Abstract
The present study was conducted to determine efficiency of green tissue-specific (pRCA) and stress-inducible promoters (pRD29A) to express E. coli beta-glucuronidase (gusA) gene in transgenic potatoes compared with constitutive promoter (35S CaMV). The promoter fragments were isolated from their original source and cloned upstream to gusA in pCAMBIA-1301 binary vector to develop plant expression constructs, i.e., pRCA-pCAMBIA and pRD29A-pCAMBIA. Agrobacterium strain GV2260 harboring recombinant plasmids were used to infect leaf discs and internodal explant of Lady Olympia cultivar. GUS histochemical analysis was performed at different stages to determine GUS activity in transgenic plants. To determine activity of stress-inducible promoter (pRD29A), transgenic plants were exposed to heat, drought and combination of both heat and drought stress. The real time (RT-qPCR) and GUS florimetric assays revealed that pRD29A promoter gets more activated under drought, heat and combination of both stresses. GUS expression levels were more than 10 folds high with pRD29A promoter compared to control. Likewise, the reduced transcripts levels of gusA gene under control of pRCA promoter were found in tuber/roots of transgenic plants compared to 35S promoter. GUS florimetric assays also showed decreased or no GUS expression in tubers. In conclusion, the results encourage the appropriate use of promoters to drive the expression of foreign gene(s) for the development of potato lines tolerant to biotic and abiotic stress while minimizing the risks of transgenic technology in potatoes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is one of the most important cultivated plants in the world in terms of its uses as both industrial raw material and direct food. Potato has the potential to solve the insufficient problems of food resources for the increasing population because of its richness in nutrient composition (Alisdair et al. 2001). Besides that, eight species of Solanum genus are suitable for consumption as human food; however, the most widely known and produced species is Solanum tuberosum (Rowe 1993).
The commercialized transgenic crops express insect or herbicide resistant gene(s) under the control of 35S cauliflower mosaic virus that is constitutively expressed in all cell types and at all developmental stages of a plant (Sunilkumar et al. 2002; Amarasinghe et al. 2006; Ahmed et al. 2017). In fact, it is one of the most preferred promoters in plant biotechnology, since its discovery (Somssich 2019). However, the literature suggests that constitutive expression of a foreign gene in plants can also result in increased metabolic burden in plants, posing potential threats of resistance development against Bt toxins or herbicides (Anayol et al. 2016; Tabashnik et al. 2017; Hussain et al. 2019). The concerns about food safety of genetically modified plants have also been raised (Conner et al. 2003). Therefore, confining expression of a foreign gene (s) in specific plant tissues or activation under a particular stress can be considered significant for the development of value added crops.
The modern technologies have opened new vistas in isolation of promoters tailored to answer specific questions in research or create new transgenic crops and products. The availability of broad spectrum promoters with the ability to regulate the spatio-temporal patterns of transgenes can increase the successful application of transgenic technology (reviewed in Potenza et al. 2004). Besides that, field of bioinformatics had led to computational analysis of primary structure and functional of a single promoter, defining enhancer function and their relationship with other motif in the genome (Reviewed in Bakhsh et al. 2011).
The targeted and confined expression of transgene(s) in specific or non-edible parts of the crop is significant and can lead to more public acceptance being less worrisome in nature (Ahmed et al. 2017). The different types of the promoters have already been isolated and characterized in this regard. Inducible promoters allow gene to be expressed when the plant is exposed to any biotic or abiotic stress; alcohol, steroid chemicals or physical factors such as light and temperature (Zhu et al. 2010).
Numerous tissue-specific promoters have been identified in plants including those involved in photosynthesis process or from seed storage genes (Mithra et al. 2017). The use of green tissue-specific promoter is ideal in crop like potato, where tubers are the edible parts. pRCA is the organ-specific and light-regulated promoter of gene encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase (RCA) in potatoes (Qu et al. 2011). Inducible promoters are responsible for the expression of associate gene (s) in response to physical, chemical and environmental cues. pRD29A promoter is cis-acting element of Arabidopsis gene (RD29A) involved in responsiveness to drought, low-temperature, or high-salt stress as established earlier by Msanne et al. (2011) in Arabidopsis.
The present study was conducted to compare the efficiency of three promoters in transgenic potatoes. The promoter fragments were amplified from their sources and fused with gusA in pCAMBIA1301 and further transferred to potato. The results showed efficient GUS expression under pRCA and pRD29A promoter establishing their suitability to develop transgenic potatoes with trait of economic importance.
