Abstract
Improvement of quality protein maize (QPM) along with high content of lysine and tryptophan had foremost importance in maize breeding program. The efficient and easiest way of developing QPM hybrids was by backcross breeding in marker aided selection. Hence, the present investigation aimed at conversion of elite maize inbred line BML-7 into QPM line. CML-186 was identified to be a donor variety as it revealed high-quality polymorphism with BML-7 for opaque-2 gene specific marker umc1066. Non-QPM inbred line BML-7 was crossed with QPM donor CML-186 and produced F1 followed by the development of BC1F1 and BC2F1 population. Foreground selection was carried out with umc1066 in F1, and selected plants were used for BC1F1 and BC2F1 populations. Two hundred plants were screened in both BC1F1 and BC2F1 population with umc1066 for foreground selection amino acid modifiers. Foreground selected plants for both opaque-2 and amino acid modifiers were screened for background selection for BML-7 genome. Recurrent parent genome (RPG) was calculated for BC2F1 population plants. Two plants have shown with RPG 90–93% in two generation with back cross population. Two BC2F2 populations resulted from marker recognized BC2F1 individuals subjected toward foreground selection followed by tryptophan estimation. The tryptophan and lysine concentration was improved in all the plants. BC2F2 lines developed from hard endosperm kernels were selfed for BC2F2 lines and finest line was selected to illustrate the QPM version of BML-7, with 0.97% of tryptophan and 4.04% of lysine concentration in protein. Therefore, the QPM version of BML-7 line can be used for the development of single cross hybrid QPM maize version.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among cereals after wheat and rice, maize is the important food crop in the world. It remained as a major staple food for millions of people in developing and developed nations. Structurally maize grain constitutes a small embryo and large endosperm with 90% of starch and 10% of proteins, respectively. Around 70% of this protein was composed of various alcohol soluble prolamins called zeins (Pirona et al. 2005). The distribution of zeins in maize is distinctive with a predominance of four types namely α, β, γ, and δ zein. Nevertheless, the distribution of two essential amino acids, i.e., tryptophan and lysine remained deficient. Consumption of normal maize kernals by Children’s and adults generally leads to nutritional disorders like marasmus and kwashiorkor (Vasal 2000). In Africa, Asia, Central, and South America millions of people rely on maize for their daily food supplement. Even high content of maize in diet produces a condition known as wet malnutrition, i.e., the person receives sufficient calories of energy but suffers from malnutrition due to lack of protein content.
The lysine and tryptophan levels were better in quality protein maize (QPM) than the conventional maize, whereas maize opaque-2 genotype resulted in lower yields due to its soft-chalky kernels, making it more prone to insects and fungal attacks. In addition, taste and appearance of kernel dissatisfied the consumers, who ultimately rejected the usage of enhanced-protein varieties in the market. However, by means of mutant opaque-2(o2) variety, breeders developed maize with rich in lysine and tryptophan by means of normal kernels in vitreous appearance (Krishna et al. 2012; Ortega and Bates 1983). More precisely, CIMMYT breeders improved high lysine maize from opaque-2 genotypes by selecting phenotypic characteristics like normal endosperm texture and increased levels of lysine and tryptophan. These modified opaque-2 maize lines were designated as “Quality Protein Maize” lines (Vasal et al. 1980). Due to advantages like vitreous/hard nature of endosperm, resistance to insect damage, kernel texture, and high yield, there was a huge and significant need to develop QPM hybrids. For this purpose, molecular tools can be used to identify the high lysine and tryptophan lines after screening the potential germplasm that can help the breeder in constructing hybrids using Marker assisted selection. QPM is a genotype in which opaque-2 mutant gene has been integrated along with associated modifiers or enhancers. Modifier genes help in converting hard endosperm to soft endosperm while enhancers confer higher tryptophan and lysine content. Thus, QPM breeding program requires manipulation of three different genetic systems (Krivanek et al. 2007).
Marker-assisted advanced backcross breeding can greatly accelerate the introgression of opaque-2 allele into normal maize. For genotype characterization, DNA-based molecular markers hold numerous advantages over the phenotype markers. Simple Sequence Repeat (SSR) markers have efficient role among DNA-based markers because of their good reproducibility, high polymorphic, and reliable nature (Gupta et al. 1996). The SSR loci consist of 2 to 6 base pair tandem repeats. The opaque-2 trait was expressed in recessive state where mutant kernels have a soft textured starchy endosperm with low density. While the mutation in opaque-2 alters the endosperm texture by decreasing α-zeins content which, in turn, increases the lysine content. Breeders have systematically transferred the modifier genes into opaque-2 germplasm to develop normal looking maize with high lysine. Molecular tools have facilitated the breeders to transfer the opaque-2 along with modifiers into elite maize inbreeds for construction of the hybrid. The present investigation for conversion of elite maize inbred line BML-7 into quality protein maize with high lysine and tryptophan using marker-assisted breeding.
