Abstract
The present research reports a detailed in silico analysis of chromatin assembly factor-1 (CAF-1) family in human malaria parasite Plasmodium falciparum. Our analysis revealed five chromatin assembly factor-1 genes in P. falciparum (PfCAF-1) and the PfCAF-1 family was divided into two classes where, Class A belongs to the CAF-1 complex and others are kept in Class B. For comparative studies, orthologs of PfCAF-1 family were identified across 53 eukaryotic species and evolutionary relationships were drawn for different CAF-1 subfamilies. The phylogenetic analysis revealed grouping of evolutionary-related species together, although, divergence was observed in branching pattern. A detailed analysis of domain composition highlighted species-specific features viz. species-specific KDDS repeats of 84 amino acids were identified in PfCAF-1A whereas, members of CAF-1C/RbAp48 and RbAp46 subfamily exhibited least variation in size and domain composition. The qRT-PCR analysis revealed upregulation of PfCAF-1 members in trophozoite or schizont stage. Furthermore, a comparative expression analysis of the available transcriptome and proteome data along with qRT-PCR analysis revealed mixed expression patterns (coordination as well as non-coordination between different studies). Protein–protein interaction network analyses of PfCAF-1 family were carried out highlighting important complexes based on interologs. The PfRbAp48 was found to be highly connected with a total of 108 PPIs followed by PfRbAp46. The results unravel insights into the PfCAF-1 family and identify unique features, thus opening new perspectives for further targeted developments to understand and combat malaria menace.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotic cells, the nuclear DNA is wrapped around a histone octamer comprising two H2A–H2B dimers and one tetramer (H3–H4)2 and packaged into a highly ordered structure called nucleosome (Luger et al. 1997). The important role of nucleosome formation is mediated by various histone chaperones such as chromatin assembly factor 1 (CAF-1) and HIRA (Burgess and Zhang 2013). CAF-1 is one of the histone chaperones that promote nucleosome synthesis during DNA replication (Sauer et al. 2018). It is a diverse family which consists of some members involved in CAF-1 complex, while others, RbAp46 (retinoblastoma associated proteins p46) and GRWD1 (glutamate-rich WD40 repeat containing 1), are exclusively found in chromatin remodeling complexes like NuRD (nucleosome remodeling deacetylase), NuRF (nucleosome remodeling factor), Sin3 (histone deacetylation complex), PRC2 (polycomb repressive complex) and pre-replication complex (Sugimoto et al. 2015a, b).
The CAF-1 complex is a highly conserved heterotrimeric protein complex responsible for normal S-phase progression and chromatin reassembly (Volk and Crispino 2015). It specifically deposits newly acetylated H3 and H4 on to the replication fork during DNA replication (Huang and Jiao 2012). The CAF-1 complex consists of three subunits i.e. p150, p60 and p48 in Homo sapiens, Cac1, Cac2 and Cac3 in Saccharomyces cerevisiae, p180, p105 and p55 in Drosophila melanogaster and FAS1, FAS2 and MSI in Arabidopsis thaliana (Verreault et al. 1996; Sauer et al. 2017). The largest subunit of the CAF-1 complex, CAF-1A (p150/CAC1/p180/Fas1) acts as a scaffold for binding of CAF-1B, CAF-1C, histones, PCNA and various DNA polymerases (Shibahara and Stillman 1999; Kim et al. 2016). The CAF-1B (p60/CAC2/p105/Fas2) subunit preferentially binds with the Asf-1, thereby recruiting Asf-1-H3/H4 complex to CAF-1 for nucleosome assembly (English et al. 2006; Takami et al. 2007; Volk and Crispino 2015). The smallest subunit CAF-1C (p48 (RbAp48-retinoblastoma-associated protein p48)/Cac3/p55/ MSI) of the complex interacts with H3 and H4, and helps in histone deposition during nucleosome assembly (Sauer et al. 2018).
Another member of CAF-1 family, RbAp46, shares 90% homology to the RbAp48 at the amino acid level in human. Both RbAp46 and 48 are the WD40 repeat proteins that were first identified in retinoblastoma-associated proteins (RbAp) (Qian et al. 1993). RbAp46 and RbAp48 are found together in HDAC (histone deacetylase complex), Sin3, NuRD and PRC2 complexes. However, only RbAp46 is found in the HAT-1 (histone acetyl transferase) complexes that promote acetylation of newly synthesized histones, whereas RbAp48 is the exclusively smallest subunit of CAF-1 complex (Murzina et al. 2008). The CAF-1 complex is also involved in various other cellular processes like cell cycle control, heterochromatin regulation and epigenetic gene silencing (Chen et al. 2008; Huang et al. 2010; Roelens et al. 2017). The loss of CAF-1 in humans results in cell cycle arrest highlighting its role in checkpoint activation (Hoek and Stillman 2003). Recently, CAF-1 is found to deposit Cse4 (CENH3) on non-centromeric nucleosomes affecting the growth and gene expression in budding yeast (Hewawasam et al. 2018). Another member of CAF-1 family is GRWD1 that binds to Cdt1 and involved in MCM loading by its histone chaperone activity. GRWD1 downregulates p53 by inducing RPL23 proteolysis and promotes tumor formation. GRWD1 yeast ortholog-RRB1 is involved in ribosome assembly (Sugimoto et al. 2015a, b; Watanable et al. 2018).
The CAF-1 family members are part of chromatin assembly and various chromatin-remodeling complexes, thus involved in regulation of many cellular and molecular processes. However, characterization of CAF-1 family in human malaria parasite P. falciparum (PfCAF-1) remains elusive except a recent study illustrating the interaction PfCAF-1A with a number of proteins involved in nucleosome formation and var gene regulation through immunoprecipitation experiments (Gupta et al. 2018). In the present study, an extensive in silico analysis of CAF-1 family in P. falciparum (PfCAF-1) was carried out revealing phylogenetic relationships, domain architecture and protein–protein interaction (PPI) networks. Subsequently, the comparative expression profiling along with qRT-PCR studies of all PfCAF-1 genes was also carried out in three different erythrocytic stages (ring, trophozoite and schizont).
Material and methods
Identification and analysis of PfCAF-1 family genes
The CAF-1 family in P. falciparum was extracted through BLASTp search using human and yeast CAF-1 sequences against the PlasmoDB database, gene text search in PlasmoDB and HMM search with Pfam seeds (PF12253, PF12265) of CAF-1 in the HMMER web server (https://www.ebi.ac.uk/Tools/hmmer/) (Eddy 1998). Each hit was further verified by SMART and Pfam databases to confirm the presence of characteristic domains (Letunic et al. 2018; El-Gebali et al. 2019). The CDS lengths and the physico-chemical properties of the proteins were extracted from PlasmoDB database. The subcellular localization of PfCAF-1 genes was predicted using different online prediction algorithms—MitoProt II-v1.101 (Claros and Vincens 1996), Euk-mPLoc 2.0 server (Chou and Shen 2010), PATS (Zuegge et al. 2001), PlasmoAP (Foth et al. 2003) and NetNES (La Cour et al. 2004).
