Abstract
Ralstonia solanacearum is an important soil-borne plant pathogen which causes bacterial wilt in a large number of crops. Bacterial Type Six Secretion System (T6SS) is known to participate in pathogenesis, bacterial interaction and inter-bacterial competition. Contribution of T6SS in the virulence of R. solanacearum on eggplant (Solanum melongena L) is studied. In this study, five T6SS gene (ompA, vgrG3, hcp, tssH and tssM) mutants have been developed by insertional mutagenesis and the virulence of the mutants was evaluated on eggplant. In general, the T6SS mutants showed significant reduction of wilt on eggplant. R. solanacearum mutant of ompA gene significantly reduced the wilt from day five through day eight in petiole inoculation. In soil drench inoculation, R. solanacearum mutant of vgrG3 gene reduced the wilt on eggplant and was significantly different throughout the experimental period. Other mutants, viz., tssH, tssM and hcp, also reduced the wilt during the initial stages of disease development. This is the first report on the role of T6SS genes, ompA, vgrG3, hcp and tssH on virulence of R. solanacearum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ralstonia solanacearum is an important soil-borne phytopathogen with wide host range and large geographical distribution (Genin and Denny 2012). It has been ranked second in the list of top ten of the most studied bacterial plant pathogens in molecular plant pathology (Mansfield et al. 2012). It causes severe economical losses in agriculturally important crops due to its destructive nature, lethality and persistence (Wicker et al. 2004). Some of reported losses include 88% of tomato in Uganda, 95% of tobacco in South Carolina, 50–100% of potato in Kenya and 100% on pepper in Ethiopia (Assefa et al. 2015; Elphinstone 2005). R. solanacearum strains show extensive diversity globally due to which it has been referred as Ralstonia solanacearum species complex (RSSC) and is classified into four phylotypes (Fegan and Prior 2005). These phylotypes represent their geographical origin: phylotype I (Asia), phylotype II (America), phylotype III (Africa) and phylotype IV (Indonesia) (Genin and Denny 2012). Safni et al. (2014) proposed taxonomic revision of the R. solanacearum species complex. Accordingly, they proposed three genospecies, viz. R. solanacearum (phylotype II strains), R. pseudosolanacearum (phylotype I and III strains) and R. syzygii (phylotype IV strains). Bacterial wilt management strategies, like soil amendments, soil solarization, use of bio-fumigants, resistant plant varieties and biocontrol agents, have shown limited success in controlling the wilt (Ramesh et al. 2016). The virulence of this pathogen is contributed by various virulence factors. These include exopolysaccharide (EPS), the type II secretory system (T2SS)-dependent plant cell wall degrading enzymes, chemotaxis, swimming motility, twitching motility and type III secretory system (T3SS) (Asolkar and Ramesh 2018a, b).
Gram negative bacteria secrete virulence proteins outside the cell to promote its survival, replication and colonization with the help of various secretion systems. These include Type I secretion systems (T1SS), Type II secretion systems (T2SS), Type III secretions systems (T3SS), Type IV secretion systems (T4SS), Type V secretion system (T5SS) and Type VI secretion systems (T6SS) (Green and Mecsas 2016). The T1SS, T2SS and T5SS secrete proteins across the bacterial envelop to the extracellular milieu. The T3SS, T4SS and T6SS deliver proteins directly across host membranes (Green and Mecsas 2016). The T2SS contributes in the virulence of R. solanacearum by secreting Plant Cell Wall Degrading Enzymes (PCWDE) and other extracellular proteins (Genin and Denny 2012). The PCWDE hydrolyse the complex carbohydrates present in the cell wall components and helps the bacterium to obtain nutrients and energy in addition to facilitating its entry and spread in the host plant (Liu et. al. 2005; Huang and Allen 2000). The T3SS enables R. solanacearum to translocate pathogenicity proteins called as ‘type III effectors (T3E)’ into the cytosol of eukaryotic host cells. The type IV pili (Tfp) help in exhibiting twitching motility and thus participate in the virulence of R. solanacearum (Liu et al. 2001). Twitching motility helps R. solanacearum to adhere to host surface and also plays an important role in natural