Abstract
microRNAs (miRNAs) play critical regulatory roles in plant growth and development. In the present study, the function of herbaceous peony (Paeonia lactiflora Pall.) miR156e-3p in the regulation of color formation has been investigated. Firstly, P. lactiflora miR156e-3p precursor sequence (pre-miR156e-3p) was isolated. Subsequently, the overexpression vector of pre-miR156e-3p was constructed and transformed into Arabidopsis thaliana. Moreover, the medium screening, GUS staining, polymerase chain reaction (PCR) of the GUS region and real-time quantitative PCR (qRT-PCR) of miR156e-3p all confirmed that the purpose gene had been successfully transferred into Arabidopsis plants and expressed, which resulted in apparent purple lateral branches. And this change in color was caused by the improved anthocyanin accumulation. In addition, expression analysis had shown that the level of miR156e-3p transcript was increased, while transcription level of target gene squamosa promoter binding protein-like gene (SPL1), encoding SPL transcription factor that negatively regulated anthocyanin accumulation, was repressed in miR156e-3p-overexpressing transgenic plants, and its downstream gene dihydroflavonol 4-reductase gene (DFR) that was directly involved in anthocyanin biosynthesis was strongly expressed, which resulted in anthocyanin accumulation of Arabidopsis lateral branches. These findings would improve the understanding of miRNAs regulation of color formation in P. lactiflora.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
microRNAs (miRNAs) are non-coding small RNAs (sRNAs) with approximately 21-nucleotides (nt) length, which are widely distributed in animals and plants and negatively regulate gene expression by either mRNA degradation or translation inhibition (Rogers and Chen 2013). Since they are discovered in Caenorhabditis elegans firstly (Lee et al. 1993), a mass of miRNAs in higher plants have been continually identified, including Arabidopsis thaliana (Sunkar and Zhu 2004), Oryza sativa (Jian et al. 2010), Triticum aestivum (Yao et al. 2007), Zea mays (Zhang et al. 2006) and so on. And the latest version of miRBase 21.0 until 2014 includes 28,645 entries hairpin precursor miRNAs (pre-miRNAs), expressing 35,828 mature miRNA products in 223 species (Kozomara and Griffiths-Jones 2014). miRNAs have an important regulatory role in plant growth and development, including root, stem, leaf, flower and fruit growth and development (Hu et al. 2014; Spanudakis and Jackson 2014; Li et al. 2010; Wang et al. 2010a, b). However, to the best of our knowledge, in color formation, only miR156, miR828, miR858 and miR778 have been reported to play important roles in anthocyanin accumulation (Shen 2015; Wang et al. 2015; Yang et al. 2013; Gou et al. 2011); miR1857 plays a major role in carotenoid accumulation (Xu et al. 2010), and miR826 and miR5090 have vital roles in chlorophyll accumulation (He et al. 2014), but their mechanisms are different.
Herbaceous peony (Paeonia lactiflora Pall.) is a traditional flower in China that belongs to the Paeoniaceae family. Because of its straight stem, elegant flower shape, rich color and beautiful appearance, P. lactiflora is loved by people at home and abroad. As far as the flower color is concerned, it can be divided into nine color categories, including black, purple, red, pink, white, blue, green, yellow and double color (Wang and Zhang 2005). In addition, in P. lactiflora flower coloration, numerous studies have examined chemical constituents, flavonoids including anthocyanins and multiform glycosides of flavones and flavonols determine its color (Zhao et al. 2014, 2016). Furthermore, many key genes regulating P. lactiflora color formation are observed on the transcription level, for example, the dihydroflavonol 4-reductase gene (PlDFR) and anthocyanidin synthase gene (PlANS) result in the shift from white to pink and red in P. lactiflora flowers (Zhao et al. 2016). And on post-transcriptional level, only our previous study reported that the yellow formation might be under the regulation of an miR156e-3p-targeted squamosa promoter binding protein-like gene (SPL1) (Zhao et al. 2017), but this result has only been obtained according to their dynamic expression patterns. However, there is no more direct evidence that P. lactiflora miR156e-3p regulates color formation. To validate the miR156e-3p regulation mechanism of the color formation in P. lactiflora, the miR156e-3p precursor sequence (pre-miR156e-3p) was isolated, and an ectopic expression analysis of its function in Arabidopsis was also conducted. These results would improve the understanding of miRNAs regulation of P. lactiflora color formation.