Identification of PfCAF-1 family orthologs across eukaryotes
For the identification of orthologs, PfCAF-1 sequences were used as query for NCBI BLASTp search against non-redundant protein sequence database. Orthologs were confirmed after manual inspection of blast parameters (e value, score, sequence coverage, percent identity) and comparative analysis of length, domain attributes, annotation of the query and the hits. The PfCAF-1 family orthologs were identified across 53 eukaryotic species covering evolutionary important taxonomic groups—Choanoflagellata (Monosiga brevicollis (Mb) and Salpingoeca rosetta (Sr), Metazoa (Trichoplax adhaerens (Tra), Homo sapiens (Hs), Danio rerio (Dr), Branchiostoma floridae (Bf), Drosophila melanogaster (Dm), Apis mellifera (Am), Caenorhabditis elegans (Ce), Brugia malayi (Bm), Nematostella vectensis (Nv), Helobdella robusta (Hr) and Daphnia pulex (Dp), Capsaspora (Capsaspora owczarzaki (Co)), Fungi (Cryptococcus neoformans (Cn), Ustilago maydis (Um), Aspergillus fumigates (Af), Schizosaccharomyces pombe (Sp), Saccharomyces cerevisiae (Sc), Neurospora crassa (Nc), Encephalitozoon cuniculi (Ec), Batrachochytrium dendrobatidis (Bd) and Spizellomyces punctatus (Spp), Apusozoa (Thecamonas trahens (Tht), Amoebozoa (Dictyostelium discoideum (Dd), Euglenozoa (Leishmania major (Lm), Leishmania infantum (Li), Trypanosoma cruzi (Tc) and Trypanosoma brucei (Tb), Ciliata (Tetrahymena thermophila (Tt) and Oxytricha trifallax (Oxt)), Apicomplexa (Plasmodium yoelii (Py), Plasmodium chabaudi (Pc), Plasmodium knowlesi (Pk), Plasmodium vivax (Pv), Plasmodium falciparum (Pf) Plasmodium berghei (Pb), Cryptosporidium parvum (Cp), Babesia bovis (Bb) and Theileria annulata (Ta), Phaeophyceae (Ectocarpus siliculosus (Es), Bacillariophyta (Thalassiosira pseudonana (Thp), Phaeodactylum tricornutum (Pht) and Aureococcus anophagefferens (Aa)), Viridiplantae (Oryza sativa (Os), Arabidopsis thaliana (At), Chlamydomonas reinhardtii (Cr), Ostreococcus tauri (Ot), Physcomitrella patens (Pp), Chlorella variabilis (Cv), Micromonas pusilla (Mp) and Selaginella moellendorffii (Sm) and Rhodophyta (Cyanidioschyzon merolae (Cm) and Galdieria sulphuraria (Gs)).
Multiple sequence alignment and phylogenetic analysis
To identify conserved features, motifs and generation of phylogenetic tree, the complete protein sequences of PfCAF-1 proteins were aligned with their homolog proteins using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the phylogenetic trees were generated using Phylip v3.625 by neighbor-joining (NJ) or maximum likelihood (ML) method with 500 bootstrap replicates (Felsenstein 2009). The trees were visualized by MEGA v5.0 (Tamura et al. 2011).
Transcriptome and proteome analysis
Microarray gene expression profile and RNA-seq data of P. falciparum at various developmental stages were obtained from Llinás et al. (2006), Le Roch et al. (2003), Otto et al. (2010), Bártfai et al. (2010) and Bunnik et al. (2013). Based on mean expression value, the expression data were normalized for each gene in all stages. Protein expression data were taken from Florens et al. (2002), Lasonder et al. (2002), Le Roch et al. (2004), Khan et al. (2005), Silvestrini et al. (2010), Solyakov et al. (2011), Treeck et al. (2011), Oehring et al. (2012), Lindner et al. (2013) and Pease et al. (2013). The heat maps for gene expression level were constructed using MeV software version 4.9.
Quantitative real time PCR validation of transcriptome data
Quantitative real-time PCR (qRT-PCR) analysis was done to validate the RNA-seq and microarray data and to check the expression patterns of PfCAF-1 genes during P. falciparum intraerythrocytic development stages (IDC)-ring (R-12 to 16 hpi (hours post invasion), trophozoite (T-30 hpi) and schizont (S-40 hpi). The expression profiles of five genes were evaluated by extracting the total RNA from R, T and S using direct-zol RNA miniprep (Zymo research) Kit according to the manufacturer’s instructions. First-strand cDNA was synthesized from 4.0 µg total RNA using thermofisher cDNA kit as per the manufacturer’s instructions. The gene-specific primers for all CAF-1 genes were designed using Primer3 software. The PCR reactions were performed using 1 µL of cDNA, 10 µL SYBER green master mix, 10 pmol each of the forward and reverse primers and 7.8 µL sterile water for a total volume of 20 µL. The PCR conditions were: holding at 50 °C for 2 min, initial denaturation at 95 °C for 10 min, followed by 35 cycles of denaturation at 95 °C for 15 s and primer annealing at 60 °C for 1 min. The seryl t-RNA synthetase gene was used as an internal control. All reactions were performed in triplicate in each experiment. Relative expression level for each gene was analyzed using 2−ΔΔCT method. Based on the transcript abundance value of five CAF-1 genes of P. falciparum in three different stages: R, T and S heat maps were constructed using MeV software.
Interaction network
Interaction network of all CAF-1 family was constructed using STRING (https://string-db.org/), BioGRID (https://thebiogrid.org/) and experimental interaction data of PfCAF-1A obtained from the literature (Gupta et al. 2018). STRING interactions based on experiment, text mining and database evidences with score between 0.150 and 0.999 were included. Protein–protein interactions of human CAF-1 genes were extracted from BIOGRID database and P. falciparum interologs were searched by BLASTp. Further, P. falciparum interologs of reported complexes of CAF-1 family in various model organisms were also traced. The graphic view of interactions was generated using Cytoscape (https://www.cytoscape.org).
