Abstract
This study examines the effect of epigallocatechin gallate (EGCG) on signaling pathways, epigenetic modulators and tumour suppressor genes in cervical cancer cells, HeLa. qRT-PCR, ELISA-based enzymatic assays and in silico studies were used to catalogue the modulation of these genes by EGCG treatment. qRT-PCR showed transcriptional modulation of several epigenetic modifiers including DNA methyltransferases and histone modifiers (DNMT1, DNMT3B, DNMT3A, AURKA, AURKC, AURKB, KDM4A, KDM5C, PRMT7, PRMT6, UBE2B, HDAC5, HDAC6, HDAC7 and HDAC11. Furthermore, ELISA-based assays showed that EGCG lowered the activity of DNA methyltransferases, histone deacetylases and histone methyltransferases (H3K9). Molecular docking results suggests that EGCG may competitively inhibit some epigenetic enzymes (DNMT1, DNMT3A, HDAC2, HDAC3, HDAC4, HDAC7 and EZH2). A functional outcome of these epigenetic alterations could be inferred from the reversal of promoter hypermethylation of tumour suppressor genes by quantitative methylation array and transcriptional re-expression of tumour suppressor genes including TP73, PTEN, SOCS1, CDH1, RARβ, and DAPK1 by qRT-PCR. Downregulation of key signaling moieties of PI3K, Wnt and MAPK pathways, cell cycle regulators, metastasis regulators and pro-inflammatory moieties including TERT, CCNB1, CCNB2, MMP2, MMP7. PIK3C2B, PIK3CA, MAPK8 and IL6 was also observed. In silico protein–protein interaction network analysis followed by KEGG analysis discerned the active participation of gene sets towards cancer pathways. This study comprehensively explains EGCG’s anti-cancer mechanism via the synchronized transcriptional alteration of several molecular targets across different signaling pathways and reversal of tumour suppressor gene silencing through modulation of epigenetic enzymes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carcinogenesis occurs due to aberrations which are genetic as well as epigenetic in nature. While normal physiology requires a functional epigenetic mechanism, any aberrant changes can alter transcriptional patterns and promote carcinogenesis. DNA methylation, histone modifications and microRNA-based regulation are the three important branches of epigenetic mechanisms. The addition of a methyl group to CpG islands in promoter regions by DNA methyltransferases (DNMTs) coupled with histone modifications result in heterochromatinization and gene silencing. Modifications of histone proteins including deacetylation, acetylation, phosphorylation, methylation, demethylation and ubiquitination catalyzed by various enzyme groups impact gene transcription. In cancer cells, tumor suppressor genes (TSGs), which control cell proliferation, cell-cycling and programmed cell death are found to be silenced due to epigenetic alterations. These alterations also influence the atypical and unsystematic activation of PI3K, Wnt and MAPK signaling pathways. This facilitates the proliferation of tumour cells, influences their migration, and enables the cells to evade apoptosis. However, epigenetic changes are reversible and therefore much attention has been focused on targeting them for therapeutic advantage. Nevertheless, currently identified drugs have been shown to be toxic with low specificity. Several studies have demonstrated the anti-cancer effect of dietary bioactive compounds, resulting in apoptosis, transcriptional modulation of molecular targets and potentiation of chemotherapeutic drugs (Kedhari Sundaram et al. 2019b; Sundaram et al. 2019a). Their anti-carcinogenic ability has also been ascribed to their capacity to influence the epigenetic machinery and reestablish TSG transcription (Sundaram et al. 2017; Kedhari Sundaram et al. 2019a). EGCG or (−)-epigallocatechin-3-gallate, a catechin present abundantly in green tea, has anti-oxidant and anti-inflammatory properties and can elicit anti-proliferative, anti-metastatic and pro-apoptotic response (Sharma et al. 2012; Liu et al. 2013; Cerezo-Guisado et al. 2015). In this report, the effect of EGCG on several molecular targets including signaling pathways and epigenetic pathways in cervical cancer cells, HeLa was explored.
Materials and methods
Cell culture
Human cervical carcinoma cells (HeLa) cells were chosen for the assay as a model of cervical cancer. HeLa cells were a kind gift from the lab of Dr. Tahir Rizvi, UAE University and maintained in Dulbecco’s Modified Eagle’s Medium (Sigma, MO, USA) fortified with fetal bovine serum (10%) (Sigma, USA) and Pen-strep (100x) (Sigma, USA). The cells were placed in an incubator at 37 °C with 5% CO2 infusion.
Preparation of EGCG
10 mM EGCG (Catalogue number E4143; Sigma, USA) was freshly prepared in serum free media and further dilutions were made using complete medium as required. EC50 was established as 100 μM EGCG following 24 h treatment using MTT assay, previously (Sharma et al. 2012). In this study, 50 µM EGCG was employed for analysis. In all assays, in the untreated control samples the same volume of media was used without EGCG.
Expression analysis of various genes involved in tumorigenesis and cancer-related pathways
Total RNA was isolated from 2 × 106 cells treated with 50 µM EGCG for 48 h as well as untreated HeLa cells using Gen Elute Mammalian Genomic Total RNA Kit (Sigma, USA). cDNA was synthesized using RT-PCR Kit (ABI, USA) using 2 μg of RNA. Selected TSGs and genes from different signaling pathways where assessed using Taq-man custom array on QuantStudio3 (Applied Biosystems, USA). Results were evaluated using the ΔΔCT method using DataAssist™ software (ThermoFisher, USA). Global normalization was performed and change in gene expression (fold change) of treated sample in comparison to untreated control was represented as relative quantity (RQ). Statistical significance was calculated as the mean of three experiments using two-tailed t test with p ≤ 0.05.
Expression analysis of the genes involved in chromatin modification
cDNA was prepared as detailed above. The expression of a panel of epigenetic enzymes involved in DNA methylation and various histone modifications was assessed using the Human Epigenetic Chromatin Modification Enzymes RT2 Profiler PCR Array (Catalogue number PAHS-085Z; Qiagen, USA). Results were evaluated using the ΔΔCT method using DataAssist™ software (ThermoFisher, USA). Endogenous gene (GAPDH) was used for normalization and change in gene expression (fold change) of treated sample in comparison to untreated control was represented as RQ. Statistical significance was calculated using the mean of three experiments using two-tailed t test with p ≤ 0.05.