transformation (Kang et al. 2002). The T6SS is one of the secretion systems in bacteria (Pukatzki et al. 2006), which plays role in the virulence of pathogenic bacteria. The T6 secretion gene cluster is widely present in the genome of many Gram-negative bacteria, especially proteobacteria (Boyer et al. 2009; Cascales and Cambillau 2012) and is involved in pathogenesis, bacterial interactions and competition. The T6SS interacts with both prokaryotic and eukaryotic cells (Silverman et al. 2012). T6SS has been identified in R. solanacearum which contributes to the virulence (Zhang et al. 2012). The T6SS secretes anti-bacterial proteins into the target cells and thus kills the neighbouring, non-immune bacterial cells by cell-to-cell contact (Cascales and Cambillau 2012). Like the T3SS, the T6SS is also involved in translocation of substrates into the recipient cells through contact-dependent manner (Silverman et al. 2012; Cornelis 2006). In eukaryotes, it secretes toxin molecules which interfere with the eukaryotic cytoskeleton and thus plays a role in pathogenesis (Cascales and Cambillau 2012).
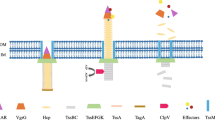
The T6SS is formed by assembly of two distinct substructures: an inverted bacteriophage-like injection apparatus and a membrane complex. The membrane complex interacts with the inverted bacteriophage-like structure and anchors it into the cell envelope. The secretion apparatus is made up of 13 core components which are supplemented with additional proteins (Cascales and Cambillau 2012). The core components are named from TssA to TssM, and many of them share homology with bacteriophage proteins (Badr et al. 2016).
In R. solanacearum, the T6SS genes, tssM and tssB, have already been proved to be involved in virulence as mutation in these genes has led to the decrease in virulence on tomato plants in comparison to the wild-type strain (Zhang et al. 2012, 2014). However, the other genes of T6SS have not been studied for their role in pathogenesis or virulence. In this study, the role of other T6SS genes, including outer membrane protein A (ompA) in the virulence of R. solanacearum, has been studied on eggplant (Solanum melongena L).
Materials and methods
Bacterial strains and Plasmids
R. solanacearum strain Rs-09–161 maintained at the culture collection of Plant Pathology laboratory, ICAR-CCARI, was used in this study to develop the mutants (Asolkar and Ramesh 2018a). The strain was isolated from wilted eggplant and is highly pathogenic (Asolkar and Ramesh 2018b). Briefly, R. solanacearum was isolated aseptically from the wilted plant by collecting the xylem ooze from the stem or root portions and streaked onto TZC medium (0.1% casein, 1% peptone, 0. 5% glucose and 1.7% agar agar amended with 0.005% v/v 2,3,5-triphenyl tetrazolium chloride). Plates were incubated at 28 + 2 °C for 48 to 72 h. Pink, fluidal colonies were purified onto TZC medium and the pure culture was maintained as glycerol stock. The bacterial strains, vectors and mutants were maintained as 30% glycerol stock at -80 °C. R. solanacearum was routinely grown on BG medium (1% peptone, 0.1% casamino acids, 0.1% yeast extract and 0.5% glucose) at 28 °C. E. coli was grown at 37 °C on Luria and Bertani medium. Ampicillin was used in the concentration 50 µg/mL while growing E. coli and R. solanacearum mutants. The details about the plasmid used in this study and the clones developed are listed in Table 1.
Development of clones in E. coli vector
The T6SS genes, viz., tssH, hcp, vgrG3, tssM and ompA, were selected in this study. These genes form the core components and accessory proteins associated with the outer membrane of the type six secretion apparatus. Primers were designed for the amplification of internal fragments of the genes using Oligo primer analysis software, version 6.4. The sequences of the primers, the expected size of band, reaction mixture and standardized PCR conditions are shown in Table 2. Mastercycler Pro (Eppendorf, GmBH) was used for product amplification and the purified fragments were cloned in the vector pTZ57R/T as per the manufacturer’s protocol (TA cloning kit-Thermo Fisher Scientific USA). The positive colonies were confirmed for the presence of insert by colony PCR using the same conditions as amplification of the internal fragment, followed by confirmation of the clones by restriction digestion (Asolkar and Ramesh 2018b).