Materials and methods
Development of expression vectors
The stress-inducible (pRD29A, accession no. AY973635.1) and green tissue-specific (pRCA, accession no. HQ259068.1) promoter fragments were amplified from Arabidopsis thaliana and Solanum tuberosum cv. Lady Olympia genomic DNA sequences, respectively, using specific primers with overhangs of KpnI and NcoI restriction enzymes. All modifications were made in binary vector pCAMBIA-1301 already available in our plant transformation laboratory. The both promoter fragments were cloned in pre-digested pCAMBIA with KpnI and NcoI restriction enzymes upstream to gusA gene (interrupted by an intronic sequence to deduce expression from eukaryotic cells). The empty vector with 35S promoter upstream to gusA gene was used as mock in all experiments to compare the efficiency of promoters. The developed constructs were maintained in JM109 at first, and were further electroporated to Agrobacterium strain GV2260 using Gene Pulser Xcell™ Electroporation Systems (Cat. No. 1652660). All DNA manipulations were performed according to the standard protocols (Sambrook and Russell 2001).
Agrobacterium-mediated potato transformation
Lady Olympia potato cultivar was used as plant material in the present study. To propagate experimental explants, tubers sprouts were subjected to surface sterilization and were multiplied by monthly subculture of single-node stem explant on basal MS medium (Murashige and Skoog 1962). The potato plantlets were then maintained in a growth room with a 16/8 h light/dark photoperiod and temperature 25 ± 2 °C. The leaf and internodal explants were excised from in vitro cultured plantlets and subjected to Agrobacterium-mediated transformation following protocol as described by Beaujean et al. (1998) with some modification adopted by Bakhsh (2020). The regeneration media consisted of BA 2 mg/L, NAA 0.2 mg/l, Trans zeatin 2 mg/l and GA3 0.1 mg/l). During all regeneration experiments, an optimized concentration (5 mg/l) of hygromycin was used to screen transformed cells. The regenerating shoots with a length of 2–3 cm were excised and transferred to magenta boxes. The data of hygromycin resistant calli and average number of shoots were calculated after 6–7 weeks after the transformation process. To determine the transformation efficiency, all plantlets were used in PCR-based screening of construct integration into plant genome. The MS salts, sucrose, plant agar and plant growth regulators used in present study were purchased from Duchefa Biochemie. Duocid (Pfizer, Istanbul, Turkey) with ingredients of ampicillin + sulbactam (1 g + 500 mg) were also added to regeneration selection medium at concentration of 300 mg/L to eliminate excessive growth of Agrobacterium.
Evaluation of transgenic plants
The primary transformants were screened for the integration and expression of gusA gene under control of different promoters. The genomic DNA was extracted using Thermoscientific GeneJET Plant Genomic DNA Purification Kit (Cat No. K0792). PCR assays were conducted using gene specific primers to amplify introduced gene. All PCR reactions were performed in a total reaction mixture volume of 20 μl containing 1 × reaction buffer, 50 ng of DNA template, 1.5 mM MgCl2, 1 mM of each of the dNTPs, 0.5 μM of each primer and one unit of Taq DNA polymerase. The primers sequences, annealing temperature and product size have been provided in Supplementary Table 1. The plasmid DNA was used as positive control, whereas DNA isolated from untransformed plants was used as negative control. Besides that, the transformants were also subjected to PCR assay with chvA gene to assess whether the primary transformants are contaminated with Agrobacterium. Following PCR assays, the amplified DNA fragments were electrophoresed on 1.0% agarose gel and visualized by ethidium bromide staining under ultraviolet (UV) light.
Analysis of promoter’s efficiency
Two sets of experiments were performed to assess the efficiency of promoters in driving expression of gusA gene. For stress-inducible promoter, PCR positive in vitro cultured plantlets expressing gusA gene under the control of pRD29A promoter were subjected to heat and drought stress in separate as well as combined stress. The control plants (transformed with mock vector containing 35S promoter to drive gusA expression) was also used as comparison. The experiments were conducted in three biological repeats under four different growth conditions (Supplementary Table 2). The temperature of growth chamber was adjusted to 35 °C day and night for 2 days for heat stress. Polyethylene Glycol (PEG) 6000 (Product No: P0805.5000, Duchefa Biochemie) was used to induce drought stress. In vessels (GA-7), 50 ml MS0 medium was supplemented with 20% PEG [(optimized concentration in our earlier experiments modified from Pino et al. (2013) and Gopal and Iwama (2007)]. As we were interested to assess the efficiency of pRCA promoter in tubers of transgenic plants; therefore, the other experiment was conducted in green house. The transformants expressing gusA gene under the control of pRCA were shifted to 75% peat and 25% perlite along with control plants (transformed with mock vector containing 35S promoter). The samples were collected from leaf and root/ tubers.