Materials and methods
Plant materials
Paeonia lactiflora cv ‘Jinhui’ was grown in the germplasm repository of Horticulture and Plant Protection College, Yangzhou University, Jiangsu Province, China (32°30′N, 119°25′E), and the ground plants grew well with sufficient light and water supply. The young petal samples in initiating bloom stage were used for the isolation of pre-miR156e-3p. Seeds from transgenic and wild-type Arabidopsis (Col-0) sown in half-strength Murashige-Skoog (MS) medium were maintained at 4 °C for 3 days in the dark to break residual dormancy; the germinated seedlings were then grown in a culture room at 22 °C under a 14/10 h light/dark photoperiod and 70% relative humidity. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C until further analysis.
Isolation of pre-miR156e-3p
Small RNA was extracted by TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Japan), and the cDNA was, respectively, synthesized using PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time) (TaKaRa, Japan). pre-miR156e-3p was isolated by polymerase chain reaction (PCR) technology with specific primers (forward primer: 5′-CGAAGAAGAGAAAGAAATGTTGAC-3′, reverse primer: 5′-ACCCTTTAGCTGATCCCGGGTTGTG-3′). PCR products were separated by 1% agarose gel electrophoresis and sequenced.
Vector construction, transformation and transgenic plants identification
pre-miR156e-3p was amplified by PCR and using primers with restriction enzyme cutting sites of BamH I and Kpn I (forward primer: 5′-CGCGGATCCACGAAGAAGAGAAAGAAATGTTGAC-3′, reverse primer: 5′-CGGGGTACCACCCTTTAGCTGATCCCGGGTTGTG-3′). After double enzymes restriction, pre-miR156e-3p was inserted into the pCAMBIA1301 vector (p1301) by T4 DNA Ligase (TaKaRa, Japan). Luria–Bertani (LB) solid medium containing 0.05% kanamycin (Kan) was used to cultivate Escherichia coli DH5a cells (Trans, China), and the plasmid extracted from the positive monoclonal coliform bacteria strains was used for double digestion with the restriction enzymes. Additionally, the final binary vector pCAMBIA1301-Pre-miR156e-3p was transferred into Agrobacterium tumefaciens (EHA105) cells using the freezing/heat shock method. And the transformation of Arabidopsis was performed by the floral dip method using EHA105 cells (Clough and Bent 1998). Seeds from transgenic Arabidopsis and Col-0 were harvested from individual plant and sown again.
To identify transgenic plants, the screening medium [1/2 MS + 30 g/L sucrose + 6.5 g/L agar + 25 mg/L ampicillin (Amp) + 25 mg/L hygromycin (Hyg), pH 5.8] and GUS staining method were used to screen transgenic Arabidopsis, and when the screening transgenic Arabidopsis grew to the bolting stage, leaves were collected for GUS region amplification (forward primer: 5′-CTGCGTTTCGATGCGGTCAC-3′, reverse primer: 5′-CTCCCTGCTGCGGTTTTTCA-3′).