DNA methyltransferase activity assay
Nuclear extract from untreated HeLa cells was obtained using the EpiQuikTM nuclear extraction kit (Catalogue number OP-0002; Epigentek, USA) following the manufacturer’s protocol. The change in activity of DNA methyltransferase (DNMT) enzymes following incubation with EGCG (50 µM) was assessed using the ELISA-based Epiquik DNMT activity assay kit (Catalogue number OP-0002; Epigentek, USA). The percentage of inhibition in comparison to the untreated control was then calculated and plotted as a graph.
Histone deacetylase activity assay
Nuclear extract from untreated HeLa cells was obtained using the EpiQuik™ nuclear extraction kit (Catalogue number OP-0002; Epigentek, USA) following the manufacturer’s protocol. The change in activity of histone deacetylase (HDAC) enzymes following incubation with EGCG (50 µM) was assessed using the ELISA based Epiquik HDAC activity assay kit (Catalogue number P-4002; Epigentek, USA). The percentage of inhibition compared to the untreated control was then calculated and plotted as a graph.
Histone methyltransferase–H3K9 activity assay
Nuclear extract from untreated HeLa cells was obtained using the EpiQuikTM nuclear extraction kit (Catalogue number OP-0002; Epigentek, USA) following the manufacturer’s protocol. The change in activity of histone methyltransferase (HMT) enzymes following incubation with EGCG (50 µM) was assessed by using the ELISA based Epiquik Histone methyltransferase (HMT)-H3K9 activity assay kit (Catalogue number P-4001; Epigentek, USA). The percentage of inhibition in comparison to the untreated control was then calculated and plotted as a graph.
Molecular modelling studies of DNMT, HDAC and EZH2 proteins
Docking of EGCG with DNMT1, DNMT3A, HDAC2, HDAC3, HDAC4, HDAC7, HDAC8 and EZH2 was performed and analyzed as described by the author’s earlier publication (Kedhari Sundaram et al. 2019a; Sundaram et al. 2019b). Full fitness values were used to rank the docked structures and the one with least energy was accepted as the best docked pose.
Global DNA methylation assay
DNA was extracted from 2 × 106 HeLa cells treated with 50 μM EGCG (24 and 48 h) using GenElute Mammalian Genomic DNA Miniprep Kit (Catalogue number G1N70; Sigma, USA). The quantum of methylated DNA in the treated and untreated samples was measured using MethylFlash™ Methylation DNA Quantification kit (Catalogue number: P-1034; Epigentek, USA) which utilizes a 5-mC antibody in an ELISA based assay. The optical density readings obtained were relative to the quantity of methylated DNA. Methylation level was expressed as percentage of control and represented in a graph.
Detection of promoter methylation using methylation array
DNA was isolated using GenElute Mammalian Genomic DNA Miniprep Kit (Catalogue number G1N70; Sigma, USA), following treatment of HeLa cells with 50 µM EGCG for 48 h and restriction digestion was carried out using EpiTect II DNA Methylation Enzyme Kit (Catalogue number: 335452; Qiagen, USA). The assay uses two restriction enzymes with differential action on methylated cytosine in the recognition sequence. The digest was set up without any enzyme, each enzyme separately and both together. These four digestion products were utilized separately to perform qPCR using Human Tumor Suppressor Genes EpiTect Methyl II Signature PCR Array (Catalogue number EAHS-551Z; Qiagen, USA). Methylation level of each gene was calculated using the Ct values from all four digests according to the manufacturer’s protocol and formula.
In silico network analysis
STRING Cytoscape app (https://apps.cytoscape.org/apps/stringapilized) was used for in silico network analysis of differentially expressed genes. The application was based on the identification of protein–protein interaction (PPIN) among input genes. It provides high level information about input genes, interacting genes, types of interactions (viz. co-occurrence, co-functional, expression, etc.), network and functional enrichment statistics and detailed evidences of the interaction. Based on gene expression data (logFC), K-means clustering of the input genes was performed. This mode of clustering allows sub-structuring of data into groups that behave in similar manner. Differentially expressed genes were categorized into clusters sharing similar biological function (co-functional) and are under similar control logic (co-regulated). Furthermore, NetworkAnalyzer (https://apps.cytoscape.org/apps/analyzer) computes topological characteristic of the complete network. The topological analysis identifies the hubs (essential nodes) for the maintenance of network robustness and stability. The topological parameters of a network provide a useful insight to the biological process. (1) Degree distributions—node degree of a node n is the number of edges linked to n. A node with a high degree is referred to as hub. (2) Betweeness—fraction of the number of shortest path that pass through each node, which measure how often nodes occur on the shortest path between other nodes. The shortest past is calculated by measuring the length of all the edges from or the vertices in the network.
Gene enrichment and KEGG pathway analysis
FunRich, user friendly bioinformatics software for functional enrichment and interaction analysis was utilized for Gene set enrichment analysis of input query genes. It is a customized platform supporting proteomics, genomics, transcriptomics, lipidomics and metabolomics datasets. It provides interactive visualization of enrichment analysis in terms of biological processes, molecular function, protein domains, site of expression and cellular component. KAAS-KEGG (Kyoto Encyclopedia of Genes and Genomes) automatic annotation server ensures pathway analysis of input genes to explore the underlying systematic biological pathway connectivity to the expressed genes on treatment with a catechin, epigallocatechin-3-gallate (EGCG).
Statistical analysis
All the statistical data have been expressed as mean ± SD of three experiments. One-way ANOVA followed by two-tailed t test was performed (* represents p ≤ 0.05) during the analysis.
Results
EGCG modulates various signaling pathways and cellular regulators
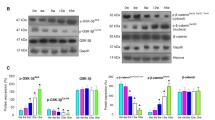
EGCG modulated the gene expression of crucial cell cycle regulators and genes involved in proliferation, migration, inflammation and signal transduction pathways as evidenced by qRT-PCR. EGCG downregulated the transcriptional expression of genes promoting the cell cycle namely CCNB1 (RQ = 0.51, p = 0.0261), CCNB2 (RQ = 0.45, p = 0.0365), CCND1 (RQ = 0.50, p = 0.046), CCND2 (RQ = 0.54, p = 0.036), and CDKN1A (RQ = 0.54, p = 0.0488). TERT (RQ = 0.41, p = 0.0501) expression which is important for telomere increase was found to be reduced alongwith the expression of VCAM (RQ = 0.54, p = 0.0367) (Fig. 1a). The expression of CDH1 (RQ = 9.76, p = 0.034) which restricts migration was found to be increased by EGCG, while simultaneously expression of pro-migration genes COL1A1 (RQ = 0.44, p = 0.05), MMP1 (RQ = 0.54, p = 0.0363), MMP10 (RQ = 0.54, p = 0.0363), MMP2 (RQ = 0.46, p = 0.05) and MMP7 (RQ = 0.54, p = 0.0363) was decreased. PIK3C2B (RQ = 0.56, p = 0.0402), PIK3CB (RQ = 0.53, p = 0.044) and PIK3CA (RQ = 0.53, p = 0.0382) which are critical genes in the PI3K pathway were found to be downregulated by EGCG possibly, initiating an inhibition of the PI3K pathway. Amongst the MAPK pathway genes, two key genes ELK1 (RQ = 0.54, p = 0.0344) and MAPK8 (RQ = 0.50, p = 0.047) were found to be downregulated by EGCG. An inhibitor of the pathway, PTPRR (RQ = 1.56, p = 0.05), was observed to have increased expression. EGCG lowered the expression of WNT2 (RQ = 0.50, p = 0.0204) gene, a key moiety of the Wnt pathway as well the expression of three pro-inflammatory genes, IL1A (RQ = 0.16, p = 0.0307), IL2 (RQ = 0.54, p = 0.0363) and IL6 (RQ = 0.38, p = 0.0467).