Results and discussion
Extraction of CAF-1 genes in P. falciparum
A total of five CAF-1 genes were identified in P. falciparum (Table 1). All the PfCAF-1 proteins possess CAF-1 domain (CAF-1A/CAF-1C) along with WD40 repeats as identified by Pfam and SMART databases (Fig. 1). However, the CAF-1 domain (CAF-1A) in PF3D7_0501800 was identified through manual inspection using multiple sequence alignment (Supplementary Fig. 1). All the five genes were distributed on different chromosomes as shown in Table 1. The protein length and molecular weight of PfCAF-1 family ranged from 428 aa and 50,174 Da (PF3D7_1433300) to 1076 aa and 125,355 Da (PF3D7_0501800), while pI of the PfCAF-1 proteins ranged from 4.4 to 5.6 with all the members being highly acidic, specifying their interactions with histones which are basic (pI H2A-10.77, H2B-9.76, H3-11.02, H4-11.3) (Table 1). As predicted through various online softwares, all the proteins were predicted to be localized in the nucleus. Only one protein was exclusively present in nucleus (PF3D7_0501800), while others were predicted to be localized in cytoplasm also along with nucleus. Notably, only three proteins comprised the nuclear export signal as predicted by NetNES server. Out of five PfCAF-1 genes, only one gene contained introns i.e. PF3D7_1433300 (Table 1). Further, number of CAF-1 genes was found to be constant at five in all the explored strains of P. falciparum available at PlasmoDB (Supplementary Table 1).
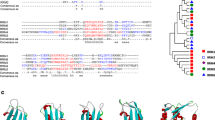
a Domain architecture analysis of PfCAF-1 family. The protein length (aa) and domain size of all the proteins was scaled as per the bar given at the top. Specific regions are abbreviated as: AD-acidic domain, KER-highly charged domain comprising lysine (K), glutamic acid (E) and arginine (R) residues, CC-coiled coil, KDDS repeats. b The multiple sequence alignment of PfCAF-1A (PF3D7_0501800) with its orthologs in other organisms highlighting PfCAF-1A specific KDDS repeats. (Bb: B. bovis, Tt: T. thermophila, Pf: P. falciparum, Pc: P. chaubadi, Pb: P. berghei, Py: P. yoelli, Pk: P. knowlesi, Pv: P. vivax, Sc: S. cerevisiae, Ce: C. elegans, At: A. thaliana, Dm: D. melanogaster)
Based on the human orthologs, the genes were classified into two different classes: Class A and Class B (Fig. 1). Class A includes proteins which belong to CAF-1 complex i.e. PF3D7_0501800, PF3D7_1329300 and PF3D7_0110700 named as PfCAF-1A, PfCAF-IB and PfCAF-1C/ PfRbAP48, respectively, whereas Class B includes two proteins that are not the part of the CAF-1 complex, PF3D7_1433300 and PF3D7_0816000, named as PfRbAP46 and PfGRWD1, respectively (Table 1). Notably, we were able to re-annotate two genes PF3D7_0110700 and PF3D7_1433300 as PfCAF-1C/ PfRbAP48 and PfRbAP46, respectively. For intergenomic analysis of abundance of CAF-1 genes, we extracted the total number of CAF-1 genes across 17 species namely C. parvum, P. vivax, P. berghei, P. knowlesi, T. thermophila, L. infantum, N. crassa, B. bovis, T. brucei, D. discoideum, S. cerevisae, C. elegans, D. melanogaster, O. sativa, A. thaliana, D. rerio and H. sapiens (Fig. 2, Supplementary Table 2). The number of CAF-1 genes varied from a minimum of four to a maximum of eight (arabidopsis and rice). Tripathi et al. (2015) showed that Arabidopsis possesses multiple copies of AtCAF-1C and rice harbors multiple copies of OsCAF-1A and OsCAF-1C genes, highlighting gene duplication events in CAF-1 family. Further, orthologs of PfCAF-1 family were retrieved across 53 eukaryotes by BLASTp and compiled in Table 2 and Supplementary Table 3. The members of CAF-1 complex were found to be well conserved throughout evolution (Table 2). Further, in 30 out of 53 organisms, there was found to be only 1 ortholog of either PfCAF-1C/RbAP48 or RbAP46. It has been reported that one protein can be orthologous to many (Tatusov et al. 1997). For example in D. melanogaster, p55 is the only ortholog of both RbAp48 and 46 (Murzina et al. 2008). It was difficult to assign orthologs to PfCAF1-C/RbAp48 and RbAp46 of an organism as both the proteins were significant hits against either RbAp48 or RbAp46 with very slight variations in blast parameters. However, the first hit confirmed by reciprocal blast was compiled as ortholog to a particular protein in Table 2.
Further, to access the evolutionary relationships of CAF-1 complex proteins in apicomplexan and model organisms a combined phylogenetic tree was constructed (Supplementary Fig. 2). For this, aligned protein sequences were used to generate a NJ tree. The tree was broadly classified into three groups: CAF-1A, CAF-1B and CAF-1C. Plasmodium species CAF-1B did not cluster with CAF-IB of other organisms, but shared the sister clade with CAF-1C members.
Chromatin assembly factor-1 subunit A (CAF-1A)
CAF-1A mediates chromatin assembly during S-phase DNA replication, nucleotide excision repair (NER), double strand break (DSB) pathways (Volk and Crispino 2015). In P. falciparum, the largest subunit of CAF-1 complex, PfCAF-1A consists of 4 × WD repeats, a highly charged KER region that facilitates interaction with histones, two coiled coil (CC) domains and a conserved CAF-1A domain (Fig. 1a). After the CAF-1 domain, PfCAF-1A also possesses 84 amino acid long stretches of KDDS repeats, not found in any other organism (Fig. 1b). Its human homolog CHAF1A/p150 revealed the presence of CAF-1A domain, coiled coil, KER region and CAF1-p150 domains. PfCAF-1A protein is larger in size as compared to its human homologue (1076 aa vs. 956 aa) and the presence of WD repeats in CAF-1A subunit is unique to P. falciparum (Fig. 3b). PfCAF-1A interacts with a number of proteins involved in heterochromatin maintenance, chromatin assembly and DNA damage repair processes (Gupta et al. 2018).