Qualitative and quantitative analysis of flavonoids
The lateral branches of each plant (0.1 g fresh weight) were extracted with 1.0 mL of acidic methanol solution (70: 0.1: 29.9; v/v/v, CH3OH: HCl: H2O) at 4 °C for 24 h. Qualitative and quantitative analysis of flavonoids was performed using high-performance liquid chromatography–electrospray ionization–ion trap mass spectrometry (HPLC–ESI–MSn) (LCQ Deca XP MAX, Thermo) coupled with photodiode array and mass spectrometry detectors (HPLC–PDA–MS, Thermo company) with a three-dimensional quadrupole ion trap mass spectrometer. The HPLC column was TSK gel ODS-80Ts QA (4.6 × 250 mm) (Tosoh, Japan). The specific conditions were the same as the report of Zhao et al. (2014). Each peak area of anthocyanins and anthoxanthins detected under 525 and 350 nm was recored. The amount of total anthocyanins and anthoxanthins was calculated in milligrams per gram fresh weight (as a quantity of Malvidin-3,5-di-O-glucoside (Mv3G5G) mg/g and as a quantity of Rutin mg/g, respectively). Additionally, total contents of flavonoids were the sum of anthocyanins and anthoxanthins.
Gene expression analysis
The transcript levels of genes were analyzed using real-time quantitative PCR (qRT-PCR) with a BIO-RAD CFX96™ Real-Time System (Bio-Rad, USA). Total RNA and small RNA were extracted according to TaKaRa RNAiso Plus (Total RNA extraction) and TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Japan), respectively. Subsequently, the cDNA was, respectively, synthesized using PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time) (TaKaRa, Japan) and miRNA First Strand cDNA Synthesis Kit with the specific primer of miR156e-3p for inverse transcription (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGATGAC-3′) (Sangon Biotech, China). All gene-specific primers for qRT-PCR are shown in Table 1. qRT-PCR was performed using the TransStart® Tip Green qPCR SuperMix (Trans, Chin8ia), and the relative expression levels were calculated by the 2−△△Ct comparative threshold cycle (Ct) method (Schmittgen and Livak 2008), and the expression level of Col-0 was used as the control.
Sequence and statistical analysis
Sequence analysis was performed by DNAMAN 5.0 software. All data were average values of three replicates at least with standard deviations. The results were analyzed for variance using the SAS/STAT statistical analysis package (version 6.12, SAS Institute, Cary, NC, USA).
Results
In our previous study, ‘jinhui’ was used for the small RNA sequence to identify miRNAs related to yellow formation (Accession numbers in NCBI: SRX1996909 and SRX1996992), and miR156e-3p was screened as the key miRNA, its sequence and precursor sequence were 21 bp (5′-GCTCTCTCTTCGTCTGTCATC-3′) and 63 bp (5′-ACGAAGAAGAGAAAGAAATGTTGACAGAAGAGAGGGAGCACAACCCGGGATCAGCTAAAGGGT-3′), respectively. Based on this, Pre-miR156e-3p was isolated, and an approximately 70-bp band was obtained by PCR technology with specific primers (Fig. 1a). This band was later extracted and sent to be sequenced, and the result showed that this band was 63 bp and exhibited 100% similarity with the small RNA sequencing result (Fig. 1b). Subsequently, the extracted product of Pre-miR156e-3p was cloned into vector p1301 and transformed into competent E. coli DH5a cells, and the screening positive transformants were later identified by double digestion with the restriction enzymes. Figure 1c shows that the successful construction of the recombinant plasmid had been cut into an approximately 70-bp band and a more than 15,000-bp band, but the negative control only had a more than 15,000-bp band. The constructed vector diagram of pCAMBIA1301-Pre-miR156e-3p is shown in Supplemental Fig. 1, and this vector was used for subsequent Agrobacterium tumefaciens-mediated transformation of Arabidopsis plants.
