):251. https://doi.org/10.1007/s13205-017-0878-3
Ba-Abbad R, Arno G, Carss K, Stirrups K, Penkett CJ, Moore AT, Michaelides M, Raymond FL, Webster AR, Holder GE (2016) Mutations in CACNA2D4 cause distinctive retinal dysfunction in humans. Ophthalmology 123(3):668–671. https://doi.org/10.1016/j.ophtha.2015.09.045
Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, Yu F (2012) An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinf 13:8. https://doi.org/10.1186/1471-2105-13-8
Cheng J, Fu J, Zhou Q, Xiang X, Wei C, Yang L, Fu S, Khan MA, Lv H, Fu J (2019) A novel splicing mutation in the PRPH2 gene causes autosomal dominant retinitis pigmentosa in a Chinese pedigree. J Cell Mol Med 23(5):3776–3780. https://doi.org/10.1111/jcmm.14278
Cheng J, Peng J, Fu J, Khan MA, Tan P, Wei C, Deng X, Chen H, Fu J (2020) Identification of a novel germline BRCA2 variant in a Chinese breast cancer family. J Cell Mol Med 24(2):1676–1683. https://doi.org/10.1111/jcmm.14861
Fu J, Li L, Lu G (2002) Relationship between microdeletion on Y chromosome and patients with idiopathic azoospermia and severe oligozoospermia in the Chinese. Chin Med J 115(1):72–75
Fu JW, Ma L, Cheng JL, Yang LS, Wei CL, Fu SY, Lv HB, Chen R, Fu JJ (2018) A novel, homozygous nonsense variant of the CDHR1 gene in a Chinese family causes autosomal recessive retinal dystrophy by NGS-based genetic diagnosis. J Cell Mol Med 22(11):5662–5669. https://doi.org/10.1111/jcmm.13841
Fu J, Cheng J, Zhou Q, Wei C, Chen H, Lv H, Fu J (2019) A novel missense variant c.G644A (p.G215E) of the RPGR gene in a Chinese family causes X-linked retinitis pigmentosa. Biosci Rep 39:10. https://doi.org/10.1042/BSR20192235
Fu J, Shen S, Cheng J, Lv H, Fu J (2020a) A case of Usher syndrome type IIA caused by a rare USH2A homozygous frameshift variant with maternal uniparental disomy (UPD) in a Chinese family. J Cell Mol Med 24(14):7743–7750. https://doi.org/10.1111/jcmm.15405
Fu J, Zhou B, Zhang L, Balaji KS, Wei C, Liu X, Chen H, Peng J, Fu J (2020b) Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol Biol Rep 47(6):4383–4392. https://doi.org/10.1007/s11033-020-05478-4
Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. https://doi.org/10.1038/nature09534
Goodloe AH, Evans JM, Middha S, Prasad A, Olson TM (2014) Characterizing genetic variation of adrenergic signalling pathways in Takotsubo (stress) cardiomyopathy exomes. Eur J Heart Fail 16(9):942–949. https://doi.org/10.1002/ejhf.145
Gustafson K, Duncan JL, Biswas P, Soto-Hermida A, Matsui H, Jakubosky D, Suk J, Telenti A, Frazer KA, Ayyagari R (2017) Whole genome sequencing revealed mutations in two independent genes as the underlying cause of retinal degeneration in an Ashkenazi Jewish Pedigree. Genes 8:9. https://doi.org/10.3390/genes8090210
Hamel CP (2014) Gene discovery and prevalence in inherited retinal dystrophies. CR Biol 337(3):160–166. https://doi.org/10.1016/j.crvi.2013.12.001
Hartong DT, Berson EL, Dryja TP (2006) Retinitis pigmentosa. Lancet 368(9549):1795–1809. https://doi.org/10.1016/S0140-6736(06)69740-7
Hoppa MB, Lana B, Margas W, Dolphin AC, Ryan TA (2012) alpha2delta expression sets presynaptic calcium channel abundance and release probability. Nature 486(7401):122–125. https://doi.org/10.1038/nature11033
Huang XF, Huang F, Wu KC, Wu J, Chen J, Pang CP, Lu F, Qu J, Jin ZB (2015) Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genetics Med 17(4):271–278. https://doi.org/10.1038/gim.2014.138