Phylogenetic and domain architecture analysis of PfCAF-1A with its orthologs in 53 eukaryotic species. a The unrooted NJ tree. The full-length protein sequences of CAF-1A family were used to perform multiple sequence alignment by Clustal omega software. Subsequently, an unrooted NJ tree was constructed using Phylip v3.695 and visualized by MEGA v5. The numbers on the nodes are the percentage bootstrap values based on 500 iterations. Different colour codes were used for different taxonomic divisions: cyan; choanoflagellata, magenta; metazoa, yellow; capsaspora, bright blue; fungi, mustard; amoebozoa, blue; euglenozoa, dark red; ciliata, bright red; apicomplexa, black; phaeophyceae, grey; bacillariophyta, light green; viridiplantae, dark green; rhodophyta. b Domain architecture. Domain organization of the representative CAF-1A members from different subclasses based on possession of different domains. Various domains are abbreviated as TM transmembrane domain, WW WWP repeating motif
To investigate the evolutionary relationships of CAF-1A subunit, NJ tree was constructed from the alignment of full-length protein sequences of 53 different eukaryotic species as mentioned in the methods (Fig. 3). Broadly, CAF-1A family was clustered into nine different clades: euglenozoa, apicomplexa, fungi, metazoans, choanoflagellates, plantae (three clades), and bacillariophyta. Notably, some clades are grouped as per eukaryotic phylogenetic classification (Morgan et al. 2008). For example, choanoflagellates shared the sister branch with metazoans. Fungi separated as monophyletic group before metazoans. However, apicomplexa shared the sister clade with ophisthokonts. Euglenozoa separated as outgroup before bacillariophyta. In eukaryotic phylogenetic classification, C. owczarzaki has been shown to share the sister group to choanoflagellates and metazoans. It is the closest unicellular relative of metazoans besides choanoflagellates and thus occupies an important phylogenetic position (Shalchian-Tabrizi et al. 2008). However, in CAF-1A subfamily phylogenetic analysis, Capsaspora shared the sister clade with chlorophytes.
Further, we explored the domain organization across 53 eukaryotic species as per SMART and Pfam databases (Fig. 3b, Supplementary Fig. S3). Importantly, 9 different domain combinations of CAF-1A subunit were found across 53 eukaryotic species. There found large variation in length of CAF-1A i.e. 140–1894 aas (Supplementary Fig. S3, Supplementary Table 3). CAF-1A domain was found to be well conserved in all the explored organisms. The domain may exist alone or in combination with other domains. Based on type of domain composition, CAF-1A family was divided into ten subclasses. Subclass i has only CAF-1A domain with eight organisms. Subclass ii has 29 members having a combination of CAF-1A domain with coiled coils (CC). L. major CAF-1A has a combination of CAF-1A, coiled coil and a transmembrane domain (TM) (Subclass iii). This domain combination is not present in any other organism. H. robusta and B. floridae CAF-1A possess CAF-1_p150+CAF-1A+CAF-1p150_C2+CC domain combination (Subclass iv). Subclass v has two organisms having a combination of CAF-1_p150_N, CAF-1_p150, CAF-1A, CAF-1p150_C2 along with coiled coils (CC). Subclass vi has five organisms which contain WD40 repeats and coiled coil domains. Subclass vii has two organisms having a combination of CAF-1A, WW and coiled coils domains. D. melanogaster and A. mellifera CAF-1A belong to subclass viii that contains CAF-1A, CAF-1p150_C2 and coiled coil domains. The CAF-1p150_N, CAF-1_p150 and CAF-1p150_C2 domains are specific features found only in some metazoans and are absent in P. falciparum. T. trahens CAF-1A has a combination of CAF-1A and SANT domain (Subclass ix), and C. variabilis CAF-1A has a combination of CAF-1A and Homeobox domain (Subclass x).
Chromatin assembly factor-1 subunit B (CAF-IB)
CAF-1B, the second subunit of CAF-1 complex, along with CAF-1A is responsible for the deposition of H3, H4 heterodimer on newly synthesized DNA (Cheloufi et al. 2015). The PfCAF-1B consists of CAF-1C_H4bd domain and WD repeats, whereas its human homologue in addition to WD repeats contains a C-terminal CAF-1_p60C domain required for direct binding to Asf-1 (Fig. 4). PfCAF-1B is shorter in length as compared to its human counterpart (582 aa vs 956 aa).
Phylogenetic and domain architecture analysis of PfCAF-1B and its orthologs in eukaryotic organisms. a The maximum likelihood tree. The tree was constructed by Phylip v3.695 using the alignment of full-length protein sequences and visualized in MEGA v5. The numbers on the nodes are the percentage bootstrap values based on 100 iterations. b Domain architecture. Domain architecture of the representative CAF-1B members from each subclass and model organisms. CAF-1B members were classified into different subclasses based on domain composition. Various domains are abbreviated as PD40 WD40-like beta propeller repeat, HIRA_B HIRA B motif, Ima1_N Ima1 N-terminal domain, ANAPC4_WD40 anaphase promoting complex subunit 4 WD40 domain
To study phylogeny of CAF-1B, both ML (Fig. 4a) and NJ (Supplementary Fig. S4) trees were constructed with the bootstrap percentages at the node of each branch. We observed better evolutionary relationships in ML tree and are explained by the same. The CAF-1B members were clustered into six different groups: metazoa, fungi and choanoflagellata, alveolata (apicomplexa and ciliate), euglenozoa, plantae (rhodophyta and viridiplantae), stramenopiles (bacillariophyta and phaeophyceae). Notably, Plasmodium species did not cluster with other apicomplexans and separated as outgroup before metazoans, thus exhibit distant relationships. Choanoflagellates CAF-1B is more closely related to fungi (shared sister clade) instead of metazoans. Straemenopiles shared sister clade with viridiplantae and euglenozoa. The apicomplexans along with ciliates separated as monophylectic group excluding Plasmodium species.
Domain architecture analysis revealed ten different domain combinations in CAF-1B family (Fig. 4b, Supplementary Fig. S4). WD 40 repeats are the conserved feature of CAF-1B family. First, 34 organisms contain only WD40 domain (Subclass i). Subclass ii has two organisms having a combination of WD40 domain with coiled coils (CC). D. discoideum CAF-1B has a combination of WD40, coiled coil and ANAPC4_WD40 (Subclass iii). This domain combination is not present in any other organism. Subclass iv has six organisms having a combination of WD40 domain with ANAPC4_WD40. A. fumigatus CAF-1B possesses ANAPC4_WD40+WD40+HIRA_B domain combination (Subclass v). U. maydis CAF-1B has a combination of WD40, ANAPC4_WD40 and PD40 domains (Subclass vi). Subclass vii has two organisms having a combination of CAF-1_p60_C and WD40 domains. Subclass viii has four organisms exhibiting a combination of WD40 repeats and CAF-1C_H4-bd domains. G. sulphuraria CAF-1B has a combination of WD40, Ima1_N and transmembrane domain (Subclass ix) and T. annulata CAF1-B has a combination of WD40, DNA_pol_alpha_N domain with a transmembrane domain (Subclass x). The protein length of CAF-1B members was found to vary from 293 to 951 aas.
Chromatin assembly factor-1 subunit C (CAF-IC)/RbAp48 and RbAP46
CAF-1C/RbAP48 and 46 are WD40-containing proteins involved in maintenance and regulation of chromatin structure (Lejon et al. 2011). These proteins are exclusive members of different chromatin complexes. RbAp48/CAF-1C is the smallest subunit of CAF-1 complex, whereas RbAP46 is the member of HAT complex. Both RbAp48 and 46 are found together in HDAC, NuRD and SIN3 complexes (Murzina et al. 2008). Further, Human RbAp48 and 46 proteins are highly homologous sharing 89% identity, whereas P. falciparum RbAp48 and 46 showed 30% identity. The percentage identity of RbAp48 and 46 across 53 eukaryotic organisms is compiled in Supplementary Table 4.