GUS histochemical assays were performed to test the functionality of gusA gene under the control of 35S, pRD29A and pRCA promoters. The assay was conducted as described by Jefferson et al. (1987). To accomplish the task, GUS buffer containing 10 mg/L X-Gluc, 10 mM EDTA, 100 mM NaH2PO4, 0.1% Triton X-100 and 50% methanol (Sigma-Aldrich Chemicals Co., Merck) was prepared. The pH of the buffer was maintained at 8.0. The putative transgenic plants in both experiments were incubated in GUS buffer for 4–6 h. Furthermore, samples were destained by adding 70% ethanol to 2 mL Eppendorf tubes. Later on, image was taken under a computer-dependent (Leica M165C) microscope.
Real time PCR assays were conducted to investigate the transcripts levels of gusA gene under the control of different promoters in both experiments. For total RNA extraction, AMRESCO RiboZol™ RNA Extraction Reagent was used. cDNA was synthesized using Fermentas cDNA synthesis kit. RT-qPCR content included total Mix (2 ×) (Qiagen), F Primer (0.2 μM), R Primer (0.2 μM), RNase-free water. RT-qPCR temperature cycle was set up as 95 °C for 15 min, 40 cycles of 95 °C for 10 s, 58 °C for 15 s, 72 °C for 20 s and melting curve analysis was performed by incubation at 99 °C to 70 °C with a transition rate of 1.0 °C/min. For normalization, elongation factor 1-α (ef1α) was selected as reference gene for the purpose of quantifying the expression of genes as earlier used by Nicot et al. (2005). Each reaction was set up with three replicates. The threshold values of samples in target gene expression analysis were analyzed by software of Rotor-Gene Q (Qiagen) RT-PCR instrument. According to the results, the standard deviations of Ct values of the samples were calculated using Microsoft Excel program and the expression level of the genes was determined according to the 2-ΔΔCt proportional calculation method (Livak and Schmittgen 2001).
The fluorogenic assays were conducted to estimate the expression of GUS protein in both experiments to understand the efficiency of promoters under study as described by Bottino (2018). In both experiments, the transgenic plants were subjected to total protein isolation. Both leaves and root/tubers were collected from transgenic plants for the purpose of total protein isolation to determine efficiency of pRCA promoter along with the control. According to the growing conditions from transgenic plants, samples were collected in liquid nitrogen as leaf or root-tuber. Assay buffer was prepared by adding 1 mM MUG in the extraction buffer (17.6 mg of 4-Methylumbelliferyl-β-D-glucuronide (MUG) was added in 50 ml of extraction buffer). Taking 50 µl of the extracted proteins, 0.5 ml of assay buffer was added on it and incubated at 37 °C (3 replicates were prepared from each transgenic plant). The reaction was stopped by adding 0.2 M Na2CO3 stop buffer incubated at + 37 °C for 1 hour, 3 hours, and overnight. The image was taken by examining the results of the stopped reaction under UV lights.
Statistical analysis
The data of GUS protein activity and gene expression analysis results was analyzed using Statistix 8.1 program Statistix 8.1 (Analytical Software 2005). Significance of variance was determined after the one-way ANOVA (p < 0.05) followed by analysis at 5% LSD (least significant difference) method as multiple comparisons (post-hoc).
Results
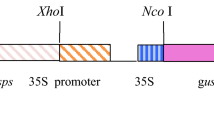
The cloning of pRD29A and pRCA fragment DNA sequences upstream to gusA gene in pCAMBIA-1301 was achieved successfully to compare their efficiency with constitutive promoter (35S). Following confirmation by standard molecular analyses, the recombinant plasmids were names as pRD29A-pCAMBIA, pRCA-Pcambia, as shown in Fig. 1.
Schematic images of different expression vectors developed by cloning promoters (pRCA and rd29A) in pCAMBIA-1301. a Original pCAMBIA1301 vector used as mock, b pRCA promoter was cloned upstream to gusA gene and named as pRCA-pCAMBIA c pRD29A promoter was cloned upstream to gusA gene and was named as pRD29A-Pcambia, respectively. The vector contains Hygromycin phosphotransferase (hptII) to encode resistance against Hygromycin that was used a plant selectable marker, whereas it contained Kanamycin for bacterial selection
Using optimized Agrobacterium-mediated transformation protocol as described by Bakhsh (2020), leaf and internodes of Lady Olympia were used as explants for genetic transformation. As shown in Fig. 2, callus induction from both explants varied with different constructs. When we compared leaf and internode explants, induction of calli (60–78%) was higher in internodal explants compared to leaf (40–55%) although it varied between different constructs. Likewise, the average number of shoot in internodal explants (5.8–6.9) was also higher compared to leaf explants (3.8–6.18), whereas leaf explants transformed with pRD29A-pCAMBIA construct showed encouraging response compared to other constructs. The regenerated shoots of certain size were transferred in larger magenta boxes and were allowed rooting. All regenerated plants showed 100% rooting on a medium supplemented with optimized concentration of hygromycin.