PCR amplification of miR156e-3p precursor and vector construction. a PCR amplification of miR156e-3p precursor. b Sequence comparison of miR156e-3p precursor. c Double digestion of the recombinant plasmid miR156e-3p. M1: 50 bp DNA Ladder Marker (Dye Plus). 1: PCR product of miR156e-3p precursor. M2: 20 bp DNA Ladder Marker. 2, 3: Double digestion of the recombinant plasmid. 4: negative control
To identify transgenic Arabidopsis plants, the harvested seeds were firstly sowed on the screening medium with antibiotics. As shown in Fig. 2a, Col-0 plants grew well in the medium without antibiotics, and the successful miR156e-3p-overexpressing transgenic plants and p1301 transgenic plants all retained robust growth and were green, while non-transgenic plants showed yellowing and death. Furthermore, the identified transgenic plants according to the screening medium were randomly selected for GUS staining. As shown in Fig. 2b, Col-0 plants remained white, and the successful miR156e-3p-overexpressing transgenic plants and p1301 transgenic plants all turned blue in the GUS staining buffer. In addition, when screening transgenic plants grew at the bolting stage, DNA extracted from the leaves was used as the template to amplify the GUS region of the p1301 vector, and the result showed that one clear and bright band could be observed in miR156e-3p-overexpressing transgenic plants and p1301 transgenic plants, whereas there was no specific band in the Col-0 plants (Fig. 2c), which was consistent with the GUS staining result.
When Arabidopsis plants grew to the bolting stage, there were no obvious differences between miR156e-3p-overexpressing transgenic plants, p1301 transgenic plants and Col-0 plants with respect to morphology, but the color of their lateral branches had dramatically changed. As shown in Fig. 3, the lateral branches of miR156e-3p-overexpressing transgenic plants had an apparent purple, whereas Col-0 plants were still green, and p1301 transgenic plants were slightly red. Subsequently, qualitative and quantitative analysis of flavonoids in the lateral branches was performed using HPLC–ESI–MSn. On the basis of the ultraviolet–visible absorption characteristics, anthocyanins and anthoxanthins were detected under the wavelength of 525 and 350 nm, respectively. As shown in Fig. 4, there were several differences in the chromatographic peaks of miR156e-3p-overexpressing transgenic plants, p1301 transgenic plants and Col-0 plants. At 525 nm, a very small amount of anthocyanin was found in Col-0 plants and p1301 transgenic plants, whereas there were obvious peaks in miR156e-3p-overexpressing transgenic plants. At 350 nm, the main peaks were largely identical in the three plants and only several minor differences existed. The statistic on the flavonoid content indicated that the highest anthocyanin content was in miR156e-3p-overexpressing transgenic plants, which was approximately 24 times that of the p1301 transgenic plants, and it was largely undetected in Col-0 plants (Fig. 5). However, when anthoxanthin and flavonoid contents were concerned, the tree plants had similar contents, which were below the very significant levels.
HPLC chromatograms of anthocyanins (detected at 525 nm) and anthoxanthins (detected at 350 nm) in transgenic Arabidopsis lateral branches. a Anthocyanin content. b Anthoxanthin content. Green line: HPLC chromatograms of Col-0 plants. Black line: HPLC chromatograms of p1301 transgenic plants. Red line: HPLC chromatograms of miR156e-3p-overexpressing transgenic plants
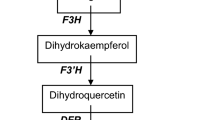
To identify the expression pattern of miR156e-3p in Arabidopsis lateral branches, the expression analysis of miR156e-3p was first performed using qRT-PCR (Fig. 6). The results revealed that the miR156e-3p transcript was detected in all the plants, but the expression levels were different from each other. Its expression level in miR156e-3p-overexpressing transgenic plants was very significantly higher than that of the Col-0 plants and p1301 transgenic plants, and the former was approximately 2.6 times that of the latter two. Based on this information and to further clarify the role of miR156e-3p in the color formation of Arabidopsis lateral branches, the expression pattern of miR156e-3p corresponding target gene was subsequently studied. In a previous study, Cui et al. (2014) reported that the miR156-SPL9-DFR pathway coordinated anthocyanin accumulation in Arabidopsis. We found an AtSPL1 (NM_180137) in Arabidopsis, which had the highest homology with miR156e-3p corresponding target gene PlSPL1, and only an AtDFR (NM_123645) in Arabidopsis. Regarding the single gene expression level, the expression level of AtSPL1 in miR156e-3p-overexpressing transgenic plants was very significantly lower than that of the Col-0 plants and p1301 transgenic plants, whereas the expression level of AtDFR presented an opposite tendency. Additionally, the expression levels of miR156, AtSPL1 and AtDFR in Col-0 plants and p1301 transgenic plants were largely the same, and there were no very significant differences between them.