a Effect of EGCG on various genes involved in carcinogenesis RQ plot of tumour suppressor genes and genes involved in cell cycle regulation and cell-signaling pathways in HeLa cells after treatment with 50 µM of EGCG for 48 h is shown. Fold change was calculated by ΔΔCT analysis in comparison to untreated HeLa cells after global normalization. Values are means ± SD of three independent experiments (p ≤ 0.05). b Effect of EGCG on genes involved in chromatin modification RQ plot of genes involved in the chromatin modification whose expression in HeLa cells is modulated following treatment with 50 µM EGCG for 48 h. Fold change was calculated by ΔΔCT analysis in comparison to untreated HeLa cells after global normalization. Values are means ± SD of three independent experiments (p ≤ 0.05)
EGCG modulates the expression of various enzymes and chromatin modifiers involved in the epigenetic pathway
To assess the effect of EGCG on epigenetic enzymes and to assess their role in the anti-cancer activity of EGCG, qRT-PCR analysis of various epigenetic enzymes was performed. 50 μM EGCG treatment for 48 h decreased the expression of AURKC (RQ = 0.25, p = 0.0502), AURKB (RQ = 0.37, p = 0.0516), KDM5C (RQ = 0.41, p = 0.0543), AURKA (RQ = 0.42, p = 0.0541), DNMT3B (RQ = 0.48, p = 0.054), DNMT1 (RQ = 0.49, p = 0.0302), PRMT7 (RQ = 0.49, p = 0.052), DNMT3A (RQ = 0.50, p = 0.05), PRMT6 (RQ = 0.50, p = 0.0376), HDAC11 (RQ = 0.54, p = 0.054), HDAC5 (RQ = 0.54, p = 0.0510), UBE2A (RQ = 0.54, p = 0.0487), UBE2B (RQ = 0.56, p = 0.0331), HDAC7 (RQ = 0.58, p = 0.0542), KDM4A(RQ = 0.59, p = 0.05) and HDAC6 (RQ = 0.70, p = 0.0517). EGCG elevated the expression of SETD7 (RQ = 1.61, p = 0.0063). Taken together, it appears that these genes could alter the expression profile of the cells and may explain the anti-cancer effect of EGCG on HeLa cells. The fold changes in comparison to untreated control is shown in Fig. 1b.
EGCG decreases the activity of DNMT, HDAC and HMT H3K9 enzymes
Relative to the untreated control, incubation of HeLa cell nuclear extract with 50 μM EGCG for 1.5 h reduced the activity of DNMTs by 50.42% (p = 0.0221) and HMT H3K9 enzymes by 72.61% (p = 0.0023). Whereas, in comparison to the untreated control incubation of HeLa cell nuclear extract with 50 μM EGCG for 1 h reduced the activity of HDACs by 55.31% (p = 0.0029) (Fig. 2).
Molecular docking analysis of sulforaphane with epigenetic enzymes and biochemical inhibition of enzyme activity EGCG was docked with the crystal structures of various enzymes using the docking server, Swiss Dock and analyzed using UCSF Chimaera. The substrate binding site of the enzymes was identified using literature resources and cavity predicting software. The substrate-binding cavity is shown in hydrophobic representation in cyan colour with the catalytic residue coloured green. The predicted interaction of EGCG (blue) with the active site residues (cyan) in the substrate binding cavity of a DNMT1, b DNMT3A, c HDAC2, d HDAC3, e HDAC4, f HDAC7 g EZH2 is shown. The bound results were then compared to known crystal structures of inhibitors for validation. h Effect of EGCG on activity of DNMT, HDAC and HMT-H3K9 in HeLa cells. Nuclear extract of HeLa cells was treated with 50 µM EGCG to observe the direct inhibition of enzymatic activity of DNMTs, HDACs and HMT-H3K9s using an ELISA-based assay. EGCG demonstrated significant inhibition of the activity of all enzyme families. Values are represented in comparison to untreated control and are means ± SD of three independent experiments. (*p ≤ 0.05)
EGCG interacts with DNMT family and could function as a competitive inhibitor
To assess the molecular interaction of EGCG with DNMT enzymes and to explain the results of the biochemical assay detailed above, molecular docking studies were performed. Molecular docking results suggest that EGCG may competitively inhibit DNMT1 and DNMT3A enzymes by binding within the catalytic cavity. The docked pose of EGCG in the catalytic cavity of these proteins are shown in Fig. 2 and the residues within 5 Å of EGCG are listed in Table 1.
Interaction of EGCG with HDAC proteins indicate inhibition of activity
To assess the molecular interaction of EGCG with HDAC enzymes and to explain the results of the biochemical assay detailed above, molecular docking studies were performed. Molecular docking studies shows that HDAC2, HDAC3, HDAC4 and HDAC7 may be competitively inhibited by EGCG. The docked poses of EGCG in the catalytic cavity of these proteins are shown in Fig. 2 and the residues of these protein within 5 ֯A of EGCG are listed in Table 1.
Interaction of EGCG with EZH2 protein indicates inhibition of activity
To assess the molecular interaction of EGCG with HMT-H3K9 enzymes and to explain the results of the biochemical assay detailed above, molecular docking was performed. The predicted interaction of EGCG with EZH2 is shown in Fig. 2 and was found to be similar to the interaction of the known inhibitor. is the co-crystallized inhibitor of EZH2 as observed in the structure. This is suggestive of an inhibitory effect of EGCG interaction with EZH2 as well. the residues of these protein within 5 °A of EGCG are listed in Table 1.