Summary of transformation experiments in Lady Olympia. Callus formation (%) was calculated out of total leaf and internodal explants used in study. The shoot formation shows average number of shoots per explants (calli) in all experiments. The regenerated plants showed 100% rooting in media supplemented with hygromycin. pRCA, pRD29A and 35S here show the construct as pRCA-pCAMBIA, pRD29A-pCAMBIA and 35S-pCAMBIA, respectively
PCR reactions showed the proper integration of T-DNA within host genomes using different primers (data shown only with gusA gene). PCR positive plants were selected for further studies (Fig. 3). No any amplification was observed in non-transgenic control plants. The results of chv gene showed absence of any Agrobacterium contamination. Only PCR positive plants were subjected to stress incubation assays to evaluate the efficiency of tissue-specific (pRCA) and stress-inducible promoter (pRD29A) in comparison with constitutive promoter (35S).
PCR assays to detect gusA gene in transgenic plants a Amplification of gusA gene in plants transformed with pRD29A-pCAMBIA, Lane 1: 1 Kb plus DNA ladder (Thermo), Lane 2, non-transgenic control, Lane 3, plasmid control, Lane 4–11: primary transformants, b amplification of gusA gene in plants transformed with pRCA-pCAMBIA, Lane 1: 1 Kb plus DNA ladder (Thermo), Lane 2: non-transgenic control, Lane 3: plasmid control, Lane 4–13: primary transformants c confirmation of plants transformed with mock (35S-pCAMBIA), Lane 1: DNA ladder Mix (Thermo), Lane 2–3: non-transgenic control, Lane 4: plasmid control, Lane 5–14: primary transformants
Histochemical GUS analysis is a cost effective and fast method to screen primary transformants. The primary transformants subjected to heat and drought stress experiment exhibited robust GUS activity (Fig. 4). Transgenic plants driving expression of gusA gene under the control of green tissue-specific pRCA promoter also showed high GUS activity in leaves when observed under microscope. While very less or no GUS activity was recorded in tubers (Fig. 4f) compared to the tubers of transgenic plants transformed with 35S-pCAMBIA (Fig. 4e). No any GUS activity was detected in non-transgenic control plants. According to histochemical GUS and PCR analysis, total transformation efficiency was determined as 0.98% in Lady Olympia potato variety.
Histochemical GUS analysis of transgenic potato plants expressing gusA gene under control of 35S, pRD29A and pRCA promoters, a Non-transgenic shoot, b transgenic plant expressing gusA gene under the control of 35S promoter, c transgenic plant expressing gusA gene under the control of pRD29A promoter, d transgenic plant expressing gusA gene under the control of pRCA promoter, e tuber of transgenic plants transformed with gusA gene under the control of 35S promoter, f tuber of transgenic plants transformed with gusA gene under the control of pRCA promoter
The plantlets expressing gusA gene under the control of pRD29A promoter were cultured under drought, heat and both drought + heat stress. The quantitative real time data showed the increased accumulated transcript levels of gusA indicating approximately 10 times in drought and 15 times more in high temperature (heat stress) compared to control indicating robust activity of stress-inducible promoter pRD29A. Interestingly, transcript levels of gusA increased more, where both stresses (drought and high temperature in combination) were applied especially in plant No. 8 (Fig. 5b). According to the qRT-PCR data, it was observed that the expression of the gusA gene under the control of pRCA in transgenic potatoes increased 4–5 times more in leaves compared to tubers. No any gusA expression was observed in tubers of Plant No. 7 and very less in Plant No 8, where gusA gene was driven by pRCA promoter. Comparing with pRCA promoter, 35S transgenic potatoes showed 5 folds more expression of gusA gene in leaves (Fig. 6b).