The phylogenetic tree of RbAp48 is divided into eight groups: i.e. viridiplantae, metazoa, ciliata, bacillariophyta, rhodophyta, fungi, apicomplexa and euglenozoa (Fig. 5a). Evolutionary-related species are grouped together in the CAF-1C NJ tree. However, there is a divergence in branching as compared to the evolutionary history of eukaryotes (Fig. 5a). Based on type of domain composition, CAF1-C family is divided into three subclasses. First, 50 organisms comprise CAF1C_H4-bd domain along with WD40 repeats (Subclass i). Subclass ii has two organisms having a combination of CAF1C_H4-bd, WD40 and coiled coils (CC). O. tauri CAF-1C has a combination of CAF1C_H4-bd, WD40 and ANAPC4_WD40 domain (Subclass iii). The CAF-1C members exhibited least variation in size as compared to other CAF-1 members. Most of CAF-1C members are of same size i.e. around 400 aas.
Phylogenetic and domain architecture analysis of PfCAF-1C/RbAp48 (a) and PfRbAp46 (b) with its orthologs in different organisms. The NJ trees were constructed using Phylip v3.695. The numbers on the nodes are the percentage bootstrap values based on 500 iterations. Domain architecture of representative organisms from each subclass was drawn as per the scale. Various domains are abbreviated as ANAPC4_WD40 anaphase promoting complex subunit 4 WD40 domain, eIF2A eukaryotic translation initiation factor eIF2A
The phylogenetic analysis classified RbAp46 into five distinct groups (Fig. 5b). Members of plantae are grouped together in cluster 1 followed by euglenozoa, fungi, metazoa and apicomplexa. C. owczarzaki, the closest unicellular relative of metazoans, shared the sister branch with metazoans. Fungi separated before metazoans as per eukaryotic evolutionary history. The length of RbAp46 was found to vary from 297 to 648 aa. Based on domain architecture, RbAp46 family is divided into four subclasses. Subclass i has two members possessing only WD40 repeats. Subclass ii has 20 organisms having a combination of CAF1C_H4-bd along with WD40 domain. S. cerevisiae RbAp46 contains CAF1C_H4-bd+WD40+eIF2A domain combination (Subclass iii). T. thermophila RbAp46 has a combination of CAF1C_H4-bd, WD40 and coiled coil (Subclass iv).
Glutamate rich WD40 repeat protein 1 (GRWD1)
GRWD1 is an evolutionary conserved cdt1-binding protein that functions in MCM loading, ribosome biogenesis; histone binding and nucleosome assembly processes (Sugimoto et al. 2015a, b). PfGRWD1 is 491 aa long and possess N-terminal CAF-1C domain, an acidic-rich region involved in protein–protein binding, a coiled-coil domain and C-terminal WD40 repeats (Fig. 1). Its human homologue HsGRWD1 is comparatively shorter (446 aas) in length and possesses fewer WD40 repeats (Fig. 6).
Phylogenetic and domain architecture analysis of PfGRWD1 with its orthologs in other organisms. a The alignment of full-length protein sequences of GRWD1 family was used to generate an unrooted NJ tree The numbers on the nodes are the percentage bootstrap values based on 500 iterations. b Domain architecture. Domain architecture of the representative GRWD1 members from each subclass. Various domains are abbreviated as TM transmembrane domain, adh_short short chain dehydrogenase
The orthologs of PfGRWD1 were identified in a variety of different species using BLASTp and phylogenetic relationships were drawn (Fig. 6a). Intriguing evolutionary relationships were observed in GRWD1 family. Phylogenetic tree of GRWD1 family was divided into eight clusters: apicomplexa, bacillariophyta, euglenozoa, plantae, metazoa, choanoflagellata, fungi and ciliata. This clustering is in accordance with evolutionary grouping of the eukaryotes. The fungi separated as monophylectic group before choanoflagellates and metazoans. Importantly, capsaspora was positioned between fungi and chanoflagellates; however, it shared the sister clade with fungi. In plantae, chlorophyta shared sister clade with streptophyta. Apicomplexans are separated before bacillariophyta as per eukaryotic phylogeny. Further, baccilariophyta and euglenozoa shared the sister groups. Overall, phylogenetic-related species group together in GRWD1 family NJ tree.
The final sub-grouping in phylogenetic analysis is in accordance with domain composition (Supplementary Fig. S5). We observed six different domain combinations in GRWD1 family (Fig. 6b). The combination of CAF-1C domain and WD repeats is conserved in all species except one, i.e. A. anophagefferens where DEXDc+HELICc is present along with WD repeats (Subclass vi). The length of this protein is the longest (974 aa) in the analyzed GRWD1 members. GRWD1 of D. pulex harbors a combination of the transmembrane domain and CAF-1C on N-terminal and WD40 repeats on its C-terminal (Subclass iv). Five organisms belonging to ciliata and euglenozoa groups contained coiled coil along with CAF-1C and WD40 repeats (Subclass-iii). However, the function of coiled coil domain in GRWD1 is unknown. T. trahens contained an additional domain besides CAF-1C and WD40, i.e. adh_short-a short chain dehydrogenase (Subclass v) involved in oxidoreductase activity. The length of GRWD1 varied from 365 to 974 aas (Supplementary Fig. S5).
Expression analysis
To gain insight into the expression profiles of PfCAF-1 family during different development stages, we investigated the transcriptome and proteome data present at PlasmoDB. A heat map was generated from the microarray data of Derisi (Llinás et al. 2006), Winzeler (Le Roch et al. 2003) and RNA-seq data of Barfai (Bártfai et al. 2010), Otto (Otto et al. 2010), and Bunnik (Bunnik et al. 2013) (Fig. 7a). We used the log2 ratios of RMA and FPKM value for microarray and RNA-seq data to generate the heat map. Further, to validate the available transcriptome data, we carried out the qRT-PCR experiments for all the five genes at different intraerythrocytic development stages (IDC): ring (R), trophozoite (T) and schizont (S) and the fold change in expression over ring stage are represented as bar graphs (Fig. 7b). The qRT-PCR primers are enlisted in Supplementary Table 5. In addition to this, proteome profile of the PfCAF-1 family (Fig. 7c) was examined from the data available at PlasmoDB-Florens et al. (2002), Lasonder et al. (2002), Le Roch et al. (2004), Khan et al. (2005), Silvestrini et al. (2010), Solyakov et al. (2011), Treeck et al. (2011), Oehring et al. (2012), Lidner et al. (2013), Pease et al. (2013) and compared with the transcriptome data.