Imani S, Cheng J, Mobasher-Jannat A, Wei C, Fu S, Yang L, Jadidi K, Khosravi MH, Mohazzab-Torabi S, Shasaltaneh MD, Li Y, Chen R, Fu J (2018a) Identification of a novel RPGRIP1 mutation in an Iranian family with leber congenital amaurosis by exome sequencing. J Cell Mol Med 22(3):1733–1742. https://doi.org/10.1111/jcmm.13454
Imani S, Ijaz I, Shasaltaneh MD, Fu S, Cheng J, Fu J (2018b) Molecular genetics characterization and homology modeling of the CHM gene mutation: a study on its association with choroideremia. Mutat Res 775:39–50. https://doi.org/10.1016/j.mrrev.2018.02.001
Kerov V, Laird JG, Joiner ML, Knecht S, Soh D, Hagen J, Gardner SH, Gutierrez W, Yoshimatsu T, Bhattarai S, Puthussery T, Artemyev NO, Drack AV, Wong RO, Baker SA, Lee A (2018) alpha2delta-4 Is required for the molecular and structural organization of rod and cone photoreceptor synapses. J Neurosci 38(27):6145–6160. https://doi.org/10.1523/JNEUROSCI.3818-16.2018
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Liu X, Cheng J, Mei Z, Wei C, Khan MA, Peng J, Fu J (2020) SCAR marker for identification and discrimination of specific medicinal Lycium chinense Miller from Lycium species from ramp-PCR RAPD fragments. 3 Biotech 10(8):334. https://doi.org/10.1007/s13205-020-02325-y
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E, Consortium C (2009) Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2(1):73–80. https://doi.org/10.1161/CIRCGENETICS.108.829747
Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506(7487):185–190. https://doi.org/10.1038/nature12975
Qin N, Yagel S, Momplaisir ML, Codd EE, D’Andrea MR (2002) Molecular cloning and characterization of the human voltage-gated calcium channel alpha(2)delta-4 subunit. Mol Pharmacol 62(3):485–496
Rahbaran M, Hassani Doabsari M, Salavitabar S, Mokhberian N, Morovvati Z, Morovvati S (2019) A novel frameshift mutation in the EDA gene in an Iranian patient affected by X-linked hypohidrotic ectodermal dysplasia. Cell Mol Biol Lett 24:54. https://doi.org/10.1186/s11658-019-0174-9
Salvo J, Lyubasyuk V, Xu M, Wang H, Wang F, Nguyen D, Wang K, Luo H, Wen C, Shi C, Lin D, Zhang K, Chen R (2015) Next-generation sequencing and novel variant determination in a cohort of 92 familial exudative vitreoretinopathy patients. Invest Ophthalmol Vis Sci 56(3):1937–1946. https://doi.org/10.1167/iovs.14-16065
Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, Project NES (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337(6090):64–69. https://doi.org/10.1126/science.1219240
Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28(12):1248–1250. https://doi.org/10.1038/nbt1210-1248
Valencia CA, Husami A, Holle J, Johnson JA, Qian Y, Mathur A, Wei C, Indugula SR, Zou F, Meng H, Wang L, Li X, Fisher R, Tan T, Hogart Begtrup A, Collins K, Wusik KA, Neilson D, Burrow T, Schorry E, Hopkin R, Keddache M, Harley JB, Kaufman KM, Zhang K (2015) Clinical Impact and Cost-Effectiveness of Whole Exome Sequencing as a Diagnostic Tool: A Pediatric Center’s Experience. Front Pediatr 3:67. https://doi.org/10.3389/fped.2015.00067
Van Den Bossche MJ, Strazisar M, De Bruyne S, Bervoets C, Lenaerts AS, De Zutter S, Nordin A, Norrback KF, Goossens D, De Rijk P, Green EK, Grozeva D, Mendlewicz J, Craddock N, Sabbe BG, Adolfsson R, Souery D, Del-Favero J (2012) Identification of a CACNA2D4 deletion in late onset bipolar disorder patients and implications for the involvement of voltage-dependent calcium channels in psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 159B(4):465–475. https://doi.org/10.1002/ajmg.b.32053