Development of R. solanacearum mutants
The electro competent cells were prepared as mentioned in Asolkar and Ramesh (2018b). Briefly, overnight grown culture of R. solanacearum isolate Rs-09-161 was inoculated into BG medium to an initial OD600nm of 0.15 and grown at 28 °C at 160 rpm. Actively growing cells were harvested at 0.5 OD600nm and electro-competent cells were prepared under ice cold conditions as follows: A series of centrifugations were carried out at 8000 rpm for 10 min to reconstitute the active cells in equal volume of cold MilliQ water, half volume of MilliQ water, one-fourth volume of 10% glycerol and finally in 1/100th volume of 10% glycerol solution, respectively. Electroporation was performed out using MicroPulser (Biorad) at 2KV using 2 µg of plasmid and plated on BG medium with ampicillin. The T6SS mutants were confirmed with specifically designed diagnostic primer located 1000 bp upstream of the forward primer of the target gene and M13 primers for each target gene depending upon the orientation of the insertion. The sequences of the diagnostic primers and standardized PCR conditions are shown in Table 3. The confirmed mutants were maintained as 30% glycerol stock at – 80 ºC.
Virulence assay of R. solanacearum mutants on eggplant
Virulence of the R. solanacearum mutants was assayed on eggplant by soil drench and petiole inoculation method as described by Asolkar and Ramesh (2018b). Highly susceptible eggplant cultivar, Agassaim, was used in this study. Seedlings were raised and maintained in the greenhouse at 30 °C during the day with 60% relative humidity, 16 h of light and 8 h of dark period. Thirty-day-old seedlings grown in the potting mixture containing red soil: FYM: sand (2:1:1) were used for inoculation studies. Wild-type strain Rs-09-161 was used as a positive control and Rs-HrcV− (Asolkar and Ramesh 2018b) was used as a non-pathogenic control in each of the experiment. In soil drench 108 CFUmL−1 of inoculum was used, while in petiole inoculation, 2000 cells were introduced on the third leaf petiole from the shoot apex by making a cut 2 cm away from the stem. The seedlings were observed for wilt symptoms for up to 15 days post-inoculation. Three experiments were carried out in petiole inoculation method and two experiments were carried out in soil drench method. The percent wilt incidence data obtained were analysed statistically by WASP software (https://ccari.res.in/wasp2.0/index.php).
Results
Development of clones in E. coli
The internal region of selected T6SS genes of R. solanacearum was amplified (Supplementary Fig. 1), the fragments were cloned in the vector pTZ57R/T and the positive clones were identified by blue and white selection. The presence of internal gene fragment in the colonies was confirmed by colony PCR of Ec-TssM, Ec-TssH, Ec-Hcp, Ec-VgrG3 and Ec-OmpA which amplified specific bands at 672, 694, 421, 719 and 566 bp, respectively (Supplementary Fig. 2). Restriction digestion of tssMpTZ57R/T, tssHpTZ57R/T, hcppTZ57R/T, vgrG3pTZ57R/T and ompApTZ57R/T further confirmed the insert in pTZ57R/T (Supplementary Fig. 3). The restriction profile of the recombined plasmids TssMpTZ57R/T, TssHpTZ57R/T, HcppTZ57R/T, VgrG3pTZ57R/T and OmpApTZ57R/T were calculated using NEBcutter V2.0 and the sizes of different fragments is given in Supplementary Table 1.