Materials and methods
The QPM germplasm line CML-186 collected from VPKAS (Vivekananda Parvatiya Krishi Anusandhan Sansthan), Almora. Parental inbred line BML-7 was used as the recipient for the breeding program. For identification of polymorphism between donor and recipient inbred lines DNA, opaque-2 gene specific SSR markers viz, umc 1066, Phi057 and Phi112 were used. Donor and recipients were also screened with six amino acid modifier-linked SSR markers, viz, mmc0241, umc1216, phi072, bnlg1633, bmc1382, and phi075. Simple Sequence Repeats (SSRs) marker information was taken from maize database (http://www.maizegdb.org).
DNA isolation
Leaf sampling was done by taking 2 g leaf from 3-week-old seedlings of each genotype. Sample was wrapped in marked aluminum foil and then frozen in liquid nitrogen before storing in −80 °C. Pre-chilled plant leaves (2 g) at −80 °C were crushed in liquid nitrogen with a mortar and pestle. A pre-heated (65 °C) extraction buffer (2 ml) containing 100 mM Tris–HCl (pH:8); 1.4 mM NaCl; 20 mM EDTA; 0.1% 2-mercaptoethanol GR (Merck); and 2% CTAB powder was added to crushed leaf material. The mixture was incubated at 65 °C for 45 min in water bath (Lab Tech) followed by addition of equal volume of Chloroform:Isoamyl alcohol (24:1) (Thermo Fisher Scientific India Pvt. Ltd) mixture and the homogenate incubated for 15 min at room temperature. Centrifugation was done for 10 min at a speed of 10,000 rpm in a centrifuge machine (Eppendorf) at room temperature. The upper aqueous phase was again removed carefully into a new tube and an equal volume of ice-cold isopropanol was added to this to precipitate the DNA. This mixture was centrifuged at maximum speed for 10 min at room temperature to collect the DNA pellet at the bottom of the tube. The DNA pellet was washed by adding 2 ml 70% ethanol and centrifuged at maximum speed of 10,000 rpm for 2 min. The pellet was dried in a vacuum centrifuge (Speed Vac) and re-suspended in 100 µl sdH2O (sterile distilled water).
The isolated DNA sample was quantified with spectrophotometer in the photometric mode which was adjusted with wavelengths set at 260 and 280 nm. Normalization of DNA samples was done to equalize the concentrations of all the DNA samples that were to be run in PCR.
Polymerase chain reaction (PCR)
DNA sample (50 ng) was used per PCR to which 23 μl master mixture (PCR reaction mix.) was added. The PCR mix was centrifuged at 1000 rpm for 1 min and loaded in a 96-well thermal cycler of PCR (Eppendorf). The program comprises of the initial denaturation at 94 °C for 4 min. which was further followed by 35 cycles 94 °C for 1 min, 55–63 °C for 30 s. and 72 °C for 1 min. Final extension was carried out at 72 °C for 7 min, and maintained at 4 °C for ever. The amplified products were used for electrophoresis.
Marker-assisted backcross breeding
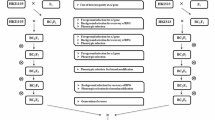
The advanced backcross breeding program was carried out at the Maize Research Centre (MRC), ARI, Rajendranagar, and Hyderabad. BML-7 inbred line was used as female parent and QPM line CML-186 used as male parent. Backcross populations were developed from the crosses between the BML-7 × CML-186. Crosses were affected during Kharif-2010 for F1 Seed production. F1 seed was subjected to foreground selection with opaque-2-specific SSR marker umc1066. These foreground selected F1 plants were backcrossed with recurrent parent to produce BC1F1 during rabi-2010. The BC1F1s were then backcrossed with recurrent parent (BML-7) to produce BC2F1 during summer-2010. BC2F1 plants were selfed to produce BC2F2 plants (Fig. 1).
Foreground selection
Fore ground selection for opaque-2 gene at backcross population is carried out with gene-specific marker Umc1066. Two hundred plants were screened for foreground selection in each F1, BC1F1, and BC2F1 populations, respectively. Foreground selection for BML-7 × CML-186 cross BC2F1 population with amino acid modifier linked SSR marker bnlg1633.
Background selection
Whole-genome background screening was done for recurrent parent BML-7 using 750 SSR markers. Back ground selection was carried out both BC2F1 population with identified 160 SSR markers. The details of SSR markers and their chromosome number were given in Table 1.