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. https://doi.org/10.1093/nar/gkq603
Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, Bowne SJ, Sullivan LS, Luo H, Zhao L, Wang X, Zaneveld JE, Salvo JS, Siddiqui S, Mao L, Wheaton DK, Birch DG, Branham KE, Heckenlively JR, Wen C, Flagg K, Ferreyra H, Pei J, Khan A, Ren H, Wang K, Lopez I, Qamar R, Zenteno JC, Ayala-Ramirez R, Buentello-Volante B, Fu Q, Simpson DA, Li Y, Sui R, Silvestri G, Daiger SP, Koenekoop RK, Zhang K, Chen R (2014) Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet 133(3):331–345. https://doi.org/10.1007/s00439-013-1381-5
Weeke P, Muhammad R, Delaney JT, Shaffer C, Mosley JD, Blair M, Short L, Stubblefield T, Roden DM, Darbar D, National Heart L, Blood Institute GOESP (2014) Whole-exome sequencing in familial atrial fibrillation. Eur Heart J 35(36):2477–2483. https://doi.org/10.1093/eurheartj/ehu156
Wei C, Xiao T, Cheng J, Fu J, Zhou Q, Yang L, Lv H, Fu J (2020) Novel compound heterozygous EYS variants may be associated with arRP in a large Chinese pedigree. Biosci Rep 40:6. https://doi.org/10.1042/BSR20193443
Wycisk KA, Budde B, Feil S, Skosyrski S, Buzzi F, Neidhardt J, Glaus E, Nurnberg P, Ruether K, Berger W (2006a) Structural and functional abnormalities of retinal ribbon synapses due to Cacna2d4 mutation. Invest Ophthalmol Vis Sci 47(8):3523–3530. https://doi.org/10.1167/iovs.06-0271
Wycisk KA, Zeitz C, Feil S, Wittmer M, Forster U, Neidhardt J, Wissinger B, Zrenner E, Wilke R, Kohl S, Berger W (2006b) Mutation in the auxiliary calcium-channel subunit CACNA2D4 causes autosomal recessive cone dystrophy. Am J Hum Genet 79(5):973–977. https://doi.org/10.1086/508944
Zhang Q, Xu M, Verriotto JD, Li Y, Wang H, Gan L, Lam BL, Chen R (2016) Next-generation sequencing-based molecular diagnosis of 35 Hispanic retinitis pigmentosa probands. Sci Rep 6:32792. https://doi.org/10.1038/srep32792
Zhou B, Wei C, Khan MA, Chen H, Fu J (2019) Characterization and molecular cloning of novel isoforms of human spermatogenesis associated gene SPATA3. Mol Biol Rep 46(4):3827–3834. https://doi.org/10.1007/s11033-019-04825-4
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31701087), the Special Training Program for Young Science and Technology Talents from Southwest Medical University (00031726), the Joint Research Foundation of Luzhou City and Southwest Medical University (2018LZXNYD-YL01).
Author information
Authors and Affiliations
Contributions
JF was in charge of the idea. J.F. and S.A. supervised the project. JC, JF, LZ, and CW performed DNA extraction, PCR, sequencing and data analysis. HL and QZ recruited the clinical patients and were in charge of the clinical assessment. JF and SK wrote the manuscript, and JF, SA and JC revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study has the Ethical Committees approval granted by the Southwest Medical University. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent for publication
Written informed consent was obtained from all participants.
Competing interests
The authors declare no conflict of interests.
Rights and permissions
About this article
Cite this article
Cheng, J., Zhou, Q., Fu, J. et al. Novel compound heterozygous missense variants (c.G955A and c.A1822C) of CACNA2D4 likely causing autosomal recessive retinitis pigmentosa in a Chinese patient. 3 Biotech 11, 208 (2021). https://doi.org/10.1007/s13205-021-02761-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02761-4