Development of R. solanacearum mutant
The recombined plasmids from Ec-TssM, Ec-TssH, Ec-Hcp, Ec-VgrG3 and Ec-OmpA were used as the vectors for developing mutants in R. solanacearum by insertional mutagenesis. The diagnostic primer designed approximately 1000 bp upstream of the gene located in the genome pairs with either of the M13 primers (based on the orientation of the inserted plasmid) amplified the fragment that indicated the disruption of the gene at the desired site. Rs-TssM− was confirmed by the presence of 1101 bp band with M13 forward primer, Rs-TssH− by 1203 bp with M13 reverse primer, Rs-Hcp− by 1014 bp with M13 forward primer and Rs-OmpA− by 980 bp with M13 forward primer, respectively (Supplementary Fig. 4).
Virulence assay of R. solanacearum mutants on eggplant
The virulence of R. solanacearum T6SS mutants: Rs-TssM−, Rs-TssH−, Rs-Hcp−, Rs-OmpA− and Rs-VgrG3− was assayed on eggplant seedlings by petiole and soil drench inoculation. The T3SS mutant Rs-HrcV- was used as a non-pathogenic control.
In petiole inoculation, wilting of seedlings was observed on fourth day of inoculation (DAI) in wild-type and the T6SS mutants. Wild-type strain Rs-09–161 exhibited aggressive wilting of the susceptible eggplant seedlings. On day five, more than 50% of the seedlings were wilted, while, on day six, more than 94% wilt was observed. Wild type caused 100% wilt on tenth DAI. In case of T6SS mutants, less than 37% of wilt was observed on fifth DAI and displayed statistically different grouping pattern. This trend continued on day six where the wilt incidence in mutant inoculated plants was in the range of 56–77% except in Rs-TssH−. On days seven and eight, although the mutants displayed reduced wilt compared to wild type, statistically significant difference was observed only in case of Rs-OmpA−. Among the T6SS mutants, less wilt was observed in Rs-OmpA− (92.7%) and Rs-TssM− (94.5%) after 9 days of inoculation (Table 4, Supplementary Fig. 5).
In soil drench inoculation, wilting of the seedlings was initiated on fifth DAI. The wild-type strain Rs-09-161 caused more than 50% wilt on seventh DAI and more than 90% wilt was observed on tenth DAI. In comparison to the wild type, there was significant reduction in the wilt causing ability of the T6SS mutants. The lowest wilt was observed with Rs-VgrG3− and was significantly different throughout the experiment. At the end of the experiment, wild type caused 97% wilt, while Rs-VgrG3− showed maximum of 74.2% wilt. Significant reduction in wilt was also observed in case of Rs-TssH− from day seven through day twelve and in Rs-TssM− from day nine through day twelve. At the end of the experiment, wilt incidence rates observed in Rs-TssM− and Rs-TssH− were 82.8 and 88.5%, respectively. In case of Rs-Hcp− and Rs-OmpA−, the wilting was much lesser than the wild type although it was not significantly different after day nine (Table 5, Supplementary Fig. 6).
Discussion
In R. solanacearum, genes coding for the virulence factors, like chemotaxis, flagella-driven swimming motility, pili-associated twitching motility, the extracellular polysaccharide (EPS), the Type Two Secretory System (T2SS)-dependent cell wall degrading enzymes and the Type III Secretory system (T3SS), have been studied (Genin and Denny 2012). The mutants of T2SS, chemotaxis, swimming motility and twitching motility displayed reduced virulence and mutants of EPS and T3SS are non-pathogenic (Saile et al. 1997, Tans-Kersten et al. 2004, Meng et al. 2011, Brito et al. 2002). T6SS has been reported in R. solanacearum (Zhang et al. 2012), which is known to be involved in inter-bacterial interaction, bio-film formation and eukaryotic cell interaction (Lossi et al. 2013). Two genes of the T6SS, the tssB and tssM, have been studied and reported to contribute towards the virulence of R. solanacearum in tomato (Zhang et al., 2012, 2014). Since there is a lack of information regarding the T6SS of R. solanacearum and the role played by its core and the accessory genes in virulence, this study was taken up. R. solanacearum wild-type Rs-09-161 which is highly virulent on solanaceous vegetables, including eggplant, (Asolkar and Ramesh 2018b) and its genome sequence is available (Ramesh et. al. 2014). Mutants of five genes of T6SS of Rs-09-161, viz., tssM, tssH, hcp, ompA and vgrG3, were developed by insertional mutagenesis. They form the core components and accessory components of the T6SS. Insertion mutagenesis is a simple, efficient method that has been routinely used for the development of mutants for a number of genes in R. solanacearum, like hrpB, hrcV, (Asolkar and Ramesh 2018b) phcA, phcB and hrpG (Lin et al. 2008). The vector pTZ57R/T lacks the oriC for R. solanacearum and hence cannot replicate in R. solanacearum Rs-09-161. When the recombined vector containing an internal fragment is transferred into the R. solanacearum cell, due to the sequence similarity shared by the cloned insert fragment and the target gene, the plasmid integrates itself into the target gene by homologous recombination. This leads to its disruption by insertional mutagenesis.