Expression profiling of PfCAF-1 genes. a Transcriptome analysis. Heat map of PfCAF-1 from microarray data (Llinas et al. 2006; Le Roch et al. 2003) and RNA-seq data (Otto et al. 2010; Barfai et al. 2010; Bunnik et al. 2013). Calorimetric representation used for heat maps of transcriptome data is green–red (green, low expression; black, medium expression; red, high expression). b RT-PCR analysis. Relative gene expression of five PfCAF-1 genes was analysed by qRT-PCR. Seryl t-RNA synthetase was used as internal control. The Y-axis indicates the relative expression level and error bars represent standard deviation calculated based on three technical replicates. c Proteome expression profiling. Heat map proteome and phosphoproteome data obtained from Florens et al. (2002), Lasonder et al. (2002), Le Roch et al. (2004), Khan et al. (2005), Silvestrini et al. (2010), Solyakov et al. (2011), Treeck et al. (2011), Oehring et al. (2012), Lidner et al. (2013), Pease et al. (2013). X-axis (Fig. 4a, c) represents different developmental stages of Plasmodium life cycle abbreviated as: ER and LR early and late rings, ET and LT early and late trophozoites, ES and LS early and late schizonts, M merozoites, G gametocytes, Sp sporozoites, Gt gamete, EG early gametocyte, MG mature gametocyte, OOC oocyst, ODS oocyst-derived sporozoites, SGS salivary gland sporozoites, phosEnr phospho-enriched, phosDep phospho-depleted, Nuc nuclear, cyt cytoplasmic. Numerals on X-axis represents 0–48 h post invasion (hpi)
As per both microarray and RNA-seq studies, PfCAF-1A was found to be upregulated at late T and S and downregulated at R. Expression of PfCAF-1A was also found to be upregulated at T and S by qRT-PCR with a significant higher expression at T (~ sixfold) as compared to R. As per proteome data, PfCAF-1A was found to be expressed in R, T, S, M, G and Gt. A quantitative proteome analysis of IDC by Pease et al. (2013) study showed relatively higher expression in R followed by T and S. This shows non-coordination between peak expression at mRNA and protein level.
Transcript of PfCAF-1B was downregulated in R and upregulated in T and S as per Derisi and RNA-seq studies. RT-PCR analysis also showed upregulation in T and S. PfCAF-1B protein was found to exist in R, T, S and G as per the compiled proteome datasets. However, discrepancies were observed in detection of PfCAF-1B by different proteome studies.
The PfCAF-1C was observed to have high transcript abundance throughout IDC, G, Sp with peak expression in T followed by S by microarray and RNA-seq datasets. The qRT-PCR analysis also confirmed upregulation of CAF-1C expression in T and S. Accordingly, its protein was found through all the developmental stages of P. falciparum (R, T, S, M, G and SGS) except Sp. The PfRbAP46 transcript is down regulated in T and upregulated in S by Derisi and qRT-PCR analyses. RNA-seq datasets also showed upregulation of RbAp46 in S. Accordingly, its protein was found to be more abundant in S. RbAP46 protein was detected in R, T, S, M and SGS. Out of the proteome studies involving different IDC stages, only four studies (Le Roch et al. 2004; Treeck et al. 2011; Lidner et al. 2013; Pease et al. 2013) detected RbAp46 highlighting discrepancies between different datasets. This may be because of variations in method of sampling and sensitivity among different mass spectrometry methods.
The PfGRWD1 transcript was found to be downregulated in S by all the datasets (microarray, RNA seq and qRT-PCR). Derisi dataset showed peak mRNA expression at R, whereas qRT-PCR showed peak expression at T. PfGRWD1 protein and was detected in R, T and S by Pease et al. (2013), whereas other proteome studies detected PfGRWD1 only in S.
Out of five predicted PfCAF-1s, only three (PfCAF-1A, PfCAF-1B, PfCAF-1C) were detected to be nuclear by Oehring et al. 2012 (Fig. 7c). These three CAF-1 complex subunits were also found to be phosphorylated by Treeck et al. (2011) and Pease et al. (2013) (Fig. 7c), thus confirming their regulatory role. The CAF-1A, B and C were found to be co-expressed through most of the developmental stages with peak expression in R and G. This co-existence of all the subunits of CAF-1 complex ensures the formation of CAF-1 complex. None of the PfCAF-1 was found to be stage specific. Overall, we observed mixed relationships (linear as well as non-linear) between the PfCAF-1 family transcriptome and proteome datasets. Broadly, there was a good coordination between transcriptome datasets; however, discrepancies were observed between different proteome studies. The qRT-PCR data were found in well coordination with earlier transcriptome data except few variations discussed above.
Interaction network
To get further insight into the functional diversity of PfCAF-1 family, we constructed a network of all PfCAF-1 PPIs by mining string database, interologs datasets and experimental interaction evidence based on literature (Fig. 8). The interolog dataset was deduced based on the human interaction data for all CAF-1 family proteins from BIOGRID database as described in methods. We identified potential P. falciparum interologs by tracing the orthologs of the proteins of human CAF-1 family interactome in human malaria parasite. A total of 334 PPIs were identified for PfCAF-1 family (Supplementary Table 6). Out of these, 271 PPIs were extracted from the string, 115 from the interolog datasets and ten from the immunoprecipitation data (Gupta et al. 2018) (Supplementary Table 5). There were 52 overlaps i.e. 50 were common between STRING and interolog dataset and 2 were common between STRING and Co-IP. The extent of connectivity of PfCAF-1 proteins ranged from a minimum of 24 (GRWD1) to a maximum of 108 PPIs (CAF-1C) (Supplementary Table 5).We considered all the interactions from string based on experiment, text mining and database evidences ranging from low to high confidence scores. Out of 271 string interactions, 57 PPIs (21.033%) were having high confidence scores (S > 0.7), 196 associations (72.32%) of medium confidence scores (0.4 ≤ S < 0.7) and 18 associations (6.64%) having low confidence scores (S < 0.4).