EGCG decrease global DNA methylation in HeLa cells
Treatment of HeLa cells with 50 µM EGCG significantly decreased global DNA methylation in a time-dependent manner to 40% after 24 h (p = 0.0053) and to 21.16% after 48 h (p = 0.0145), in comparison to untreated control (Fig. 3).
Effect of EGCG on global DNA methylation in HeLa cells a The global methylation level of DNA in HeLa cells treated with 50 µM EGCG for 24 h and 48 h was assessed using an ELISA-based assay. DNA from untreated HeLa cells were used as control. EGCG significantly decreased the levels of global DNA methylation in HeLa cells in a time-dependent manner. The decrease is methylation level is represented as a percentage of the untreated control. Values are means ± SD of three independent experiments. (*p ≤ 0.05). b Effect of EGCG on gene specific DNA methylation in DNA extracted from HeLa cells treated with 50 µM EGCG for 48 h was subjected to methylation-dependent restriction digestion. The products of the digest were used as template for a PCR reaction using Human Tumor Suppressor Genes EpiTect Methyl II Signature PCR Array, to determine the 5´ CpG island promoter methylation level of tumour suppressor genes. EGCG significantly decreased the promoter methylation levels in HeLa cells in comparison to untreated control. Values are means ± SD of three independent experiments (*p ≤ 0.05)
EGCG treatment reduces the promoter methylation of tested TSGs
A functional outcome of reduction in the gene expression as well as enzyme activity of DNMTs can be expected as alteration in methylation levels of promoter 5′CpGs. To measure alterations in promoter methylation, the methyl II PCR array was carried out following differential digestion of DNA with methylation-sensitive and methylation-dependent restriction enzymes. EGCG reduced the promoter 5′CpG methylation of several TSGs namely APC (1.81%, p = 0.0006), CDH1 (57.16%, p = 0.036), CDH13 (7.06%, p = 0.007), DAPK1 (32.46%, p = 0.033), FHIT (15.62%, p = 0.034), GSTP1 (75.16%, p = 0.05), MGMT (3.21%, p = 0.0012), MLH1 (6.47%, p = 0.030), PTEN (8.68%, p = 0.0007), RARB (39.31%, p = 0.007), RASSF1 (16.68%, p = 0.05), SOC51 (49.41%, p = 0.004), TIMP3 (16.49%, p = 0.0006) and VHL (11.30%, p = 0.012) (Fig. 3).
EGCG restores the expression of tumour suppressor genes
To correlate changes in gene expression following alteration of promoter methylation, qRT-PCR of several TSGs was performed. It was observed that transcription levels of TSGs namely, TP53 (RQ = 1.62, p = 0.0322), PTEN (RQ = 2.04, p = 0.05), SOCS1 (RQ = 2.16, p = 0.05), CDH1 (RQ = 9.76, p = 0.034), RARβ (RQ = 1.532, p = 0.052) and DAPK1 (RQ = 1.521, p = 0.051) were upregulated following EGCG treatment (Fig. 1). This finding correlated with the decrease in promoter methylation levels.
Network analysis
Clustering of differentially expressed genes enables sub-structuring of data into groups of functionally similar input genes (Fig. 4a). STRING Cytoscape App enables visualization of clusters into different colours sharing either similar functions or regulated by similar control mechanism. Prior to partitioning of the data, scaling of gene expression data was performed based on mean expression (low drop counts) and logFC. Mean variance of the expression values computed optimal number of clusters as 7 for the data categorization. Evaluation of the identified clusters was performed by estimating an average silhouette width as 9. Silhouette value describes the accountability of gene relatedness to a particular cluster as compared to other clusters. If average silhouette width is high, then numbers of clusters is valid. Furthermore, topological parameters computed by NetworkAnalyzer are enlisted in Table 2.
a Protein–protein interaction network protein–protein interaction network of genes which are differentially modulated following EGCG treatment of HeLa cells is shown using k-means clustering. STRING Cytoscape App was used after scaling of gene expression data; evaluation of the clusters was performed by estimating an average silhouette width as nine. b KEGG pathway enrichment analysis KEGG pathway enrichment analysis of genes differentially modulated following EGCG treatment of HeLa cells predicts relationship between biological response variable and metabolic pathway based on linear model computing p-statistic score
Gene ontology and KEGG pathway analysis
Gene ontology-based analysis calculates p value and false discovery rate (FDR) of each GO term using a modified Fisher’s exact test and Benjamini–Hochberg (BH) method. Enriched terms with p value < 0.05 represent enrichment analysis in terms of (1) biological process (2) cellular component and (3) molecular functions. Automated organization functionality of the differentially expressed genes was explored by GO enrichment analysis (Table 2). KAAS-KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation server predicts relationship between biological response variable and metabolic pathway based on linear model computing p-statistic score (Fig. 4b).
Discussion
Carcinogenesis is the outcome of atypical alterations in signaling pathways driven by both genetic and epigenetic alterations. Epigenetic alterations are increasingly being acknowledged as an early and ongoing alteration that aids in cancer initiation and progression (Sundaram et al. 2020). The anti-carcinogenic property of EGCG has been established earlier in our laboratory and the EC50 in 24 h in HeLa cells for EGCG was found to be 100 μM (Sharma et al. 2012). In this study, further exploration into their modulation of various molecular targets was conducted using the sub-lethal dose of 50 μM EGCG which reduced cell viability to approximately 25% in 24 h and 50% in 48 h.