One of the common and the best methods to assess the role of any gene in the virulence of R. solanacearum is to assess the disease causing potential of the mutant compared to wild type in terms of percent wilt and delay in causing the wilt (Liu et al. 2005; Huang and Allen 2000). Researchers used different methods of inoculation to assess the virulence of R. solanacearum (Liu et al. 2005; Lin et al. 2008; Tans-Kersten et al. 2004).
In petiole inoculation study, reduced wilt incidence by the mutants was observed till sixth day after inoculation and thereafter no significant difference except in case of Rs-OmpA− (till day eight). OmpA forms an accessory gene of the T6SS, which forms a translocation pore for translocation of type six effectors (Pautsch and Schulz 1998). It is found associated only in virulent organisms along with sciE and sciT genes and is present in all species of E. coli, Yersinia, Pseudomonas, Acinetobacter, Burkholderia, Xanthomonas, Ralstonia, Hahella and Mesorhizobium (Pautsch and Schulz 1998; Shrivastava and Mande 2008). OmpA protein was also reported to be involved in the pathogenesis of Acinetobacter baumannii by invasion of epithelial cells and death of dendritic cells (Choi et al. 2008; Lee et al. 2010).
Any delay in causing wilt in the plants can also be considered as an important criterion in assessing the role of a particular gene (Liu et al. 2001) as petiole inoculation allows the bacterium to directly enter and colonize the internal tissues without facing any host defence barrier. Unless knockout of a gene makes the bacterium avirulent as in the case of hrcV mutant (Asolkar and Ramesh 2018b) in this study or makes the bacterium close to avirulent as in the case of sdpD gene reported by Liu et al. (2005), it would not be possible to see a complete lack of virulence function. This might be the reason why a difference in wilt incidence was not observed between the T6SS mutants and the wild type in the later stages of infection in petiole inoculation method (Table 4). Petiole inoculation has been used in virulence assay of phcA, phcB, hrpG, fliC and flhDC gene mutants (Tans-Kersten et al. 2004; Lin et al. 2008).
In soil drench inoculation assays, clear cut difference in the wilt incidence was observed between wild type and the mutants. Significantly reduced wilt incidence was observed till 12 DAI in the mutants compared to wild type. Soil drench inoculation is considered as the natural way of infection by R. solanacearum as the inoculated pathogen to undergo all the colonization process, to overcome the host resistance and competing microflora. Hence, the difference of wilt incidence between wild type and its T6SS mutants was clearly visible. Generally this method of inoculation is preferred to study the role of virulence genes in R. solanacearum (Liu et al. 2005; Lin et al. 2008). Rs-TssM− and Rs-OmpA− showed a delay of two days to cause 50% wilt, while Rs-TssH− showed a delay of three days to cause 50% wilt. Maximum delay of 5 days was observed in case of Rs-VgrG3−. On day nine, more than 85% wilt was observed in wild type. In case of mutants, the wilt was in the range of 40–55%.