Transgenic plants expressing gusA gene under control of pRD29A promoter were subjected to heat and drought stress separately and in combination (a), Coefficients of gusA transcript levels determined by real time PCR in transgenic plants transformed with pRD29A-pCAMBIA relative to the internal standard. For normalization of the data, EF1α was used and the expression levels were expressed as relative fold change (b)
Transgenic plant expressing gusA gene under control of pRCA promoter along with control (35S) (a), Coefficients of gusA transcript levels determined by real time PCR in transgenic plants transformed with pRCA-pCAMBIA relative to the internal standard. For normalization of the data, EF1α was used and the expression levels were expressed as relative fold change. The letters indicate the statistical difference of GUS gene expression (p ≤ 0.05) (b)
To further confirm the efficacy of pRD29A and pRCA promoters, GUS protein was estimated using GUS fluorogenic assay as described by Bottino (2018). The GUS protein activity was completely absent in non-transgenic control potato plant (Fig. 7a), whereas pRD29A plant in control conditions showed very little activity (Fig. 7a). Overall, GUS protein activity in plants transformed with pRD29A-pCAMBIA increased under drought and heat stress in all tested plants establishing the activity of promoter under abiotic stress conditions. Plant No. 8 showed highest GUS protein under combination of heat and drought stress compared to other plants. Whereas GUS protein in the leaves of transgenic plants transformed with mock (35S promoter) and pRCA-pCAMBIA were found high (Fig. 7b). The mock transgenic plants showed robust GUS activity in root/tuber, whereas no any activity was found in tubers of plants carrying gusA gene under pRCA revealing the confinement of gene expression to green part of the plants. From the results of GUS fluorogenic analysis, we transferred the data to the number using color picker program according to the brightness of the samples incubated at 37 °C. According to data, the reduced or no GUS protein activity was detected in control, pRCA root/tuber and pRD29A transgenic potatoes samples grown in optimum conditions. The level of GUS activity varied among different samples as revealed from the data (Fig. 7v).
GUS fluorogenic assays of transgenic plants expressing gusA gene under pRD29A promoter (a), primary transformants expressing gusA gene under the control of pRCA promoter (b). GUS proteins data from the results of GUS fluorogenic analysis with the color picker program. The letters indicate the statistical difference of GUS protein activity (p ≤ 0.05)
Discussion
The current developments in genomics have assisted plant biotechnologists to identify new plant promoters to drive the expression of targeted genes spatio-temporally in crop plants (Porto et al. 2014). The knowledge about the choice and efficiency of a promoter is handy for researchers especially using new genome editing techniques like RNA interference and Crispr-Cas-9. The commercialized crop express insect or herbicide resistant gene(s) under the control of 35S CaMV; a widely used constitutive promoter (Ho et al. 1999; Bakhsh et al. 2016). There are different concerns about the use of virus based promoters in genetically modified crops (Ho et al. 1999; Podevin and Du Jardin 2012; Khabbazi et al. 2018) that provides impetus for the use of plant origin promoters in next generation genetically modified (GM) or non GM crops. Considering importance of promoters, the present study was conducted to understand the efficiency of two plant origin promoters, i.e., stress-inducible promoter RD29A and green tissue-specific promoter pRCA to drive the expression of a reporter gene (gusA) in transgenic potatoes in comparison with widely used 35S promoter.
To obtain transgenic potatoes, we proceeded with Agrobacterium-mediated transformation of Lady Olympia potato cultivar using protocol as described by Bakhsh (2020). The overall transformation efficiency in present study was recorded as 0.98% that is less when compared to earlier report of potato transformation (Veale et al. 2012; Hameed et al. 2017; Ahmed et al. 2017; Bakhsh 2020). That might be attributed to different factors, such as the type of vector, Agrobacterium strain, explant type, and varietal genetic background (Bakhsh et al. 2014; Heidari Japelaghi et al. 2018; Dönmez et al. 2019). The transformation efficiency was calculated according to the PCR positive plants out of total explants used and detection of GUS activity as described previous studies (Ahmed et al. 2018; Bakhsh et al. 2020). Few of plants that survived on hygromycin selection pressure in regeneration selection media could not show amplification of gusA gene in PCR assays; were discarded (Fig. 3). The selective agents are incorporated in regeneration selection medium during in vitro culture for selecting preferentially transformed cells by the introduction of gene (s) within T-DNA that encode resistance to antibiotic or herbicide resistance (Dandekar and Fisk 2005). Although hygromycin offers good selection system after kanamycin during regeneration process; escapes were recorded in present study contrary to the results obtained by Zuraida et al (2013) who described hygromycin as effective selectable marker in rice to inhibit growth and development of non-transformed embryogenic calli and somatic embryos. Although it might be due to the difference of species as we optimized different concentration of hygromycin in potatoes.