The observation of cell cycle dysregulation is supported by changes in the expression of the genes that govern it (Fig. 1). CCNB1 and CCNB2 are required for the cell to cross G2-M checkpoint (Huang et al. 2013). EGCG showed overall disturbances in the cell cycle and downregulated CCNB1, CCNB2 which monitor G2-M cell cycle checkpoint as well as CCND1 and CCND2. CDKN1A is downregulated facilitating the onset of apoptosis. VCAM helps in cell adhesion and also influences proliferation; HDAC inhibitors like TSA are known to suppress its expression (Hebbel et al. 2010). Its decrease should facilitate reduction in both cancer hallmarks. Replicative immortality is achieved by the overexpression of telomerase reverse transcriptase (TERT), which limits telomere length and enables continued proliferation and evasion of apoptosis (Wang et al. 2015b). EGCG is known to restrict migration as evidenced through the scratch wound assay (Sharma et al. 2012). To understand the molecular changes behind these observations, various genes involved in this hallmark were tested for transcriptional alterations (Fig. 1). Migration is initiated and aided by the activity of the matrix metalloproteases MMPs. Several MMPs were downregulated by EGCG namely, MMP1, MMP10, MMP2 and MMP7. Transcriptional elevation of CDH1 is mediated by EGCG; EGCG decreased the expression of COLIA1, which are further likely to suppress migration. The key properties acquired by cancer cells such as proliferation, invasion and migration, evasion of apoptosis is mediated by dysregulation of signaling network. Therefore, the PI3K pathway is considered therapeutically important in alleviating cancer with PI3KCA, PI3KCB, and the pathway inhibitor, PTEN being crucial entities. PIK3C2B, PIK3CA, PIK3CB are downregulated by EGCG with concurrent upregulation of the PI3K pathway inhibitor PTEN suggesting a therapeutic inhibition of this pathway. EGCG inhibited the proliferation and induced apoptosis in PANC-1 cells, by upregulating PTEN and downregulating mTOR and AKT (Liu et al. 2013). The MAPK pathway has been associated with promoting tumour proliferation. It is noteworthy that two key genes of this pathway ELK1, MAPK8 were found to be downregulated by EGCG. Furthermore, EGCG was found to restore the expression of PTPRR. In cervical cancer cells, PTPRR a MAP kinase pathway inhibitor, is often hypermethylated. EGCG has been documented to involve the ERK1/2 and MAPK signaling pathway to induce apoptosis in human colon cancer cells (Cerezo-Guisado et al. 2015). Progression of cervical carcinogenesis and metastasis is aided by the Wnt signaling pathway; wherein the WNT2 gene is usually overexpressed EGCG downregulated the expression of WNT2 gene (Fig. 1). A similar inhibition of growth via blocking of the Wnt pathway by EGCG was reported in lung cancer stem cells (Zhu et al. 2017). The influence of inflammatory molecules in the development and progression of cancer is well documented marking it as a possible therapeutic target. They are also responsible for the reduced efficacy of therapeutic agents. Three inflammatory genes, IL1A, IL2, IL6 are steeply downregulated by treatment with EGCG (Fig. 1).
DNA methyltransferases (DNMTs), histone deacetylases (HDACs), histone acetylases (HATs), histone methyltransferases (HMTs) and ubiquitinases are key enzymes that modulate the epigenetic responses and govern gene expression. Aberrant epigenetic processes affect all stages of carcinogenesis by silencing the expression of TSGs (Soto et al. 2017).
DNA methylation is strongly correlated to transcriptional silencing gene specific methylation is found to be increased and abnormal hypermethylation results in transcriptional silencing of tumour suppressor genes. All three DNMTs (DNMT1, DNMT3A and DNMT3B) are overexpressed in cervical cancer cells and its expression levels are correlated to disease progression (Piyathilake et al. 2017). EGCG downregulated the transcript levels of all three DNA methyltransferases DNMT1, DNMT3A and DNMT3B (Fig. 1b). Additionally, EGCG was able to exert an inhibitory effect and lowered DNMT activity by approximately 50% of untreated control (Fig. 2). Further, in silico docking results suggests that EGCG may inhibit DNMT1 and DNMT3A competitively by binding within the catalytic cavity of the enzymes (Fig. 3). DNMT1 and DNMT3A structures co-crystalized with their natural ligand, DNA was retrieved and was found to occupy the region defined as the catalytic cavity (Song et al. 2011; Zhang et al. 2018). The binding zone of EGCG on DNMT1 overlaps with the well-known co-crystallized inhibitor sinefungin (PDB ID 3SWR) (Valente et al. 2014). Dhe-Paganon et al. opine that DNMT1 activity could be impaired by drugs that target this region (Dhe-Paganon et al. 2011). This strengthens the suggestion that EGCG could function as a DNMT inhibitor.
Furthermore, the polycomb repressor protein, EZH2 recruits DNMT to target sites (Chen et al. 2016). It is interesting, therefore, that the current study predicts that EGCG may bind to EZH2 at the DNMT1 recruitment site and potentially inhibit it (Fig. 2). EGCG occupies the same cavity as the EZH2 inhibitors 74D (PDB ID: 5LS6), A9G (PDB ID: 5WG6) as well as the byproduct of methylation, S-adenosyl homocysteine (SAH) (Vaswani et al. 2016; Bratkowski et al. 2018). Furthermore, PI3K-AKT and Wnt pathways stabilize DNMT1 protein thereby, contributing to its activity (Feinberg 2015). EGCG may inhibit the activity of these pathway (Fig. 1) and further indirectly contribute to reduced DNMT activity. Decreased DNMT expression and activity promotes restoration of TSG expression and lowered cell proliferation and cell death.
Nucleosomal histone proteins undergo several chemical modifications including methylation, acetylation, ubiquitination, and phosphorylation. Histone modifications influences transcription of various genes (both activation and repression) involved in various cell regulatory pathways such as proliferation, migration and differentiation.
Amongst the histone modifications known to influence carcinogenesis, histone deacetylation is of central importance. Addition of lysine residues causes de-condensation of chromatin and promotes transcription, while removal of acetyl groups causes chromatin condensation and transcriptional repression. In addition to silencing of TSGs, HDACs also deacetylate non-histone proteins such as TP53, rendering them non-functional. Over-expression of HDAC1, HDAC2, HDAC3 and HDAC6 in cancer cells have been reported earlier (Movafagh and Munson 2019). EGCG downregulated HDAC5, HDAC6, HDAC7 and HDAC11 (Fig. 1). Additionally, EGCG was also found to lower the biochemical activity of HDAC (Fig. 2). Furthermore, in silico docking studies showed that EGCG may also competitively inhibit HDAC2, HDAC3, HDAC4 and HDAC7. The docking pose of EGCG on HDAC2 was found to overlap that of the co-crystallized inhibitors, IWX (PDB ID:5IWG), SAHA (PDB ID: 4LXZ) LLX, (PDB ID: 3MAX) and 2OY (PDB ID: 4LY1). Similarly, binding of EGCG to HDAC4 was found to overlap well with the co-crystallized inhibitors EBE (PDB ID: 6FYZ), TFG (PDB ID: 2VQJ), 9F4 (PDB ID: 4CBT) and HA3 (PDB ID: 2VQM). Suppression of HDACs reverses TSG silencing, lessens growth and induces apoptosis (Sundaram et al. 2019a).