In drench inoculation, the wilt causing ability of Rs-VgrG3− was greatly reduced and significantly different throughout the experimental period. The vgrG (valine-glycine repeat protein G) is a trimeric protein which helps in assembly of the TssBC sheath and stacking of Hcp hexamers and resembles gp27/gp5 which forms the tail spike in T4 (Pukatzki et al. 2009). The reduction in wilt observed by the Rs-VgrG3− mutant could be associated with interruption or knocking out of the virulence function VgrG3 protein. The vgrG mutant strain of Acinetobacter baumannii has been reported to cause reduced eukaryotic cell adherence and impaired lethality in mice (Wang et al. 2018).
Rs-TssH− and Rs-TssM− in drench inoculation showed a delay of four days to cause 75% wilt as compared to wild-type Rs-09-161. Wilt caused by these two mutants was considerably lesser than the wild type throughout the experiment and was significantly different from day nine through day twelve. This is the period when R. solanacearum cells spread throughout the plant in systemic manner and impair the transport of water. The gene tssH belongs to the AAA+ super family of ATPases and a subfamily ClpV and plays an important role in assembly mechanism of the T6SS (Cascales and Cambillau 2012). It has been reported that TssH plays a role in release of type six effectors by contraction of the tail sheath (Gallique et al. 2017) and mutation of tssH gene could have reduced virulence in R. solanacearum.
The importance of tssM gene in biofilm formation and virulence on tomato has been reported by Zhang et al. (2012). These results are in agreement with them as Rs-TssM− has exhibited reduction in wilt in drench as well as petiole (day five and six) assay. Colonization and proliferation studies on tomato with tssM deletion mutant in R. solanacearum have displayed significantly reduced bacterial cell numbers in the root and stem tissue (Zhang et al. 2012).
Reduction in wilt was also observed in mutants Rs-Hcp− and Rs-OmpA−; however, it was not at par with Rs-TssM−, Rs-TssH− and Rs-VgrG3−. Reduction in wilt was observed from day seven through day thirteen in case of Rs-Hcp− and Rs-OmpA−. Rs-Hcp− and Rs-OmpA− caused a delay of 2 days to cause 75% wilt in drench inoculated plants as compared to wild-type Rs-09-161. Hcp (haemolysin co-regulated protein) is secreted as substrate along with vgrG by T6SS (Pukatzki et al. 2009). The attenuation in wilt displayed by Rs-Hcp− mutant could be probably attributed to one of the roles played by Hcp proteins. In case of petiole inoculation, Rs-OmpA− depicted significant reduction in wilt; however, this was not observed in drench inoculation. Since petiole inoculation gives a direct entry in the vascular tissue, ompA gene probably plays an important role in systemic invasion of the R. solanacearum in the vascular tissue by translocation of type six effectors and not involved in the primary stages of plant infection.
In this study, the role of five genes of the T6SS of R. solanacearum in the virulence has been investigated by developing mutants and testing the mutants on eggplant. All the mutants delayed the wilt incidence on eggplant, when inoculated by drench method. However, the T6SS mutants except Rs-OmpA− did not indicate any delay in wilting in the petiole inoculation. Earlier reported work on T6SS in R. solanacearum is limited to the gene tssM and tssB. To best of the authors knowledge, this is the first report proving the role of tssH, hcp, ompA and vgrG3 in the virulence of R. solanacearum.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- T6SS:
-
Type Six Secretion System
- RSSC:
-
Ralstonia solanacearum Species complex
- EPS:
-
Extracellular Polysaccharide
- T2SS:
-
Type Two Secretion System
- T3SS:
-
Type three Secretion system
References
Asolkar T, Ramesh R (2018a) Identification of virulence factors and type III effectors of phylotype I, Indian Ralstonia solanacearum strains Rs-09-161 and Rs-10-244. J Genet 97:55–66. https://doi.org/10.1007/s12041-018-0894-z