Discussion
Regulation of plant color formation by miRNAs has been partially reported in a number of studies. Overexpression of miR828 could reduce anthocyanin accumulation in Arabidopsis (Yang et al. 2013). miR858 up-regulated the expression of target genes MYB12 and Sly-myb-like in transgenic tomato plants, and flavonoid biosynthetic genes expressions were also up-regulated, which resulted in anthocyanin accumulation (Shen 2015). Overexpression of miR778 moderately enhanced anthocyanin accumulation under Pi deficient conditions (Wang et al. 2015). In addition, miR156 regulating plant color formation had been deeply studied (Gou et al. 2011). miR156 was first observed in Arabidopsis with 20 nucleotides in length, and it was considered to be the most conservative miRNA and existed in monocotyledons, dicotyledons, ferns and mosses (Franco-Zorrilla et al. 2007). A previous study revealed that SPL family was the target gene of miR156 (Rhoades et al. 2002), 11 of 17 SPL in Arabidopsis were putative targets of miR156 and 11 of 19 SPL in O. sativa were putative targets of miR156, and these results all indicated that miR156 played a crucial role in SPL family expression and regulation (Chen et al. 2015; Schwab et al. 2005). In our previous study, miR156e-3p was screened as the key miRNA related to color formation in P. lactiflora, and its target gene was SPL1 (Zhao et al. 2017). In addition, an increasing number of studies suggested that the miR156 and SPL family were involved in many processes of growth and development, metabolism regulation, and abiotic stresses, and they had been become the regulatory hub of growth and development of plants (Lei and Liu 2016). In the present study, pre-miR156e-3p was isolated and introduced into transgenic Arabidopsis to validate that the color formation was under the regulation of miR156e-3p-targeted SPL1. This method had not only been widely used in model plants, including Arabidopsis, Nicotiana tabacum and O. sativa, but had also been successfully applied in Brassica campestris (Jiang 2014) and Lycopersicon esculentum (Jia et al. 2013). Subsequently, the medium screening, GUS staining, PCR of the GUS region and qRT-PCR of miR156e-3p all confirmed that the purpose gene had been successfully transferred into Arabidopsis plants and expressed, which indicated that this method could also be used to verify the function of P. lactiflora miRNAs.
In the anthocyanin accumulation, Gou et al. (2011) constructed overexpression and silencing expression vectors (Pro35S:MIR156, Pro35S:MIM156) to study the miR156 function in Arabidopsis and observed that increased miR156 activity promoted anthocyanin accumulation, whereas reduced miR156 activity resulted in high levels of flavonols. Our study obtained similar results, i.e., miR156e-3p-overexpressing transgenic plants resulted in apparent purple lateral branches, which were caused by the accumulation of anthocyanins, and anthocyanins were largely undetected in Col-0 plants. Moreover, Cui et al. (2014) found that the anthocyanin accumulation was under the regulation of miR156-targeted SPL, and SPL negatively regulated anthocyanin accumulation by directly preventing the expression of DFR. In this study, miR156e-3p was expressed most heavily in miR156e-3p-overexpressing transgenic plants, and the expression level of its target gene SPL1 was clearly decreased, and its downstream gene DFR was strongly expressed, which resulted in increased anthocyanin accumulation of lateral branches, and this result was consistent with a previous study (Cui et al. 2014). These findings would improve the understanding of the miRNAs regulation of color formation in P. lactiflora.