Histone methyltransferases (HMTs) catalyze the addition of up to three methyl groups onto specific lysine moieties in histone proteins. Depending on the lysine residue involved, these marks may be transcriptionally permissive or repressive. H3K4me3 marks the trimethylation of lysine 4 on histone H3 at promoter regions and is associated with transcriptional activation. Promoter region trimethylation of lysine 9 and 27 of histone 3 (H3K9 and H3K27) is found to be high in cervical cancer and are associated with repressive signaling of TSG expression. EGCG significantly reduced H3K9 methyltransferase activity (Fig. 3), which may aid in the re-expression of silenced TSGs. Expression of SETD7 was upregulated following EGCG treatment (Fig. 1). In cervical cancer, Human Papilloma Virus (HPV) downregulates the expression of the TSG, SETD7 which can activate TP53 (Hsu et al. 2012). Further, SETD7 inactivates DNMT1; this contributes to reduced protein levels of DNMT1 and consequently lowered DNA methylation levels (Wang et al. 2008). Protein arginine methyltransferases (PRMTs) methylate the arginine residue of histone and some non-histone proteins impacting several cellular functions. PRMTs are known to be upregulated in cancer and silence TSGs, making them an important therapeutic target. PRMT7 expression is downregulated by EGCG (Fig. 1). PRMT7 is involved in regulating cellular resistance to chemotherapeutics, supports the Wnt pathway and in breast cancer, augments metastasis (Baldwin et al. 2015).
EGCG decreases the expression of KDM4A and KDM5C which may limit oncogenic activation (Fig. 1). Overexpression of the oncogene, H3K9/36me3 lysine demethylase KDM4A causes copy number gains and promotes chromosomal instability (Luo et al. 2016). KDM5C is oncogenic, is overexpressed in cervical cancer, with its expression increasing with disease severity (Ianca et al. 2015). Transcript of AURKA, AURKB and AURKC were significantly reduced following EGCG treatment (Fig. 1).The expression of AURKA, AURKB and AURKC in cervical cancer is very high and aids in tumour progression (Burk et al. 2017). They contribute to cell proliferation, migration, ability to cross checkpoint at G2-M, and alongside HDAC regulate the AKT pathway (Burk et al. 2017).
Histone ubiquitination involves the addition of ubiquitin to histone proteins and regulates multiple cell functions and aids cancer development. The expression of the ubiquitinases, UBE2A and UBE2B was decreased by EGCG. Malignant transformation as well instability of chromosomes in cervical cancer may be attributed to increased expression of UBE2A (Wang et al. 2015a). UBE2B, targets the Wnt pathway as well as the tumour suppressor gene, TP53.
DNA methylation is affected by DNMT activity and expression; with reduced DNMT activity commensurate with lowered DNA methylation. EGCG reduced both DNMT expression and activity (Figs. 1, 2). EGCG mediated time-dependent decrease in global DNA methylation levels (Fig. 3). EGCG lowers global methylation in skin cancer cells (Nandakumar et al. 2011). A significant therapeutic milestone in the management of cancer is the reversal of gene specific 5′ promoter CpG methylation. Key tumour suppressor genes commonly methylated in cervical cancer were chosen and the quantum of their promoter methylation was compared before and after treatment with EGCG. EGCG was able to alter the methylation status of the promoter region of tumour suppressor genes namely APC, CDH1, CDH13, DAPK1, FHIT, GSTP1, MGMT, MLH1, PTEN, RARB, RASSF1, SOC51, TIMP3 and VHL (Fig. 3). In cervical cancer patient samples, high levels of promoter methylation correlating with disease severity has been reported in several TSGs including DAPK, PTEN, RARβ, RASSF1A, CDH13, MLH1, SOCS1, MGMT, VHL and FHIT (Bhat et al. 2016). Reduced methylation and re-expression of FHIT, DAPK, MGMT, APC, CDH1, PTEN in cervical cancer cells has been achieved resulting in cancer regression by using demethylating and deacetylating agents. The role of the TSGs whose methylation levels were lowered by EGCG are detailed below. Taken together their re-expression can contribute greatly to the overall anti-carcinogenic outcome following EGCG treatment. APC encodes a tumor suppressor protein that acts as an antagonist of the Wnt signaling pathway and takes part in processes including cell migration and adhesion, transcriptional activation and apoptosis (Ayala‑Calvillo et al. 2017). It is methylated in cervical cancer with its status linked to delocalization of β-catenin in HPV16 infected cervical cancer cells and higher methylation index with disease (Ayala‑Calvillo et al. 2017).
CDH1 (e-cadherin) is instrumental in preventing migration and metastasis and its methylation in cervical cancer also increases with disease progression (Li et al. 2017). In further support of the observations of this study, it has been reported that reduced expression of EZH2 lowers H3K27me3 levels in the promoter region of CDH1 gene and contributes to restoration of its expression (Lin et al. 2013). Another regulator of cell migration, CDH13 (H cadherin) is also frequently hypermethylated in cervical cancer (Henken et al. 2007). Reduction in methylation of CDH13 by EGCG was also evidenced in this study.
Promoter hypermethylation of DAPK1, a pro-apoptotic, serine/threonine protein kinase is correlated with disease grade (Banzai et al. 2014). FHIT is hypermethylated in cervical cancer samples and cell lines (Banzai et al. 2014). Likewise, GSTP1 is involved in xenobiotic metabolism and methylated in certain cancers (Zambrano et al. 2005). MGMT is a DNA repair enzyme that is commonly hypermethylated in cervical cancer and shows linear increase with disease severity (Banzai et al. 2014). Phosphatase and tensin-homologue (PTEN) is commonly methylated and silenced in cervical cancer patients which restricts its ability to inhibit the PI3K pathway, thereby, causing increased cell division and survival (Song et al. 2012). RARβ is involved in cell proliferation and is usually methylated in several cancers including cervical cancer which is positively correlated with disease severity (Sun et al. 2015). RASSF1A functions as cell cycle and apoptosis regulator and is silenced by promoter methylation in cervical cancer (Donninger et al. 2007). Suppressor of cytokine signaling-1 (SOCS1), another frequently silenced TSG is associated with cytokine suppression, degradation of HPV E7, increase in expression of the TSG, Rb and reduces metastatic potential (Henken et al. 2007). MLH1 regulates several processes including DNA repair and controls cell cycle through the action of both p21 and p73. Its methylation in cervical cancer correlates with higher evidence of metastasis and cancer recurrence (Spathis et al. 2011). Thus, in this study it was observed that EGCG is able to reduce the promoter methylation of several important TSGs which are normally hypermethylated in cervical cancer cells.