Asolkar T, Ramesh R (2018b) Development of T3SS mutants (hrpB− and hrcV−) of Ralstonia solanacearum, evaluation of virulence attenuation in brinjal and tomato-A pre-requisite to validate T3Es of R. solanacearum. Indian J Microbiol 58:372–380. https://doi.org/10.1007/s12088-018-0736-y
Assefa M, Dawit W, Lencho A, Hunduma T (2015) Assessment of wilt intensity and identification of causal fungal and bacterial pathogens on hot pepper (Capsicum annuum L.) in BakoTibbe and Nonno districts of west Shewa zone. Ethiopia Int J Phytopathol 4:21–28
Badr S, Li Y, Duan K (2016) Comparison of the structure, regulation and functions between type three and type six secretion system in gram-negative bacteria. J Med Microb Diagn 5:2161–2703. https://doi.org/10.4172/2161-0703.1000243
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC genomics 10:104. https://doi.org/10.1186/1471-2164-10-104
Brito B, Aldon D, Barberis P, Boucher C, Genin S (2002) A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum HRP genes. Mol Plant Microbe Interact 15:109–119. https://doi.org/10.1094/MPMI.2002.15.2.109
Cascales E, Cambillau C (2012) Structural biology of type VI secretion systems. Philos Trans Royal Soc B 367:1102–1111. https://doi.org/10.1098/rstb.2011.0209
Choi CH, Lee JS, Lee YC, Park TI, Lee JC (2008) Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol 8:216. https://doi.org/10.1186/1471-2180-8-216
Cornelis GR (2006) The type III secretion injectisome. Nat Rev Microbiol 4:811-825. https://doi.org/10.1038/nrmicro1526
Elphinstone JG (2005) The current bacterial wilt situation: a global overview. In: Prior P, Hayward AC, Allen C (eds) Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul MN, pp 9–28
Fegan M, Prior P (2005) How complex is the Ralstonia solanacearum species complex. In: Prior P, Hayward AC, Allen C (eds) Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, Madison, Wisconsin, USA, pp 449–461
Gallique M, Decoin V, Barbey C, Rosay T, Feuilloley MG, Orange N, Merieau A (2017) Contribution of the Pseudomonas fluorescens MFE01 type VI secretion system to biofilm formation. PLoS ONE 12:e0170770. https://doi.org/10.1371/journal.pone.0170770
Genin S, Denny TP (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89. https://doi.org/10.1146/annurev-phyto-081211-173000
Green ER, Mecsas J (2016) Bacterial secretion systems–an overview. Microbiol Spectr 4:1–32. https://doi.org/10.1128/microbiolspec.VMBF-0012-2015
Huang Q, Allen C (2000) Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol Mol Plant Path 57:77–83. https://doi.org/10.1006/pmpp.2000.0283
Kang Y, Liu H, Genin S, Schell MA, Denny TP (2002) Ralstonia solanacearumrequires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol Microbiol 46:427–437. https://doi.org/10.1046/j.1365-2958.2002.03187.x
Lee JS, Choi CH, Kim JW, Lee JC (2010) Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J Microbiol 48:387–392. https://doi.org/10.1007/s12275-010-0155-1
Lin YM, Chou IC, Wang JF, Ho FI, Chu YJ, Huang PC et al (2008) Transposon mutagenesis reveals differential pathogenesis of Ralstonia solanacearum on tomato and Arabidopsis. Mol Plant Microbe interact 21:1261–1270. https://doi.org/10.1094/MPMI-21-9-1261
Liu H, Kang Y, Genin S, Schell MA, Denny TP (2001) Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215–3229. https://doi.org/10.1099/00221287-147-12-3215
Liu H, Zhang S, Schell MA, Denny TP (2005) Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol Plant Microbe Interact 18:1296–1305. https://doi.org/10.1094/MPMI-18-1296
Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A (2013) The HsiB1C1 (TssB/TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheath-like structure. J Biol Chem 288:7536–7548. https://doi.org/10.1074/jbc.M112.439273
Mansfield J, Genin S, Magori S et al (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. https://doi.org/10.1111/J.1364-3703.2012.00804.X