Estimation of tryptophan and lysine content
In biochemical analysis, a random sample of 20 seeds as representative of each ear was taken for tryptophan analysis with three replications. Total protein was estimated for both the parents (BML-7 and CML-186) and selected BC2F2 plants. Tryptophan content was estimated by high-performance liquid chromatography (HPLC) method. HPLC (SCHIMADZU, Tokyo) with reversed-phase C18 column was used for analysis. Lysine content was calculated by a correlation with Tryptophan concentration according to the equation (Babu 2015).
Results and discussion
Both the parents HPLC results indicated that BML-7 contains lysine and tryptophan 2.2 and 0.41% respectively. CML-186 had lysine and tryptophan content with 4.04, 0.98%, respectively (Table 2). CML-186 was showed good polymorphism with BML-7. Hence, CML-186 was used as donor for BML-7 marker-assisted conversion (Fig. 2).
F1 population
Cross was made between BML-7 (Non QPM inbred) and CML-186 (QPM donor) during kharif -2010. Recurrent parent BML-7 as a female parent and CML-186 (donor) as male parent to produce F1 seeds. Recurrent parent BML-7 was used as a female and donor parent CML-186 was used as male parent which is a QPM line to produce F1 seeds.
Foreground selection on F1 (BML-7 × CML-186) was carried out with opaque-2 gene-specific SSR marker umc1066. All the F1 populations exhibited heterozygote for gene of interest (Fig. 3). Since SSR is a co-dominant marker, we could differentiate between heterozygous and homozygous inbred lines. Two bands with equal intensity in a genotype were considered as heterozygous, whereas in case of variable intensities and of superimposed conditions, it was considered as heterogeneous. In heterogeneous condition, band with higher intensity was used for scoring and of lower intensity was rejected.
Transfer of recessive genes by conventional breeding needs further selfing over generations next to every backcross. This procedure requires extensively long duration for improving best variety for profitable breeding purposes. Marker-aided background selection may aid in recovering the same genotype of recurrent parent within three generations which would not be achieved by the conventional phenotypic selections which require six-to-seven generations. Hence, MAS centered on SSR markers designed for conversion of normal maize lines into QPM was known to be efficient, simple, rapid, and cost-effective method as a complementary to prevailing breeding protocols (Dreher et al. 2000).
BC1F1 population
F1 (BML-7 × CML-186) seeds were sown as per year to row method during rabi 2010–11. These F1 plants were crossed with recurrent parent BML-7 eventually to produce BC1F1 seeds. Two hundred BC1F1 [(BML-7 × CML-186) × BML-7] plants were screened for opaque-2 gene. Out of 200 plants, 98 plants was shown heterozygote condition (Fig. 4).
BC2F1 population
BC1F1 seeds were sown along with recurrent inbred line BML-7 during summer 2011. Foreground selected 98 plants were back crossed with recurrent parent BML-7 (Fig. 5). Two hundred BC2F1 plants were screened with gene specific marker umc1066. Out of 200 plants, 90 plants exhibited heterozygous loci (Fig. 6). These results confirmed that SSR markers were trustworthy for opting QPM kernels and may even be extended further for recognizing completely modified QPM kernels. Identified 90 heterozygous plants were screened for amino acid modifier marker bnlg1633. Out of 90 plants, 65 plants were shown heterozygous for amino acid modifier marker bnlg1633 (Fig. 5). Though a huge number of SSR loci were identified and used for parental polymorphism, sufficient polymorphic loci were not detected, as both parental lines were isogenic but then differ in tryptophan content (Babu 2015).
Whole genome background selection
A total of 750 SSR markers were screened which are distributed all over the 10 chromosomes for polymorphism between BML-7 (Recipient) and CML-186 (Donor) for background selection. Out of 750 SSR markers, 151 SSR markers were shown polymorphism between donor (CML-186) and recipient (BML-7) (Table 1).
It has been revealed from simulation studies that cumulative sum of markers per non-carrier chromosome was essential from primary generations. In the course of applied MAS program, a great attention has to be taken to avoid sampling error and exaggerated approximations of recurrent parent genome connected with less integer of marker data points (Hospital et al. 1992). Out of 65 BC2F1 plants, three plants had shown 90.47, 92.8,5 and 90.48% Recurrent Parent Genome (RPG); three plants had shown 86.9, 85.71, and 88.9% RPG and remaining plants have shown 80–85% RPG. Plants which exhibited >80% RPG were selfed, to produce BC2F2 plants. The whole-genome background selection was to improve quickly maximum ratio of recurrent parent genome (RPG) on non target loci through DNA-based markers that were dispersed equally all over the genome (Hospital et al. 1992; Frisch et al. 1999; Visscher 1996; Young and Tanksley 1989). Simulation studies have showed that application of background selection in the present and later generation along with that of foreground selection in every back cross generations may, perhaps, be efficient (Frisch et al. 1999; Ribaut and Hoisington 2002). In the current investigation, twofold generation-based marker breeding program where whole-genome background selection is on non-target loci remained functional merely in the BC2 generation. In this study, we selected markers aligned on chromosomes, especially SSR consensus map of maize genome accessible in public domain (http://www.maizegdb.org).