Conclusion
Overexpression of P. lactiflora miR156e-3p changed the color of transgenic Arabidopsis lateral branches to purple. This change in color was caused by the improved anthocyanin accumulation, which was regulated by miR156-targeted SPL, and SPL negatively regulated anthocyanin accumulation by directly preventing the expression of DFR.
References
Chen Z, Gao X, Zhang J (2015) Alteration of osa-miR156e expression affects rice plant architecture and strigolactones (SLs) pathway. Plant Cell Rep 34:767–781
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
He H, Liang G, Li Y, Wang F, Yu D (2014) Two young microRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol 164:853–865
Hu G, Fan J, Xian Z, Huang W, Lin D, Li Z (2014) Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci 229:86–95
Jia XY, Yu ZQ, Liang JP, Tang GL, Jin LH, Zhang L, He LH, Li RZ (2013) Cloning of Arabidopsis At-pri-miR828 gene and its genetic transformation into tomato. Acta Hortic Sin 40:2419–2428 (in Chinese)
Jian XY, Zhang L, Li GL, Zhang L, Wang XJ, Cao XF, Fang X, Chen F (2010) Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 95:47–55
Jiang JX (2014) Identification and functional characterization of microRNAs involved in pollen development in Brassica campestris ssp. Chinensis. Master Dissertation. Zhejiang University, Hangzhou, China (in Chinese)
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:68–73
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lei KJ, Liu H (2016) Research advances in plant regulatory hub miR156 and targeted. Chem Life 36:13–20 (in Chinese)
Li H, Deng Y, Wu T, Subramanian S, Yu O (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153:1759–1770
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Rogers K, Chen X (2013) Biogenesis, turnover and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 36:1101–1108
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specifc effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shen J (2015) The cloning and drought response of miR858 and the mechanism for it negatively regulates anthocyanin biosynthesis in tomato. Master Dissertation. Shanxi Agricultural University, Taigu, China (in Chinese)
Spanudakis E, Jackson S (2014) The role of microRNAs in the control of flowering time. J Exp Bot 65:365–380
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Wang JG, Zhang ZS (2005) Herbaceous peonies of China. China Forestry Publishing House, Beijing in Chinese)
Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X (2010a) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62:761–773
Wang L, Mai YX, Zhang YC, Qian L, Yang HQ (2010b) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3:794–806
Wang L, Zeng HQ, Song J, Feng SJ, Yang ZM (2015) miRNA778 and SUVH6 are involved in phosphate homeostasis in Arabidopsis. Plant Sci 238:273–285
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom 11:246
Yang F, Cai J, Yang Y, Liu Z (2013) Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tiss Org Cult 2:159–167
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8:R96
Zhang BH, Pan XP, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhao DQ, Jiang Y, Ning CL, Meng JS, Lin SS, Ding W, Tao J (2014) Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.). BMC Genom 15:689
Zhao DQ, Wei MR, Liu D, Tao J (2016) Anatomical and biochemical analysis reveal the role of anthocyanins in flower coloration of herbaceous peony. Plant Physiol Biochem 102:97–106
Zhao DQ, Wei MR, Shi M, Hao ZJ, Tao J (2017) Identification and comparative profiling of miRNAs in herbaceous peony (Paeonia lactiflora Pall.) with red/yellow bicoloured flowers. Sci Rep 7:44926
Acknowledgements
This work was supported by the Natural Science Foundation of China (31,400,592), the program of key members of Yangzhou University outstanding young teachers and the Priority Academic Program Development from Jiangsu Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author declares that there is no competing interest towards the publication of this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, D., Xia, X., Wei, M. et al. Overexpression of herbaceous peony miR156e-3p improves anthocyanin accumulation in transgenic Arabidopsis thaliana lateral branches. 3 Biotech 7, 379 (2017). https://doi.org/10.1007/s13205-017-1011-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-1011-3