Meng F, Yao J, Allen C (2011) A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J Bacteriol 193:2477–2486. https://doi.org/10.1128/JB.01360-10
Pautsch A, Schulz GE (1998) Structure of the outer membrane protein A transmembrane domain. Nat Struct Mol Biol 5:1013–1017. https://doi.org/10.1038/2983
Pukatzki S, Ma AT, Sturtevant D et al (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Nat Acad Sci 103:1528–1533. https://doi.org/10.1073/pnas.0510322103
Pukatzki S, McAuley SB, Miyata ST (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol 12:11–17. https://doi.org/10.1016/j.mib.2008.11.010
Ramesh R, Gaitonde S, Achari G, Asolkar T et al (2014) Genome Sequencing of Ralstonia solanacearumBiovar 3, Phylotype I, Strains Rs-09-161 and Rs-10-244, Isolated from Eggplant and Chili in India. Genome Announcement 2:1–2. https://doi.org/10.1128/genomeA.00323-14
Ramesh R, Achari G, Asolkar T, Dsouza M, Singh N (2016) Management of bacterial wilt of brinjal using wild brinjal (Solanum torvum Sw) as root stock. Indian Phytopathol 69:2–6
Safni I, Cleenwerck I, DeVos P, Fegan M, Sly L, Kappler U (2014) Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov'. Int J Syst Evol Microbiol 64:3087–3103. https://doi.org/10.1099/ijs.0.066712-0
Saile E, McGarvey J, Schell M, Denny T (1997) Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87:1264–1271. https://doi.org/10.1094/PHYTO.1997.87.12.1264
Shrivastava S, Mande SS (2008) Identification and functional characterization of gene components of Type VI Secretion system in bacterial genomes. PLoS ONE 3:e2955. https://doi.org/10.1371/journal.pone.0002955
Silverman JM, Brunet YR, Cascales E, Mougous JD (2012) Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472. https://doi.org/10.1146/annurev-micro-121809-151619
Tans-Kersten J, Brown D, Allen C (2004) Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol Plant Microbe Interact 17:686–695. https://doi.org/10.1094/MPMI.2004.17.6.686
Wang J, Zhou Z, He F, Ruan Z, Jiang Y, Hua X, Yu Y (2018) The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS ONE 13:e0192288. https://doi.org/10.1371/journal.pone.0192288
Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, Prior P (2004) Emerging strains of Ralstonia solanacearum in Martinique (French West Indies): a case study for epidemiology of bacterial wilt. In: Momol MT, Ji P, Jones JB (eds) Proceedings of the first International Symposium on Tomato Diseases, International Society for Horticultural Science, Bruggen, Belgium 695:145–152. https://doi.org/10.17660/ActaHortic.2005.695.16
Zhang L, Xu J, Xu J, Chen K, He L, Feng J (2012) TssM is essential for virulence and required for type VI secretion in Ralstonia solanacearum. J Plant Dis Prot 119:125–134. https://doi.org/10.1007/BF03356431
Zhang L, Xu J, Xu J, Zhang H, He L, Feng J (2014) TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb Pathog 74:1–7. https://doi.org/10.1016/j.micpath.2014.06.006
Acknowledgement
The financial support for this work was provided by Indian Council of Agricultural Research, New Delhi, India through “Outreach project on Phytophthora, Fusarium and Ralstonia diseases of horticultural and field crops”- (PhytoFuRa). The authors are grateful to Director, ICAR- Central Coastal Agricultural Research Institute, Old Goa for providing other necessary facilities.
Author information
Authors and Affiliations
Contributions
TA and RR have made major contributions to the concept and design of the study, analysis and interpretation of the data, and writing of the manuscript. TA executed the experiment.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Ethical approval
This article does not contain any studies with human or animal subjects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asolkar, T., Ramesh, R. The involvement of the Type Six Secretion System (T6SS) in the virulence of Ralstonia solanacearum on brinjal. 3 Biotech 10, 324 (2020). https://doi.org/10.1007/s13205-020-02311-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02311-4