Interaction network analysis of PfCAF-1 proteins. A PPI network of five PfCAF-1 proteins with yellow nodes was constructed using Cytoscape software. The interaction data were derived from STRING (light pink nodes, grey edges), co-IP (red nodes and edges) and BIOGRID database (dark pink nodes and edges). The interactions common between string and co-IP are highlighted by diamond shape and the interactions common between STRING and BIOGRID are indicated by triangle shape. Node size is proportional to the degree of the node. Inset shows the level of conservation of different CAF-1 proteins complexes in P. falciparum-CAF-1 complex, replication-dependent chromatin assembly complex, NuRD complex (nucleosome remodeling deacetylase), Sin3 complex (histone deacetylation complex) and pre-RC complex (pre-replication complex)
Further, we tried to trace five important protein complexes namely CAF-1 complex (Hoek and Stillman 2003), replication-dependent chromatin assembly complex (Shibahara and Stillman 1999), NuRD complex (Basta and Rauchman 2015), Sin3 complex (Kuzmichev et al. 2002) and Pre-RC complex (Tsakraklides and Bell 2010) in the PPI network of PfCAF-1 family. We identified all the complexes in P. falciparum (Fig. 8). The CAF-1 complex and replication-dependent chromatin assembly complex, both were found to be absolutely conserved in P. falciparum. However, NuRD complex, Sin3 complex and Pre-RC complex were found to be partially conserved as orthologs of one or more subunits which could not be identified (Fig. 8). These were P66, MBD and MTA1/2/3 in NURD complex, SAP30, SDS and Sin3 in Sin3 complex, and CDC6 in Pre-RC complex. We also observed that the PfCAF-1 family members, RbAp48 and 46, are part of multiple complexes and have high connectivity than other members, thus can be explored as drug target. All the PfCAFs harbored WD40 repeats justifying their high connectivity. WD40 repeats serve as a platform for the binding of other proteins; thus result in assembly of multi-protein complexes. Chahar et al. (2015) showed that the interaction network of PfWD40 proteins involved in chromatin-associated processes is composed of 203 PPIs. Overall, the PfCAF-1 proteins are highly interconnected and reside in multiple complexes.
Conclusion
In the present study, a comprehensive genome-wide analysis of CAF-1 family in P. falciparum was performed to evaluate domain composition, evolutionary relationships, expression profile and protein–protein interactions network. A total of five putative PfCAF-1 genes: three as a part of CAF-1 complex and two as part of chromatin modifying complexes were identified in P. falciparum. Mining of CAF-1 genes across model organisms and apicomplexa disclosed a minimum of four and a maximum of eight CAF-1 genes (arabidopsis and rice). Further, a comparative analysis of domain attributes of CAF-1 family across 53 eukaryotic organisms highlighted species-specific features. CAF-1 complex was found to be conserved through all the explored organisms. Phylogenetic analysis corroborated grouping of different CAF-1 genes as per evolutionary history. Broadly, closely related species are clustered together in same clade with retention of evolutionary history of branching. Clustering in different subgroups was also supported by domain features. A detailed transcriptome and proteome profiling of PfCAF-1 genes showed mixed expression pattern coordination as well as discrepancies. qRT-PCR analysis of PfCAF-1 genes confirmed differential expression in ring, trophozoite and schizont stages. Further, we mined PPIs of all PfCAF-1 genes and traced important complexes in P. falciparum. In nutshell, the present efforts delineate the specific features of the CAF-1 family in P. falciparum thus providing a platform for further studies.
References
Bártfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Françoijs KJ, Stunnenberg HG (2010) H2A.Z demarcates intergenic regions of the Plasmodiumfalciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog 6:e1001223
Basta J, Rauchman M (2015) The nucleosome remodeling and deacetylase complex in development and disease. Transl Res 165:36–47
Bunnik EM, Chung DW, Hamilton M, Ponts N, Saraf A, Prudhomme J, Florens L, Le Roch KG (2013) Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol 14:R128
Burgess RJ, Zhang Z (2013) Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20:14–22
Chahar P, Kaushik M, Gill SS, Gakhar SK, Gopalan N, Datt M, Sharma A, Gill R (2015) Genome-wide collation of the Plasmodium falciparum WDR protein superfamily reveals malarial parasite-specific features. PLoS ONE 10(6):e0128507
Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, Wesche DJ, Abazova N, Hogue M, Tasdemir N, Brumbaugh J, Rathert P, Jude J, Ferrari F, Blanco A, Fellner M, Wenzel D, Zinner M, Vidal SE, Bell O, Stadtfeld M, Chang HY, Almouzni G, Lowe SW, Rinn J, Wernig M, Aravin A, Shi Y, Park PJ, Penninger JM, Zuber J, Hochedlinger K (2015) The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528:218–224
Chen Z, Tan JL, Ingouff M, Sundaresan V, Berger F (2008) Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsisthaliana. Development 135:65–73
Chou KC, Shen HB (2010) A New method for predicting the sub-cellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE 5:e9931
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432
English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127:495–508
Felsenstein J (2009) PHYLIP—(phylogeny inference package) version 3.69. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ (2002) A proteomic view of the Plasmodiumfalciparum life cycle. Nature 419:520–526
Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI (2003) Dissecting apicoplast targeting in the malaria parasite Plasmodiumfalciparum. Science 299:705–708
Gupta MK, Agarawal M, Banu K, Reddy KS, Gaur D, Dhar SK (2018) Role of chromatin assembly factor 1 in DNA replication of Plasmodium falciparum. Biochem Biophys Res Commun 495:1285–1291
Hewawasam GS, Dhatchinamoorthy K, Mattingly M, Seidel C, Gerton JL (2018) Chromatin assembly factor-1 (CAF-1) chaperone regulates Cse4 deposition into chromatin in budding yeast. Nucleic Acids Res 46:4831
Hoek M, Stillman B (2003) Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci USA 100:12183–12188
Huang H, Jiao R (2012) Roles of chromatin assembly factor 1 in the epigenetic control of chromatin plasticity. Sci China Life Sci 55:15–19
Huang H, Yu Z, Zhang S, Liang X, Chen J, Li C, Ma J, Jiao R (2010) Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J Cell Sci 123:2853–2861
Khan SM, Franke-Fayard B, Mair GR, Lasonder E, Janse CJ, Mans M, Waters AP (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121:675–687
Kim D, Setiaputra D, Jung T, Chung J, Leitner A, Yoon J, Aebersold R, Hebert H, Yip CK, Song JJ (2016) Molecular architecture of yeast chromatin assembly factor 1. Sci Rep 6:26702
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16:2893–2905
La Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536
Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, Eling WM, Hall N, Waters AP, Stunnenberg HG, Mann M (2002) Analysis of the Plasmodiumfalciparum proteome by high-accuracy mass spectrometry. Nature 419:537–542
Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503–1508
Le Roch KG, Johnson JR, Florens L, Zhou SA, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, Yates JR 3rd, Winzeler EA (2004) Global analysis of transcript and protein levels across the Plasmodiumfalciparum life cycle. Genome Res 14:2308–2318
Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, Laue ED, Mackay JP (2011) Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48·FOG-1 complex. J Biol Chem 286:1196–1203
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496
Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, Kappe SH (2013) Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics 12:1127–1143
Llinás M, Bozdech Z, Wong ED, Adai AT, DeRisi JL (2006) Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34:1166–1173
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260
Morgan RO, Fernandez MP (2008) Molecular phylogeny and evolution of the coronin gene family. Subcell Biochem 48:41–55
Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16:1077–1085
Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS (2012) Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol 13:R108
Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Böhme U, Lemieux J, Barrell B, Pain A, Berriman M, Newbold C, Llinás M (2010) New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol 76:12–24
Pease BN, Huttlin EL, Jedrychowski MP, Talevich E, Harmon J, Dillman T et al (2013) Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. J Proteome Res 12:4028–4045
Qian YW, Wang YC, Hollingsworth RE Jr, Jones D, Ling N, Lee EY (1993) A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature 364:648–652
Roelens B, Clémot M, Leroux-Coyau M, Klapholz B, Dostatni N (2017) Maintenance of heterochromatin by the large subunit of the CAF-1 replication-coupled histone chaperone requires its interaction with HP1a through a conserved motif. Genetics 205:125–137
Sauer PV, Timm J, Liu D, Sitbon D, Boeri-Erba E, Velours C, Mücke N, Langowski J, Ochsenbein F, Almouzni G, Panne D (2017) Insights into the molecular architecture and histone H3–H4 deposition mechanism of yeast chromatin assembly factor 1. Elife 6:piie23474
Sauer PV, Gu Y, Liu WH, Mattiroli F, Panne D, Luger K, Churchill ME (2018) Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res 46:9907–9917
Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T (2008) Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE 3:e2098
Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575–585
Silvestrini F, Lasonder E, Olivieri A, Camarda G, Schaijk BV, Sanchez M, Younis SY, Sauerwein R, Alano P (2010) Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodiumfalciparum. Mol Cell Proteomics 9:1437–1448
Solyakov L, Halbert J, Alam MM, Semblat JP, Dorin-Semblat D, Reininger L, Bottrill AR, Mistry S, Abdi A, Fennell C, Holland Z, Demarta C, Bouza Y, Sicard A, Nivez MP, Eschenlauer S, Lama T, Thomas DC, Sharma P, Agarwal S, Kern S, Pradel G, Graciotti M, Tobin AB, Doerig C (2011) Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodiumfalciparum. Nat Commun 2:565
Sugimoto N, Maehara K, Yoshida K, Yasukouchi S, Osano S, Watanabe S, Aizawa M, Yugawa T, Kiyono T, Kurumizaka H, Ohkawa Y, Fujita M (2015a) Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res 43:5898–5911
Sugimoto N, Maehara K, Yoshida K, Yasukouchi S, Osano S, Watanabe S, Aizawa M, Yugawa T, Kiyono T, Kurumizaka H, Ohkawa Y, Fujita M (2015b) Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res 43:5898–5911
Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T (2007) Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell 1:129–141
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278(5338):631–637
Treeck M, Sanders JL, Elias JE, Boothroyd JC (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 10:410–419
Tripathi AK, Singh K, Pareek A, Singla-Pareek SL (2015) Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol 15:42
Tsakraklides V, Bell SP (2010) Dynamics of pre-replicative complex assembly. J Biol Chem 285:9437–9443
Verreault A, Kaufman PD, Kobayashi R, Stillman B (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95–104
Volk A, Crispino JD (2015) The role of the chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in homeostasis and disease. Biochim Biophys Acta 1849:979–986
Watanabe S, Fujiyama H, Takafuji T, Kayama K, Matsumoto M, Nakayama K, Yoshida K, Sugimoto N, Fujita M (2018) GRWD1 regulates ribosomal protein L23 levels via the ubiquitin-proteasome system. J Cell Sci 131(15):jcs213009
Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G (2001) Deciphering apicoplast targeting signals-feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280:19–26
Acknowledgements
SSG and RG acknowledge the partial support of Radha Krishnan Foundation Fund, MDU, Rohtak; University Grants Commission (UGC), Department of Science and Technology (DST) and Council of Scientific and Industrial Research (CSIR), Govt. of India, New Delhi. MK acknowledges the award of University Research Scholarship (URS) from MDU, Rohtak, Haryana. Authors are thankful to Dr. Amit Sharma, ICGEB, New Delhi for constructive suggestions.
Author information
Authors and Affiliations
Contributions
RG and SSG conceived and designed the experiments. MK and AN performed the experiments. MK and RG analyzed the data. MK, RG and SSG wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2020_2096_MOESM1_ESM.pptx
Supplementary file1. S1 Fig. Multiple sequence alignment of CAF1A domain of PfCAF-1A in comparison to its human ortholog (HsCAF-1A) and other model organisms (Sc: S. cerevisae, Ce: C. elegans, At: A. thaliana, Dm: D. melanogaster). Clustal omega was used to perform the alignment. S2 Fig. Phylogenetic analysis of CAF-1 family from apicomplexans and model organisms. Tree was constructed with 500 bootstrap value using Phylip v3.625 and visualized by MEGA v5.0. S3 Fig. Phylogenetic and domain architecture analysis of CAF-1A subfamily. The unrooted NJ tree was constructed and domain architecture of all organisms is drawn as per SMART and Pfam databases. Different domain combinations of CAF-1A subfamily are highlighted with green box and red star. S4 Fig. Phylogenetic analysis of CAF-1B subfamily along with domain architecture. The unrooted NJ tree of CAF-1B family was constructed and domain architecture of all organisms is drawn as per SMART and Pfam database. Specific domain combinations of PfCAF-1B family are highlighted with green box and red star. S5 Fig. Phylogenetic and domain architecture analysis of GRWD1 subfamily. The unrooted NJ tree was constructed and domain architecture of all organisms is drawn as per SMART and Pfam database. Different domain compositions are highlighted with green box and red star. (PPTX 547 kb)
Supplementary file2. S1 Table. List of putative CAF-1 genes in 17 eukaryotic organisms. (XLSX 15 kb)
13205_2020_2096_MOESM4_ESM.xlsx
Supplementary file4. S3 Table. List of orthologs of CAF-1 family identified across 53 eukaryotic species with BLAST parameters, domain composition and protein length (aa). (XLSX 48 kb)
13205_2020_2096_MOESM5_ESM.xlsx
Supplementary file5. S4 Table. RbAp48 and RbAp46 and their percentage identity across 53 organisms of different taxonomic divisions. (XLSX 12 kb)
13205_2020_2096_MOESM7_ESM.xlsx
Supplementary file7. S6 Table. PfCAF-1 family protein–protein interaction data obtained from the STRING, BIOGRID and co-IP. (XLSX 49 kb)
Rights and permissions
About this article
Cite this article
Kaushik, M., Nehra, A., Gill, S.S. et al. Unraveling CAF-1 family in Plasmodium falciparum: comparative genome-wide identification and phylogenetic analysis among eukaryotes, expression profiling and protein–protein interaction studies. 3 Biotech 10, 143 (2020). https://doi.org/10.1007/s13205-020-2096-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-2096-7