Marker-assisted breeding (MAB) is one of the most predictable and frequently cited tools used for indirect selection in breeding programs (Semagn et al. 2006). The rapid inbred line conversion approaches bring together the noteworthy features of both marker-assisted and phenotypic selection methods like fixing the target trait from large segregating generation and recovery of maximum extent of recurrent parent genome (RPG) within two back cross generations providing abundant scope for possible desired agronomic and biochemical traits. Recurrent genotype recovery can be improved with the help of molecular markers. By the advent of molecular marker technology in third generation, detection of PCR amplified products by SNPs and fluorescent centered dyes, viz, Taqman probe and molecular beacons may assist and enhance the power and efficiency of genotyping much better in the present and future generations (Salvi et al. 2001; Tyagi et al. 1998). Results unveiled 4.04% of lysine and 0.97% of tryptophan content by BC2F2 plants. Hence, QPM version of BML-7 can be used as parent for single cross hybrids, while duration of time required for a conventional breeding program can be highly reduced with the approach of marker-assisted selection.
References
Babu PK (2015) Mapping QTLs for opaque2 modifiers influencing the tryptophan content in quality protein maize using genomic and candidate gene-based SSRs of lysine and tryptophan metabolic pathway. Plant Cell Rep 34:37–45
Dreher K, Khairallah M, Pandey S (2000) Is marker assisted selection cost effective compared to conventional plant breeding methods? The case of Quality Protein Maize. International Consortium on Agricultural Biotechnology Research (ICABR). ‘The economics of Agricultural Biotechnology’. Held in Revello, Italy, 24–28
Frisch M, Bohn M, Melchinger AE (1999) Comparison of selection strategies for marker-assisted backcrossing of a gene. Crop Sci 39:1295–1301
Gupta PK, Balyan HS, Sharma PC, Ramesh B (1996) Microsatellites in plants: a new class of molecularmarkers. Curr Sci 70:45–54
Hospital F, Chevalet C, Mulsant P (1992) Using markers in gene introgression breeding programs. Genetics 132:1199–1210
Krishna MSR, Chinnababunaik V, Sokka Reddy S (2012) Assessment of genetic diversity among the QPM lines using SSR markers. Afr J Biotech 11(98):16427–16433
Krivanek AF, De Grote H, Gunaratna NS, Diallo AO, Friesen D (2007) Breeding and disseminating quality protein maize (QPM) for Africa. Afr J Biotech 6:312–324
Ortega EI, Bates LS (1983) Biochemical and agronomic studies of two modified hard-endosperm opaque-2 maize (Zea mays L.) populations. Cereal Chem 60:107–111
Pirona R, Hartings H, Lauria M, Rossi V, Motto M (2005) Genetic control of endosperm development and of storage products accumulation on maize seeds. Maydica 50:515–530
Ribaut JM, Hoisington D (2002) Stimulation experiments on efficiencies of gene introgression by back crossing. Crop Sci 42:557–565
Salvi ND, George L, Eapen S (2001) Plant regeneration from leaf base callus of turmeric and random amplified polymorphic DNA analysis of regenerated plants. Plant Cell. 66:113–119
Semagn K, Bjornstad A, Ndjiondjop MN (2006) An overview of molecular markers methods for plants. Afr J Biotechnol 5:2540–2568
Tyagi S, Bratu DP, Kramer FR (1998) Multicolor molecular beacons for allele discrimination. Nat Biotechnol 16:49–53
Vasal SK (2000) Quality protein maize story. Proceedings of workshop on improving human nutrition through agriculture. The role of international agricultural research. IRRI, Los Baños, pp 1–16
Vasal SK, Villegas E, Bjarnason M, Gelaw B, Goertz P (1980) Genetics modifiers and breeding strategies in developing hard endosperm opaque2 materials. In: Pollmer WG, Phipps RH (eds) Improvement of quality traits of maize for grains and silage use. Nighoff, The Hague, pp 37–73
Visscher PM (1996) Proportion of the variation in genetic composition in backcrossing programs explained by genetic markers. J Hered 87:136–138
Young ND, Tanksley SD (1989) Analysis of the size of chromosomal segments retained around Tm-2 locus of tomato during back cross breeding. Theor Appl Genet 77:353–359
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors state no conflict among them.
Rights and permissions
About this article
Cite this article
Krishna, M.S.R., Sokka Reddy, S. & Satyanarayana, S.D.V. Marker-assisted breeding for introgression of opaque-2 allele into elite maize inbred line BML-7. 3 Biotech 7, 165 (2017). https://doi.org/10.1007/s13205-017-0842-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-0842-2