It has been shown that DNA methylation determines transcriptional status of genes and inhibition of this process by any agent achieves restoration of transcription (Kedhari Sundaram et al. 2019a; Sundaram et al. 2019b). To correlate the changes in promoter methylation with alterations at the transcript level, qRT-PCR was performed, and fold changes were calculated (Fig. 1). 50 μM EGCG treatment restored the expression of PTEN, SOCS1, RARβ, DAPK1 and CDH1. This shows the correlation between lowered promoter methylation and enhanced transcription levels. A few TSGs which exhibited decreased methylation levels did not demonstrate a commensurate increase in expression, this may be attributed to other routes of epigenetic silencing which are unaffected by EGCG and hence not according transcriptional permissiveness.
In silico network analysis of differentially expressed genes was performed to identify functionally similar genes. Partitioning of genes was also performed by performing k-means clustering based on similar functional annotation and regulation mechanism. The complete list of input genes was sub-structured into seven clusters (depicted as different colours viz. red, green, blue, yellow, light green, cyan, and indigo). This clustering enables delineation of downstream biological processes associated with expressed genes after treatment with EGCG. The network generated was subjected to topological analysis deciphering nodes (genes) essential for maintaining its robustness. Computation of degree of each node identifies histone deacetylases (HDAC2, HDAC3, HDAC4 and HDAC7), DNMT enzymes (DNMT1 and DNMT3A) as hubs (essential genes). HDACs participation in several biological processes and transcriptional machinery proves its specificity towards controlling global or specific programs of gene expression and disease states. Furthermore, GO ontology analysis identifies active participation of highly expressed genes towards major mechanism of transcription, replication, and regulation. Furthermore, enrichment analysis based on (1) cellular component identifies major presence in plasma membrane, cytosol, nucleoplasm and cytoplasm. (2) Enrichment analysis based on molecular functions prioritized cytokine activity, methyltransferase activity, lipid kinase, protein serine/threonine kinase activity, metallopeptidase activity, etc. (iii) Biological processes enrichment analysis predicts participation in protein metabolism, immune response, regulation of cell cycle; signal transduction, chromatin modification, etc. Furthermore, KEGG pathway analysis depicts the active participation of highly expressed genes in viral carcinogenesis, prostate cancer, chronic myeloid leukemia, thyroid hormone signaling pathway, transcriptional regulation in cancer, colorectal cancer, pathways in cancer, thyroid cancer. On treatment with EGCG, highly expressed genes affects underlying characterised KEGG cancer pathways (Fig. 4).
Conclusion
This study shows the transcriptional alteration of several key genes involved in different cell-signaling pathways and genes that govern proliferation, apoptosis and migration in cervical cancer cells, HeLa following treatment with the phytochemical, EGCG. Reversal of TSG silencing and alteration in their methylation status has been mediated by EGCG through modulation of the expression and activity of epigenetic enzymes, such as DNMTs, HDACs and HMTs. This study comprehensively catalogues the EGCG-induced altered transcriptional status of numerous genes including epigenetic genes that have not been reported previously and attempts to delineate the anti-cancer mechanism of action of EGCG on HeLa cells (Fig. 5 and Table 3). On the basis of the results of this study, EGCG may be considered a potential epigenetic-based anti-cancer agent that warrants further mechanistic studies.
References
Ayala-Calvillo E, Mojica-Vazzquez L, Garcia-Carranca A, Gonzalez-Maya L (2017) Wnt/β-catenin pathway activation and silencing of the APC gene in HPV-positive human cervical cancer-derived cells. Mol Med Rep. https://doi.org/10.3892/mmr.2017.7853
Baldwin RM, Haghandish N, Daneshmand M et al (2015) Protein arginine methyltransferase 7 promotes breast cancer cell invasion through the induction of MMP9 expression. Oncotarget 6:3013–3032. https://doi.org/10.18632/oncotarget.3072
Banzai C, Nishino K, Quan J et al (2014) Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. Int J Clin Oncol 19:127–132
Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K (2016) Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie 121:298–311. https://doi.org/10.1016/j.biochi.2015.12.018
Bratkowski M, Yang X, Liu X (2018) An evolutionarily conserved structural platform for PRC2 inhibition by a class of Ezh2 inhibitors. Sci Rep 8:9092
Burk RD, Chen Z, Saller C et al (2017) Integrated genomic and molecular characterization of cervical cancer. Nature. https://doi.org/10.1038/nature21386
Cerezo-Guisado MI, Zur R, Lorenzo MJ et al (2015) Implication of Akt, ERK1/2 and alternative p38MAPK signalling pathways in human colon cancer cell apoptosis induced by green tea EGCG. Food Chem Toxicol 84:125–132. https://doi.org/10.1016/j.fct.2015.08.017
Chen S, Ling Q, Yu K et al (2016) Dual oxidase 1: A predictive tool for the prognosis of hepatocellular carcinoma patients. Oncol Rep 35:3198–3208. https://doi.org/10.3892/or.2016.4745
Dhe-Paganon S, Syeda F, Park L (2011) DNA methyl transferase 1: regulatory mechanisms and im-plications in health and disease. Int J Biochem 2:58–66
Donninger H, Vos MD, Clark GJ (2007) The RASSF1A tumor suppressor. J Cell Sci 120:3163–3172
De FM, Cardoso S, Henrique C, Castelletti M (2017) Putative biomarkers for cervical cancer: SNVs, methylation and expression pro fi les. Mutat Res Mutat Res 773:161–173. https://doi.org/10.1016/j.mrrev.2017.06.002
Feinberg AP (2015) Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13:497–510. https://doi.org/10.1038/nrc3486.Cancer
Hebbel RP, Vercellotti GM, Pace BS et al (2010) The HDAC inhibitors trichostatin A and suberoylanilide hydroxamic acid exhibit multiple modalities of benefit for the vascular pathobiology of sickle transgenic mice. Blood 115:2483–2490. https://doi.org/10.1182/blood-2009-02-204990