The plants confirmed by PCR and histochemical assays were subjected to different analysis to evaluate the efficiency of promoters. The drought (20%PEG) and heat stress (35 °C) stresses in separate and in combination were applied to transgenic plants expressing gusA gene under control of stress-inducible promoter. GUS histochemical analysis of stressed plants showed that pRD29A promoter gets activated under stress and leads to the indication of GUS activity comparable to 35S constitutive promoter (Fig. 4c). Real time analysis of these plants under stress showed accumulated transcript levels of gusA gene indicating high gene expression under stress condition driven by pRD29A promoter in tested transgenic plants, with maximum expression in Plant No. 8 under combination of heat and drought stress (Fig. 5).
GUS gene reporter system is an invaluable tool for studying gene expression in plant related research. In present study, we followed the protocol of determining GUS activity using GUS fluorogenic assay as described by Bottino (2018). When assayed by GUS fluorogenic assays, again the plants under stress showed high GUS protein expression (Fig. 7a–c) indicating that the promoter is quite suitable for transgenic studies to drive the expression of gene(s) encoding traits of economic importance. These results are in agreement with earlier studies by Behnam et al. (2007) and Pinhero et al. (2011) that showed the activity of pRD29A promoter in Arabidopsis and potato under low temperature. However, in the present study, we were interested to determine the activity of this promoter under high temperature and drought conditions keeping in view the important of potato crop in Central Anatolian region, Turkey that contributes 60% to national potato production (Çalışkan et al. 2010).
Transgenic plants with expression of gusA driven by pRCA promoter showed high GUS activity in leaves, again comparable to the control (35S); however, no any GUS activity was detected in tuber or roots of transgenic plants (Fig. 4) showing the organ and light specific activation of introduced gene under control of pRCA. The results of RT-qPCR further showed the least expression of gusA gene in tubers and roots of transgenic plants (Fig. 6), although leaf showed high GUS expression in all three tested plants, lesser than 35S promoter. These results are in agreement with earlier reports in crops like sweet potato (Tanabe et al. (2015) and cotton (Bakhsh et al. 2012), where high transgene expression was recorded using green tissue-specific promoters with low or no expression in seed (Özcan et al. 1993) and tubers (Rahamkulov 2019). Likewise GUS protein was not detected in root and tubers of transgenic plants with pRCA promoter indicating the suitability of promoter in transgenic technology to contain transgene expression to green parts of the plant that can be ideal for many crops like potato and corn. It is important to mention here, 35S promoter driven GUS protein was high in leaves and tubers of control plants (Fig. 7). The light and organ-specific promoters of genes encoding small and large subunit of Ribulose-1,5-bisphosphate carboxylase/oxygenase have been characterized and utilized in earlier studies (reviewed in Potenza et al. 2004; Bakhsh et al. 2011). After establishing pRCA promoter role in inducing tissue-specific specific expression of gusA gene, we used this promoter to drive expression of dsRNAs of insect molting-associated Ecdysone receptor gene in potatoes against Colorado potato beetle (CPB) and confirmed the confinement of dsRNAs in green part of the plants with increased mortality of CPB (Unpublished data).
Since commercialization, advantages and disadvantages of genetically modified crops are being discussed. To minimize the biosafety concerns and to increase the acceptance of transgenic crops in public, the use of tissue-specific and stress-inducible promoters can be consummated to drive the expression of foreign gene(s) in crops.