Henken FE, Wilting SM, Overmeer RM et al (2007) Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer 97:1457–1464. https://doi.org/10.1038/sj.bjc.6604055
Hsu CH, Peng KL, Jhang HC et al (2012) The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene 31:2335–2349. https://doi.org/10.1038/onc.2011.415
Huang Y, Sramkoski RM, Jacobberger JW (2013) The kinetics of G2 and M transitions regulated by B cyclins. PLoS ONE 8:30–35. https://doi.org/10.1371/journal.pone.0080861
Ianca I, Botezatu A, Plesa A, Huica Irina Socolov D, Anton G (2015) Histone lysine demethylases as epigenetic modifiers in HPV-induced cervical neoplasia. Romanian Biotechnol Lett 20:10236–10244
Kedhari Sundaram M, Hussain A, Haque S et al (2019a) Quercetin modifies 5′CpG promoter methylation and reactivates various tumor suppressor genes by modulating epigenetic marks in human cervical cancer cells. J Cell Biochem. https://doi.org/10.1002/jcb.29147
Kedhari Sundaram M, Raina R, Afroze N et al (2019b) Quercetin modulates signaling pathways and induces apoptosis in cervical cancer cells. Biosci Rep 39:BSR20190720. https://doi.org/10.1042/bsr20190720
Li X, Tian R, Gao H et al (2017) Identification of a histone family gene signature for predicting the prognosis of cervical cancer patients. Sci Rep 7:16495. https://doi.org/10.1038/s41598-017-16472-5
Lin WC, De YM, Yu PN et al (2013) The role of Sp1 and EZH2 in the regulation of LMX1A in cervical cancer cells. Biochim Biophys Acta Mol Cell Res 1833:3206–3217. https://doi.org/10.1016/j.bbamcr.2013.08.020
Liu S, Wang X-J, Liu Y, Cui Y-F (2013) PI3K/AKT/mTOR signaling is involved in (−)-epigallocatechin-3-gallate-induced apoptosis of human pancreatic carcinoma cells. Am J Chin Med 41:629–642
Luo H, Shenoy AK, Li X et al (2016) MOF acetylates the histone demethylase LSD1 to suppress epithelial-to-mesenchymal transition. Cell Rep 15:2665–2678. https://doi.org/10.1016/j.celrep.2016.05.050
Movafagh S, Munson A (2019) Histone deacetylase inhibitors in cancer prevention and therapy. Elsevier Inc., Amsterdam
Nandakumar V, Vaid M, Katiyar SK (2011) (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p 16 INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 32:537–544
Piyathilake C, Badiga S, Borak S et al (2017) A higher degree of expression of DNA methyl transferase 1 in cervical cancer is associated with poor survival outcome. Int J Womens Health 9:413–420. https://doi.org/10.2147/IJWH.S133441
Sharma C, Nusri QelBegum AS et al (2012) (−)-Epigallocatechin-3-gallate induces apoptosis and inhibits invasion and migration of human cervical cancer cells. Asian Pac J Cancer Prev 13:4815–4822. https://doi.org/10.7314/APJCP.2012.13.9.4815
Song J, Rechkoblit O, Bestor TH, Patel DJ (2011) Structure of DNMT1–DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331:1036–1040. https://doi.org/10.1126/science.1195380
Song MS, Salmena L, Pandolfi PP (2012) The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 13:283
Soto D, Song C, McLaughlin-Drubin ME (2017) Epigenetic alterations in human papillomavirus-associated cancers. Viruses 9:248. https://doi.org/10.3390/v9090248
Spathis A, Aga E, Alepaki M et al (2011) Promoter methylation of p16INK4A, hMLH1, and MGMT in liquid-based cervical cytology samples compared with clinicopathological findings and HPV presence. Infect Dis Obstet Gynecol 2011. https://doi.org/10.1155/2011/927861
Sun Y, Li S, Shen K et al (2015) DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. Int J Clin Exp Pathol 8:14939–14945
Sundaram MK, Ansari MZ, Al MA et al (2017) Genistein induces alterations of epigenetic modulatory signatures in human cervical cancer cells. Anticancer Agents Med Chem 17:1–11. https://doi.org/10.2174/1871520617666170918142114
Sundaram MK, Raina R, Afroze N et al (2019a) Combinational use of phytochemicals and chemotherapeutic drugs enhance their therapeutic potential on human cervical cancer cells. Int J Cancer Manag. https://doi.org/10.5812/ijcm.91783
Sundaram MK, Unni S, Somvanshi P, Bhardwaj T (2019b) Genistein modulates signaling pathways and targets several epigenetic markers in HeLa cells. Genes (Basel) 10:1–20. https://doi.org/10.3390/genes10120955
Sundaram MK, Silas S, Hussain A (2020) Epigenetic aberrations in cervical cancer. In: Epigenetics and Reproductive Health. Elsevier, pp 343–370
Valente S, Liu Y, Schnekenburger M et al (2014) Selective non-nucleoside inhibitors of human DNA methyltransferases active in cancer including in cancer stem cells. J Med Chem 57:701–713
Vaswani RG, Gehling VS, Dakin LA et al (2016) Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem 59:9928–9941. https://doi.org/10.2210/PDB5LS6/PDB
Wang C, Chang J-F, Yan H et al (2015) A conserved RAD6-MDM2 ubiquitin ligase machinery targets histone chaperone ASF1A in tumorigenesis. Oncotarget. https://doi.org/10.18632/oncotarget.5011
Wang HY, Park S, Kim S et al (2015) Use of hTERT and HPV E6/E7 mRNA RT-qPCR TaqMan assays in combination for diagnosing high-grade cervical lesions and malignant tumors. Am J Clin Pathol 143:344–351. https://doi.org/10.1309/AJCPF2XGZ2XIQYQX
Wang J, Hevi S, Kurash JK et al (2008) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41:125
Zambrano P, Segura-Pacheco B, Perez-Cardenas E et al (2005) A phase I study of hydralazine to demythylate and reactivate the expression of tumor suppressor genes. BMC Cancer 5:1–12. https://doi.org/10.1186/1471-2407-5-44
Zhang Z-M, Lu R, Wang P et al (2018) Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554:387
Zhu J, Jiang Y, Yang X et al (2017) Wnt/β-catenin pathway mediates (−)-epigallocatechin-3-gallate (EGCG) inhibition of lung cancer stem cells. Biochem Biophys Res Commun 482:15–21
Acknowledgements
The authors are grateful to Dr. Kota Reddy, Academic President, Manipal Academy of Higher Education, Dubai, UAE for his constant support and encouragement. The author Dr. Shafiul Haque is grateful to Jazan University, Saudi Arabia, for providing the access of the Saudi Digital Library for this study.
Funding
No funds were received from any agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kedhari Sundaram, M., Haque, S., Somvanshi, P. et al. Epigallocatechin gallate inhibits HeLa cells by modulation of epigenetics and signaling pathways. 3 Biotech 10, 484 (2020). https://doi.org/10.1007/s13205-020-02473-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02473-1