References
Ahmed HAA, Onarıcı S, Bakhsh A, Akdoğan G, Karakoç ÖC, Özcan SF, Aydın G, Aasim M, Ünlü L, Sancak C, Naimov S (2017) Targeted expression of insecticidal hybrid SN19 gene in potato leads to enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata Say) and tomato leafminer (Tuta absoluta Meyrick). Plant Biotechol Rep 11:315–329
Ahmed HA, Barpete S, Akdogan G, Aydin G, Sancak C, Ozcan S (2018) Efficient regeneration and Agrobacterium tumefaciens mediated genetic transformation of potato (Solanum tuberosum L.). Fresenius Environ Bull 27:3020–3027
Alisdair R, Fernie R, Willmitzer L (2001) Update on tuber formation, dormancy, and sprouting molecular and biochemical triggers of potato tuber development. Plant Physiol 127:1459–1465
Amarasinghe BHR, Nitschke EF, Wu Y, Udall JA, Dennis ES, Constable G, Llewellyn DJ (2006) Genomic approaches to the discovery of promoters for sustained expression in cotton (Gossypium hirsutum L.) under field conditions: expression analysis in transgenic cotton and Arabidopsis of a RuBisCo small subunit promoter identified using EST sequence analysis and cDNA microarrays. Plant Biotechnol 23:437–450
Anayol E, Bakhsh A, Karakoç ÖC, Onarıcı S, Köm D, Aasim M, Özcan SF, Barpete S, Khabbazi SD, Önol B, Sancak C (2016) Towards better insect management strategy: restriction of insecticidal gene expression to biting sites in transgenic cotton. Plant Biotechnol Rep 10:83–94
Analytical Software (2005) Statistix 8.1 for Windows. Analytical Software, Tallahassee, Florida
Bakhsh A (2020) Development of efficient, reproducible and stable Agrobacterium-mediated genetic transformation of five potato cultivars. Food Technol Biotechnol 58(1):57–63
Bakhsh A, Qayyum RA, Shamim Z, Husnain T (2011) A mini review: rubisco small subunit as a strong, green tissue-specific promoter. Arch of Biol Sci 63:299–307
Bakhsh A, Siddique S, Husnain T (2012) A molecular approach to combat spatio-temporal variation in insecticidal gene (Cry1Ac) expression in cotton. Euphytica 183:65–74
Bakhsh A, Anayol E, Ozcan SF (2014) Comparison of transformation efficiency of five Agrobacterium tumefaciens strains in Nicotiana Tabacum L. Emir J Food Agric 26:259–264
Bakhsh A, Anayol E, Khabbazi SD, Karakoç ÖC, Sancak C, Özcan S (2016) Development of insect-resistant cotton lines with targeted expression of insecticidal gene. Arch of Biol Sci 68:773–780
Bakhsh A, Hussain T, Rahamkulov I, Ufuk D, Çalışkan ME (2020) Transgenic potato lines expressing CP4-EPSP synthase exhibit resistance against glyphosate. Plant Cell Tiss Organ Cult 140:23–34
Beaujean A, Sangwan R, Lecardonnel A, Sangwan-Norreel B (1998) Agrobacterium-mediated transformation of three economically important potato cultivars using sliced internodal explants, an efficient protocol of transformation. J Exp Bot 49:1589–1595
Behnam B, Kikuchi A, Celebi-Toprak F, Kasuga M, Yamaguchi-Shinozaki K, Watanabe KN (2007) Arabidopsis rd29A:DREB1A enhances freezing tolerance in transgenic potato. Plant Cell Rep 26:1275–1282
Bottino PJ (2018) GUS Gene assay in transformed tissues. Available at https://www.goldbio.com/documents/1053/Gus%2520Gene%2520Assay%2520Protocol.pdf. Retried on March 2020
Çalışkan ME, Onaran H, Arıoğlu H (2010) Overview of the Turkish potato sector: challenges, achievements and expectations. Potato Res 53:255–266
Conner AJ, Glare TR, Nap JP (2003) The release of genetically modified crops into the environment. Part II. Overview of ecological risk assessment. Plant J 33:19–46
Dandekar AM, Fisk H (2005) Plant transformation: Agrobacterium-mediated gene transformation. In: Pena L (ed) Transgenic plants: methods and protocols. Humana Press, New Jersey, pp 35–46
Dönmez BA, Dangol SD, Bakhsh A (2019) Transformation efficiency of five Agrobacterium strains in diploid and tetraploid potatoes. Sarhad J Agric 35:1344–1350
Gopal J, Iwama K (2007) In vitro screening of potato against water-stress mediated through sorbitol and polyethylene glycol. Plant Cell Rep 26:693–700
Hameed A, Tahir MN, Asad S, Bilal R, Van Eck J, Jander G, Mansoor S (2017) RNAi-mediated simultaneous resistance against three RNA viruses in potato. Molecular Biotechnol 59:73–83
Heidari Japelaghi R, Haddad R, Valizadeh M, Dorani Uliaie E, Jalali Javaran M (2018) High-efficiency Agrobacterium-mediated transformation of tobacco (Nicotiana tabacum). J Plant Mol Breed 6:38–50
Ho MW, Ryan A, Cummins J (1999) Cauliflower mosaic viral promoter-a recipe for disaster? Microb Ecol Health Dis 11:194–197
Hussain T, Aksoy E, Çalışkan ME, Bakhsh A (2019) Transgenic potato lines expressing hairpin RNAi construct of molting-associated EcR gene exhibit enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata, Say). Transgenic Res 28:151–164
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Khabbazi SD, Khabbazi AD, Özcan SF, Bakhsh A, Başalma D, Özcan S (2018) Expression of GNA and biting site-restricted cry1Ac in cotton; an efficient attribution to insect pest management strategies. Plant Biotechnol Rep 12:273–282
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2∆∆C(T) method. Methods 25:402–408
Mithra SVA, Kulkarni K, Srinivasan R (2017) Plant promoters: characterization and applications in transgenic technology. In: Abdin M, Kiran U, Kamaluddin Ali A (eds) Plant biotechnology: principles and applications. Springer, Singapore
Msanne J, Lin J, Stone JM, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107
Murashige T, Skoog F (1962) Revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nicot N, Hausman JF, Hoffman L, Evers D (2005) Housekeeping gene selection for real time PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Özcan S, Firek S, Draper J (1993) Selectable marker genes engineered for specific expression in target cells for plant transformation. Nat Biotechnol 11:218–221
Pinhero R, Pazhekattu R, Marangoni AG, Liu Q, Yada RY (2011) Alleviation of low temperature sweetening in potato by expressing Arabidopsis pyruvate decarboxylase gene and stress-inducible rd29A: a preliminary study. Physiol Mol Biol Plants 17:105–114
Pino MT, Ávila A, Molina A, Jeknic Z, Chen TH (2013) Enhanced in vitro drought tolerance of Solanum tuberosum and Solanum commersonii plants overexpressing the ScCBF1 gene. Cien. Inv. Agr. 40 (1): 171–184. Int J Agric Nat Resour 40:171–184
Podevin N, Du Jardin P (2012) Possible consequences of the overlap between the CaMV 35S promoter regions in plant transformation vectors used and the viral gene VI in transgenic plants. GM crops food 3:296–300
Porto MS, Pinheiro MPN, Batista VGL, dos Santos RC, de Albuquerque Melo Filho P, de Lima LM (2014) Plant promoters: an approach of structure and function. Mol Biotechnol 56:38–49
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. Vitro Cell Devel Biol Plant 40:1–22
Qu D, Song Y, Li WM, Pei XW, Wang ZX, Jia SR, Zhang YQ (2011) Isolation and characterization of the organ-specific and light-inducible promoter of the gene encoding rubisco activase in potato (Solanum tuberosum). Genet Mol Res 10:621–631
Rahamkulov I (2019) Comparison of different promoters activity in transgenic potatoes. Nigde Omer Halisdemir University, Nigde, Turkey, Page No, p 74
Rowe CR (1993) Potato health manegement. Ohio State University, USA, Department of Plant Pathology
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Somssich M (2019) A short history of the CaMV 35S promoter. PeerJ Prepr. https://doi.org/10.7287/peerj.preprints.27096v2
Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS (2002) Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol Biol 50:463–474
Tabashnik BE, Carrie`re Y (2017) Surge in insect resistance to transgenic crops and prospects for sustainability. Nat Biotechnol 35:926–935
Tanabe N, Tamoi M, Shigeoka S (2015) The sweet potato RbcS gene (IbRbcS1) promoter confers high-level and green tissue-specific expression of the GUS reporter gene in transgenic Arabidopsis. Gene 567:244–250
Veale MA, Slabbert MM, Van Emmenes L (2012) Agrobacterium-mediated transformation of potato cv. Mnandi for resistance to the potato tuber moth (Phthorimaea operculella). S Afr J Bot 80:7–74
Zhu LP, Yu Z, Zou CX, Li QL, Chuan Y (2010) Plant stress-inducible promoters and their function. Yi Chuan = Hereditas 32:229–234
Zuraida AR, Zulkifli RK, Alizah Z, Zamri Z, Aziz A (2013) Hygromycin as selective marker in Agrobacterium-mediated genetic transformation of indica rice MR 219. J Trop Agric Food Sci 41:71–79
Acknowledgements
The present study is a part of research project supported by TÜBİTAK (Grant No. 215O520), where we used pRCA promoter to drive the expression of EcR dsRNAs in green part of potato plants. The authors are highly thankful to Tübitak for providing fellowship to Mr. Ilhom Rahamkulov from the project budget. Authors are grateful to Prof. Dr. Mehmet Emin Çalışkan for providing seed tuber of Lady Olympia used in the study. Special thanks to Dr. Emre Aksoy for his technical help during experiments and interpretation of results. We are also thankful to Dr. Halil Toktay for providing access to microscope facility.
Author information
Authors and Affiliations
Contributions
The data presented in the manuscript is MS thesis work of IR who completed his studies under the supervision of AB.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rahamkulov, I., Bakhsh, A. Tissue-specific and stress-inducible promoters establish their suitability for containment of foreign gene(s) expression in transgenic potatoes. 3 Biotech 10, 426 (2020). https://doi.org/10.1007/s13205-020-02350-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02350-x